Proteomic and Bioinformatic Investigations of Heat-Treated Anisakis simplex Third-Stage Larvae
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. A. simplex L3 Larvae Collection and Identification
2.3. Protein Extraction from A. simplex L3 Larvae
- the procedure for native crude (CR) antigens of A. simplex was performed, as previously reported [42];
- the procedure for heat-treated CR antigens of A. simplex was performed by the heating of native CR antigen in a thermomixer at 100 °C for 60 min;
- the procedure for heat-sterilized CR antigens of A. simplex was performed by the autoclaving of CR antigens at 121 °C for 60 min.
2.4. Generation of Rabbit Anti-A. simplex Antiserum
2.5. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS–PAGE) and IgG Western Blot (WB)
2.6. Sample Processing and Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) Analysis
2.7. Bioinformatic Analysis
3. Results
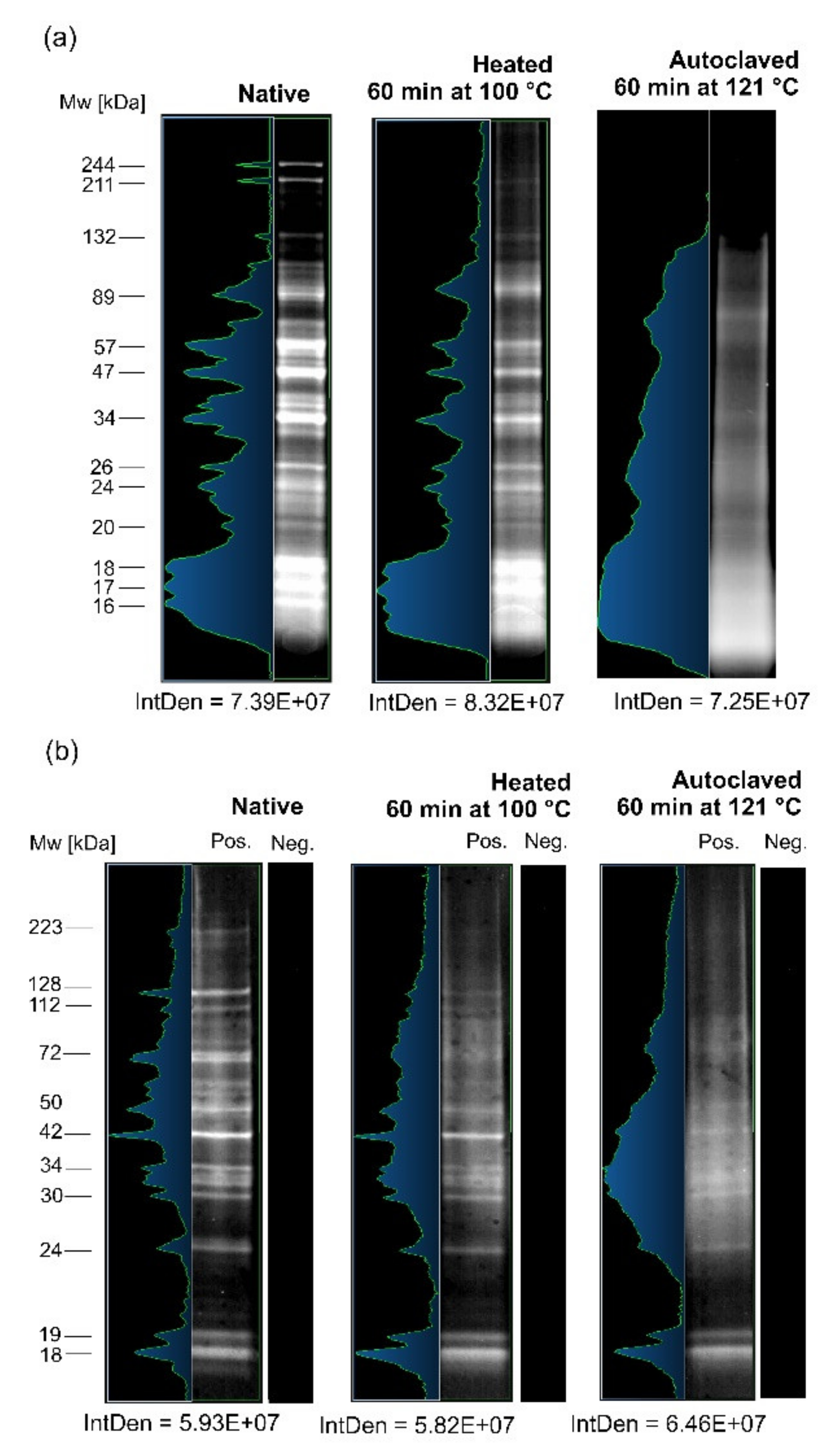
3.1. Comparative SDS-PAGE and IgG-WB Analyses of A. simplex Antigens
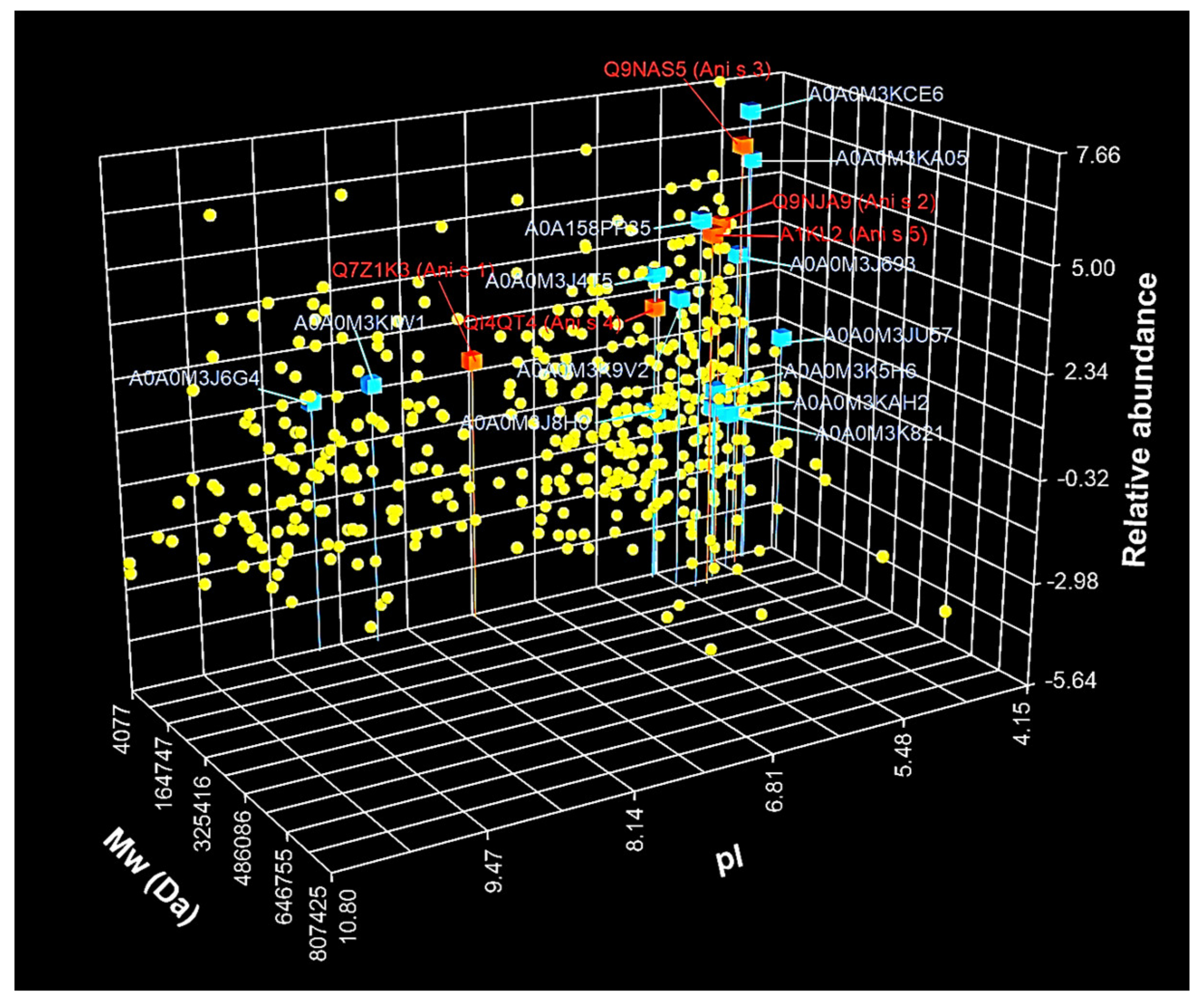
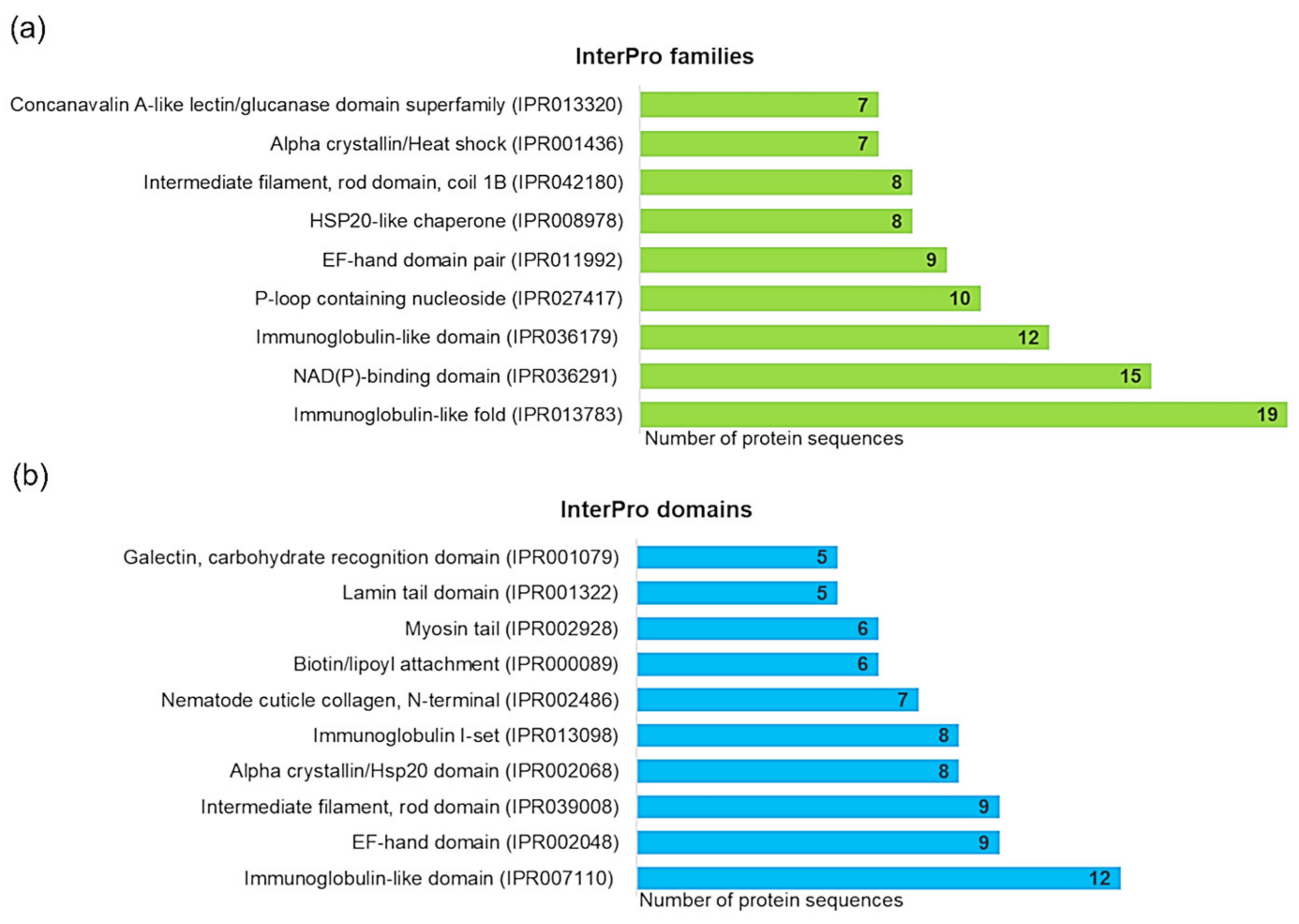
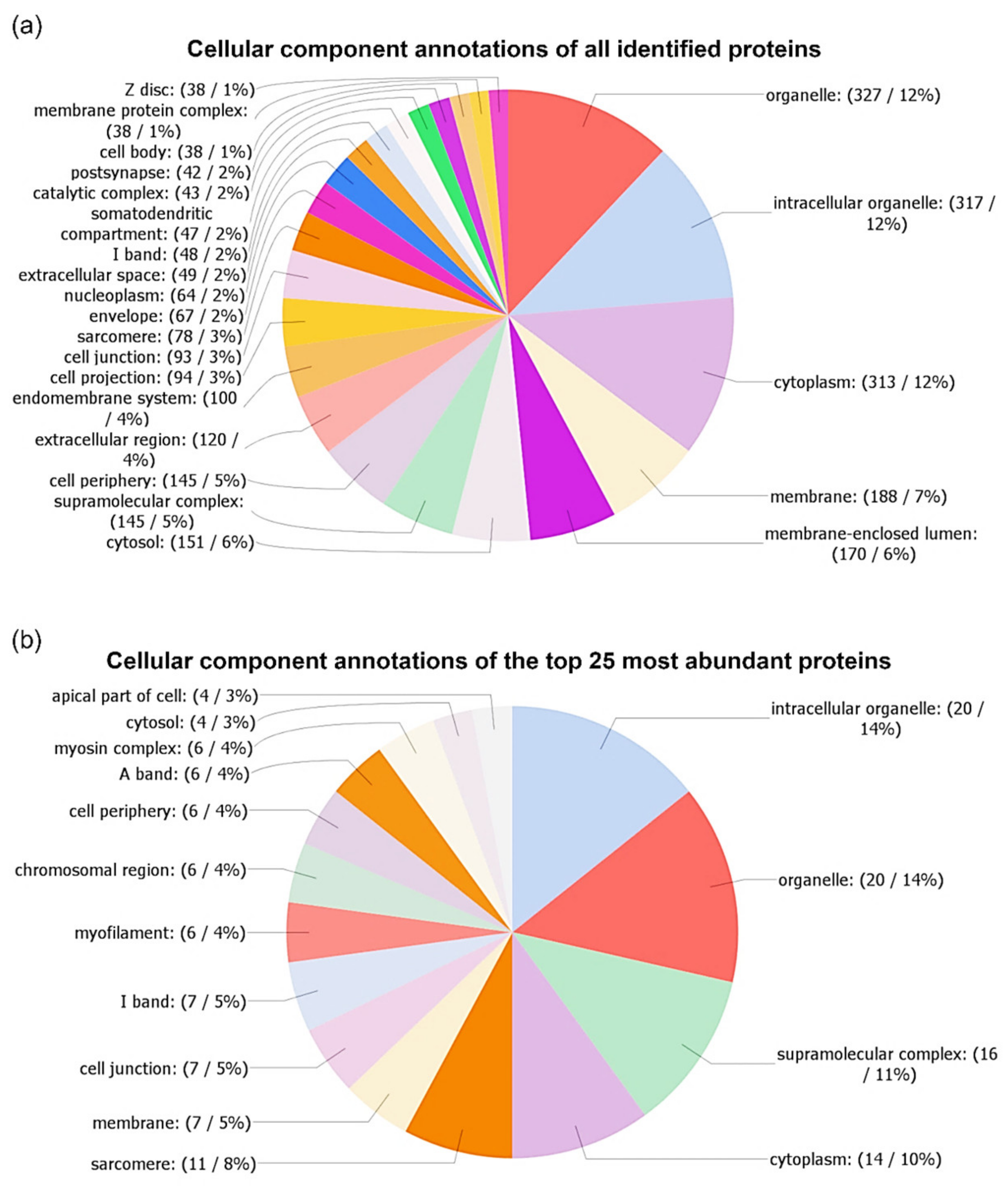
3.2. Identification and Characterization of A. simplex Proteins
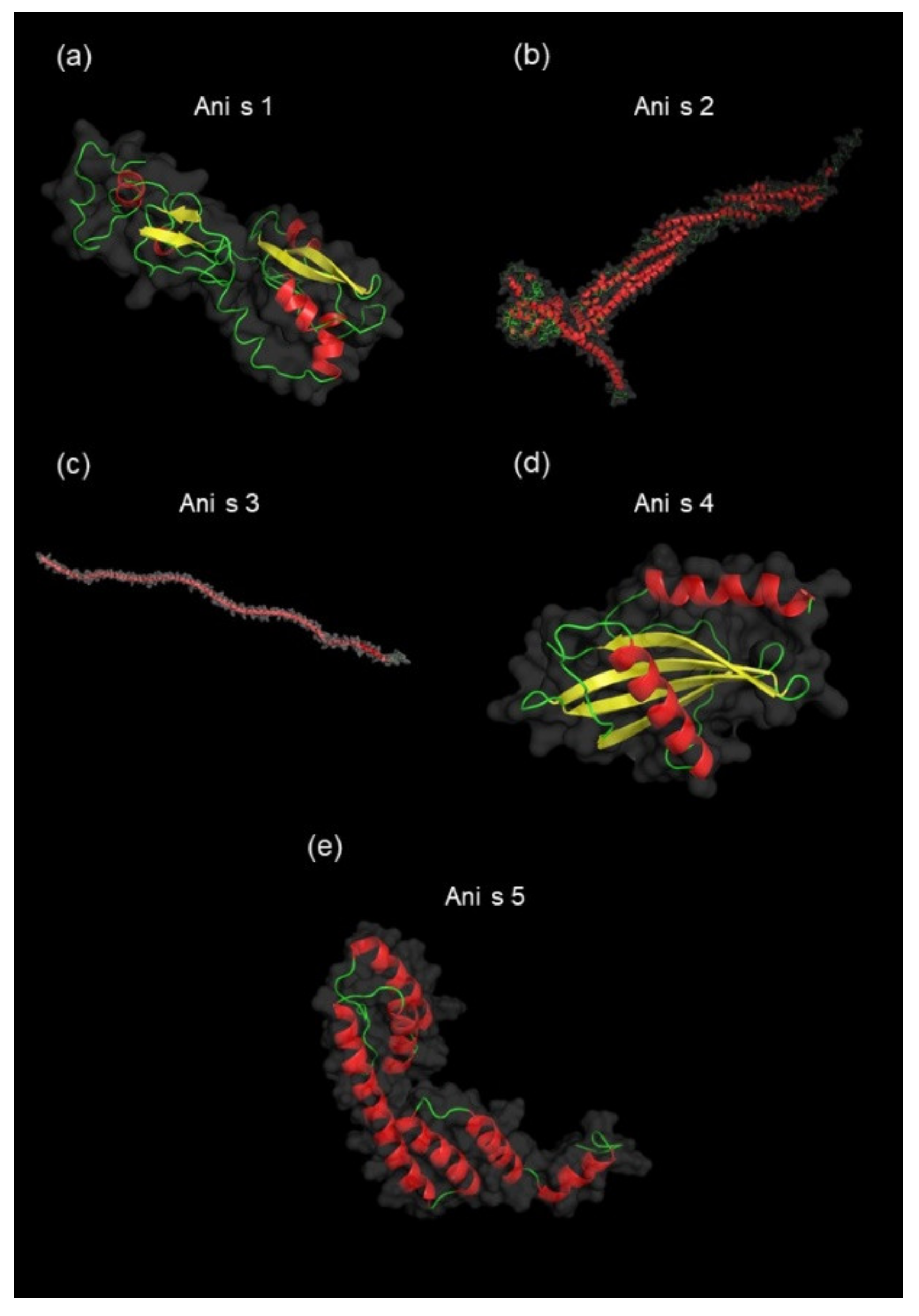
3.3. Identification and Characterization of Detected A. simplex Allergens and Potential Allergens
4. Discussion
4.1. Electrophoretic and Immunological Investigations of the Influence of High Temperature on A. simplex Antigen
4.2. Identification and Label-Free Quantification of Proteins, Allergens, and Potential Allergens of A. simplex
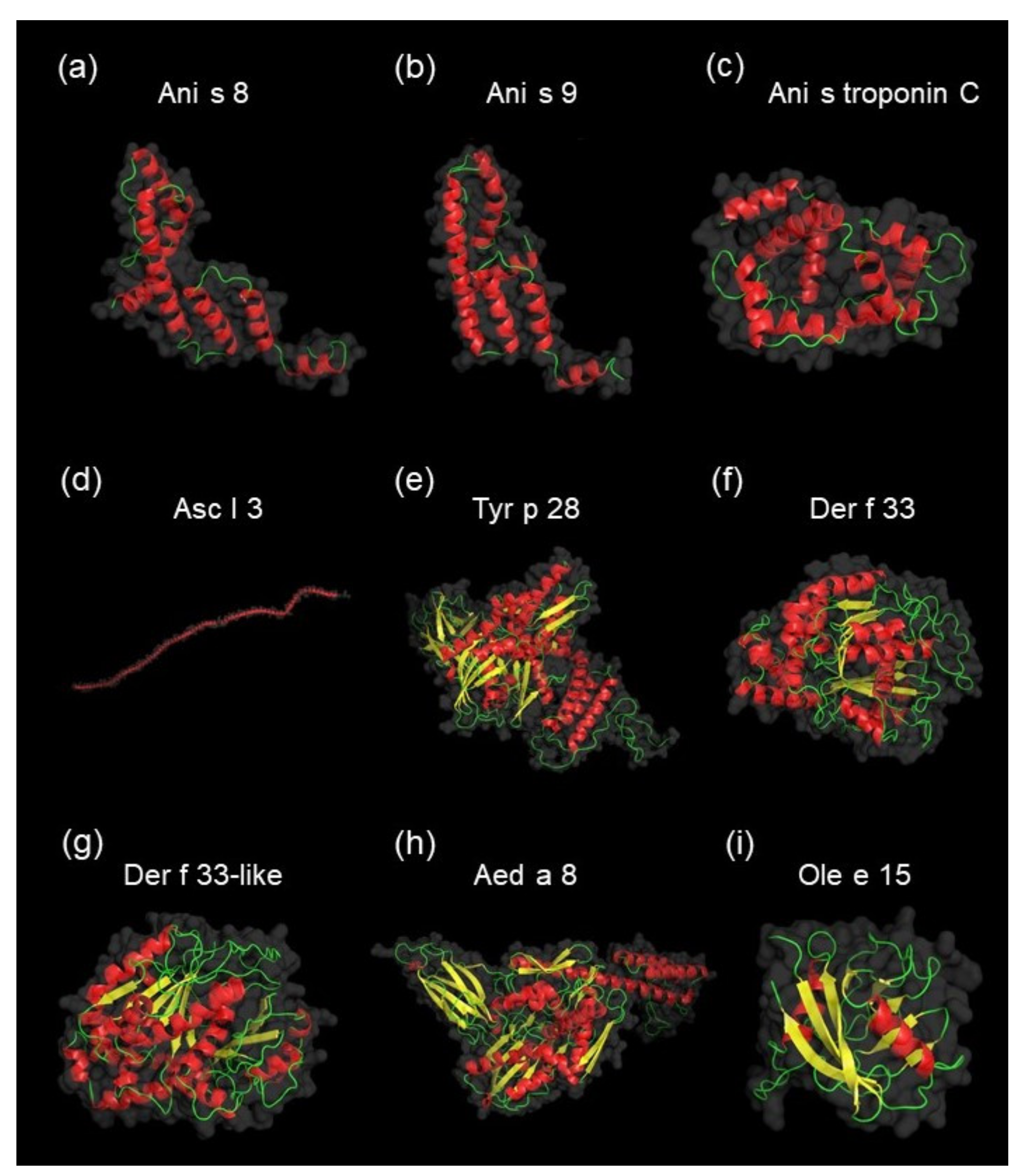
4.3. Computational Investigations of Detected Proteins, Allergens, and Potential Allergens of A. simplex
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jenkins, E.J.; Castrodale, L.J.; de Rosemond, S.J.; Dixon, B.R.; Elmore, S.A.; Gesy, K.M.; Hoberg, E.P.; Polley, L.; Schurer, J.M.; Simard, M. Tradition and transition: Parasitic zoonoses of people and animals in Alaska, northern Canada, and Greenland. Adv. Parasitol. 2013, 82, 33–204. [Google Scholar] [PubMed]
- Zolfaghari Emameh, R.; Purmonen, S.; Sukura, A.; Parkkila, S. Surveillance and diagnosis of zoonotic foodborne parasites. Food Sci. Nutr. 2018, 6, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Trevisan, C.; Torgerson, P.R.; Robertson, L.J. Foodborne Parasites in Europe: Present Status and Future Trends. Trends Parasitol. 2019, 35, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Pozio, E. How globalization and climate change could affect foodborne parasites. Exp. Parasitol. 2020, 208, 107807. [Google Scholar] [CrossRef]
- Różycki, M.; Bilska–Zając, E.; Kochanowski, M.; Grądziel-Krukowska, K.; Zdybel, J.; Karamon, J.; Wiśniewski, J.; Cencek, T. First case of Trichinella spiralis infection in beavers (Castor fiber) in Poland and Europe. Int. J. Parasitol. Parasites Wildl. 2020, 11, 46–49. [Google Scholar] [CrossRef]
- Sroka, J.; Karamon, J.; Wójcik-Fatla, A.; Piotrowska, W.; Dutkiewicz, J.; Bilska-Zając, E.; Zając, V.; Kochanowski, M.; Dąbrowska, J.; Cencek, T. Toxoplasma gondii infection in slaughtered pigs and cattle in Poland: Seroprevalence, molecular detection and characterization of parasites in meat. Parasites Vectors 2020, 13, 1–11. [Google Scholar] [CrossRef]
- Zolfaghari Emameh, R.; Kuuslahti, M.; Näreaho, A.; Sukura, A.; Parkkila, S. Innovative molecular diagnosis of Trichinella species based on β-carbonic anhydrase genomic sequence. Microb. Biotechnol. 2016, 9, 172–179. [Google Scholar] [CrossRef]
- Armentia, A.; Santos, J.; Serrano, Z.; Martín, B.; Martín, S.; Barrio, J.; Fernández, S.; González-Sagrado, M.; Pineda, F.; Palacios, R. Molecular diagnosis of allergy to Anisakis simplex and Gymnorhynchus gigas fish parasites. Allergol Immunopathol 2017, 45, 463–472. [Google Scholar] [CrossRef]
- Lim, H.; Jung, B.K.; Cho, J.; Yooyen, T.; Shin, E.H.; Chai, J.Y. Molecular diagnosis of cause of anisakiasis in humans, South Korea. Emerging Infect. Dis. 2015, 21, 342–344. [Google Scholar] [CrossRef]
- Kunz, S.; Balmer, V.; Sterk, G.J.; Pollastri, M.P.; Leurs, R.; Müller, N.; Hemphill, A.; Spycher, C. The single cyclic nucleotide-specific phosphodiesterase of the intestinal parasite Giardia lamblia represents a potential drug target. PLoS Negl. Trop. Dis. 2017, 11, e0005891. [Google Scholar] [CrossRef]
- Sánchez-Alonso, I.; Carballeda-Sangiao, N.; González-Muñoz, M.; Navas, A.; Arcos, S.C.; Mendizábal, A.; Tejada, M.; Careche, M. Pathogenic potential of Anisakis L3 after freezing in domestic freezers. Food Control 2018, 84, 61–69. [Google Scholar] [CrossRef]
- Panel on Biological Hazards. Scientific opinion on risk assessment of parasites in fishery products. EFSA J. 2010, 8, 1543. [Google Scholar] [CrossRef]
- Bao, M.; Pierce, G.J.; Strachan, N.J.; Pascual, S.; González-Muñoz, M.; Levsen, A. Human health, legislative and socioeconomic issues caused by the fish-borne zoonotic parasite Anisakis: Challenges in risk assessment. Trends Food Sci. Technol. 2019, 86, 298–310. [Google Scholar] [CrossRef]
- Guardone, L.; Armani, A.; Nucera, D.; Costanzo, F.; Mattiucci, S.; Bruschi, F. Human anisakiasis in Italy: A retrospective epidemiological study over two decades. Parasite 2018, 25, 41. [Google Scholar] [CrossRef] [PubMed]
- Orphanet. Prevalence and Incidence of Rare Diseases. Available online: http://www.orpha.net/orphacom/cahiers/docs/GB/Prevalence_of_rare_diseases_by_alphabetical_list.pdf (accessed on 12 June 2020).
- Bao, M.; Pierce, G.J.; Strachan, N.J.; Martínez, C.; Fernández, R.; Theodossiou, I. Consumers’ attitudes and willingness to pay for Anisakis-free fish in Spain. Fish. Res. 2018, 202, 149–160. [Google Scholar] [CrossRef]
- Bao, M.; Pierce, G.J.; Pascual, S.; González-Muñoz, M.; Mattiucci, S.; Mladineo, I.; Cipriani, P.; Bušelić, I.; Strachan, N.J. Assessing the risk of an emerging zoonosis of worldwide concern: Anisakiasis. Sci. Rep. 2017, 7, 43699. [Google Scholar] [CrossRef]
- Carballeda-Sangiao, N.; Rodríguez-Mahillo, A.I.; Careche, M.; Navas, A.; Caballero, T.; Dominguez-Ortega, J.; Jurado-Palomo, J.; González-Muñoz, M. Ani s 11-like protein is a pepsin-and heat-resistant major allergen of Anisakis spp. and a valuable tool for Anisakis allergy component-resolved diagnosis. Int. Arch. Allergy Immunol. 2016, 169, 108–112. [Google Scholar] [CrossRef]
- Baird, F.J.; Su, X.; Aibinu, I.; Nolan, M.J.; Sugiyama, H.; Otranto, D.; Lopata, A.L.; Cantacessi, C. The Anisakis transcriptome provides a resource for fundamental and applied studies on allergy-causing parasites. PLoS Negl. Trop. Dis. 2016, 10, e0004845. [Google Scholar] [CrossRef]
- Fæste, C.K.; Jonscher, K.R.; Dooper, M.M.; Egge-Jacobsen, W.; Moen, A.; Daschner, A.; Egaas, E.; Christians, U. Characterisation of potential novel allergens in the fish parasite Anisakis simplex. EuPA Open Proteom. 2014, 4, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Llorens, C.; Arcos, S.C.; Robertson, L.; Ramos, R.; Futami, R.; Soriano, B.; Ciordia, S.; Careche, M.; González-Muñoz, M.; Jiménez-Ruiz, Y.; et al. Functional insights into the infective larval stage of Anisakis simplex s.s., Anisakis pegreffii and their hybrids based on gene expression patterns. BMC Genom. 2018, 19, 592. [Google Scholar] [CrossRef]
- Arcos, S.C.; Ciordia, S.; Roberston, L.; Zapico, I.; Jiménez-Ruiz, Y.; Gonzalez-Muñoz, M.; Moneo, I.; Carballeda-Sangiao, N.; Rodriguez-Mahillo, A.; Albar, J.P.; et al. Proteomic profiling and characterization of differential allergens in the nematodes Anisakis simplex sensu stricto and A. pegreffii. Proteomics 2014, 14, 1547–1568. [Google Scholar] [CrossRef] [PubMed]
- Kochanowski, M.; Dąbrowska, J.; Różycki, M.; Karamon, J.; Sroka, J.; Cencek, T. Proteomic profiling reveals new insights into the allergomes of Anisakis simplex, Pseudoterranova decipiens and Contracaecum osculatum. J. Parasitol. 2020, in press. [Google Scholar]
- Anibarro, B.; Seoane, F.; Mugica, M. Involvement of hidden allergens in food allergic reactions. J Investig. Allergol. Clin. Immunol. 2007, 17, 168–172. [Google Scholar] [PubMed]
- Audicana, M.T.; Ansotegui, I.J.; de Corres, L.F.; Kennedy, M.W. Anisakis simplex: Dangerous—Dead and alive? Trends Parasitol. 2002, 18, 20–25. [Google Scholar] [CrossRef]
- Audicana, M.T.; Kennedy, M.W. Anisakis simplex: From obscure infectious worm to inducer of immune hypersensitivity. Clin. Microbiol. Rev. 2008, 21, 360–379. [Google Scholar] [CrossRef]
- Bucci, C.; Gallotta, S.; Morra, I.; Fortunato, A.; Ciacci, C.; Iovino, P. Anisakis, just think about it in an emergency! Int. J. Infect. Dis. 2013, 17, e1071–e1072. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuizen, N.; Lopata, A.L.; Jeebhay, M.F.; De’Broski, R.H.; Robins, T.G.; Brombacher, F. Exposure to the fish parasite Anisakis causes allergic airway hyperreactivity and dermatitis. J. Allergy Clin. Immunol. 2006, 117, 1098–1105. [Google Scholar] [CrossRef]
- Uña-Gorospe, M.; Herrera-Mozo, I.; Canals, M.L.; Martí-Amengual, G.; Sanz-Gallen, P. Occupational disease due to Anisakis simplex in fish handlers. Int. Marit. Health 2018, 69, 264–269. [Google Scholar] [CrossRef]
- Mazzucco, W.; Lacca, G.; Cusimano, R.; Provenzani, A.; Costa, A.; Di Noto, A.M.; Massenti, M.F.; Leto-Barone, M.S.; Lorenzo, G.D.; Vitale, F. Prevalence of sensitization to Anisakis simplex among professionally exposed populations in Sicily. Arch. Environ. Occup. Health 2012, 67, 91–97. [Google Scholar] [CrossRef]
- Barbarroja-Escudero, J.; Sanchez-Gonzalez, M.; Antolin-Amerigo, D.; Rodriguez-Rodriguez, M.; Alvarez-Mon, M. Nonoccupational airborne-induced anaphylaxis caused by Anisakis simplex. J. Investig. Allergol. Clin. Immunol. 2016, 26, 196–197. [Google Scholar] [CrossRef][Green Version]
- Meseguer, J.; Navarro, V.; Sánchez-Guerrero, I.; Bartolomé, B.; Negro Alvarez, J.N. Anisakis simplex allergy and nephrotic syndrome. Allergol. Immunopathol. 2007, 35, 216–220. [Google Scholar] [CrossRef][Green Version]
- Cuende, E.; Audicana, M.; Garcia, M.; Anda, M.; Fernández, L.C.; Jimenez, C.; Vesga, J. Rheumatic manifestations in the course of anaphylaxis caused by Anisakis simplex. Clin. Exp. Rheumatol. 1998, 16, 303–304. [Google Scholar] [PubMed]
- Moneo, I.; Carballeda-Sangiao, N.; González-Muñoz, M. New Perspectives on the Diagnosis of Allergy to Anisakis spp. Curr. Allergy Asthma Rep. 2017, 17, 27. [Google Scholar] [CrossRef] [PubMed]
- Caballero, M.L.; Moneo, I. Several allergens from Anisakis simplex are highly resistant to heat and pepsin treatments. Parasitol. Res. 2004, 93, 248–251. [Google Scholar] [CrossRef]
- Armentia, A.; Martin-Gil, F.; Pascual, C.; Martin-Esteban, M.; Callejo, A.; Martinez, C. Anisakis simplex allergy after eating chicken meat. J. Investig. Allergol. Clin. Immunol. 2006, 16, 258–263. [Google Scholar] [PubMed]
- Verhoeckx, K.C.; Vissers, Y.M.; Baumert, J.L.; Faludi, R.; Feys, M.; Flanagan, S.; Herouet-Guicheney, C.; Holzhauser, T.; Shimojo, R.; van der Bolt, N.; et al. Food processing and allergenicity. Food Chem. Toxicol. 2015, 80, 223–240. [Google Scholar] [CrossRef] [PubMed]
- Vojdani, A. Detection of IgE, IgG, IgA and IgM antibodies against raw and processed food antigens. Nutr. Metab. 2009, 6, 22. [Google Scholar] [CrossRef]
- Sathe, S.K.; Teuber, S.S.; Roux, K.H. Effects of food processing on the stability of food allergens. Biotechnol. Adv. 2005, 23, 423–429. [Google Scholar] [CrossRef]
- Pedreschi, R.; Nørgaard, J.; Maquet, A. Current challenges in detecting food allergens by shotgun and targeted proteomic approaches: A case study on traces of peanut allergens in baked cookies. Nutrients 2012, 4, 132–150. [Google Scholar] [CrossRef]
- Di Girolamo, F.; Muraca, M.; Mazzina, O.; Lante, I.; Dahdah, L. Proteomic applications in food allergy: Food allergenomics. Curr. Opin. Allergy Clin. Immunol. 2015, 15, 259–266. [Google Scholar] [CrossRef]
- Kochanowski, M.; González-Muñoz, M.; Gómez-Morales, M.Á.; Gottstein, B.; Dąbrowska, J.; Różycki, M.; Cencek, T.; Müller, N.; Boubaker, G. Comparative analysis of excretory-secretory antigens of Anisakis simplex, Pseudoterranova decipiens and Contracaecum osculatum regarding their applicability for specific serodiagnosis of human anisakidosis based on IgG-ELISA. Exp. Parasitol. 2019, 197, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Gasser, R.B.; Podolska, M.; Chilton, N.B. Characterisation of anisakid nematodes with zoonotic potential by nuclear ribosomal DNA sequences. Int. J. Parasitol. 1998, 28, 1911–1921. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Ikeda, K.; Shiomi, K. Elucidation of IgE-binding epitopes of Ani s 1: The major Anisakis simplex allergen. Mol. Biochem. Parasitol. 2010, 174, 128–131. [Google Scholar] [CrossRef]
- Garcia Alonso, M.; Caballero, M.; Umpierrez, A.; Lluch-Bernal, M.; Knaute, T.; Rodríguez-Pérez, R. Relationships between T cell and IgE/IgG4 epitopes of the Anisakis simplex major allergen Ani s 1. Clin. Exp. Allergy 2015, 45, 994–1005. [Google Scholar] [CrossRef]
- García-Mayoral, M.F.; Treviño, M.A.; Perez-Pinar, T.; Caballero, M.L.; Knaute, T.; Umpierrez, A.; Bruix, M.; Rodriguez-Perez, R. Relationships between IgE/IgG4 epitopes, structure and function in Anisakis simplex Ani s 5, a member of the SXP/RAL-2 protein family. PLoS Negl. Trop. Dis. 2014, 8, e2735. [Google Scholar] [CrossRef]
- Carballeda-Sangiao, N.; Olivares, F.; Rodriguez-Mahillo, A.I.; Careche, M.; Tejada, M.; Moneo, I.; Gonzalez-Muñoz, M. Identification of autoclave-resistant Anisakis simplex allergens. J. Food Prot. 2014, 77, 605–609. [Google Scholar] [CrossRef]
- Klapper, R.; Carballeda-Sangiao, N.; Kuhn, T.; Jensen, H.M.; Buchmann, K.; Gonzalez-Muñoz, M.; Karl, H. Anisakid infection levels in fresh and canned cod liver: Significant reduction through liver surface layer removal. Food Control 2018, 92, 17–24. [Google Scholar] [CrossRef]
- Cuadrado, C.; Sanchiz, A.; Vicente, F.; Ballesteros, I.; Linacero, R. Changes induced by pressure processing on immunoreactive proteins of tree nuts. Molecules 2020, 25, 954. [Google Scholar] [CrossRef] [PubMed]
- Lasekan, A.O.; Nayak, B. Effects of buffer additives and thermal processing methods on the solubility of shrimp (Penaeus monodon) proteins and the immunoreactivity of its major allergen. Food Chem. 2016, 200, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, C.; Cabanillas, B.; Pedrosa, M.M.; Varela, A.; Guillamón, E.; Muzquiz, M.; Crespo, J.F.; Rodriguez, J.; Burbano, C. Influence of thermal processing on IgE reactivity to lentil and chickpea proteins. Mol. Nutr. Food Res. 2009, 53, 1462–1468. [Google Scholar] [CrossRef] [PubMed]
- Viñas, M.; Postigo, I.; Suñén, E.; Martínez, J. Urticaria and silent parasitism by Ascaridoidea: Component-resolved diagnosis reinforces the significance of this association. PLoS Negl. Trop. Dis. 2020, 14, e0008177. [Google Scholar] [CrossRef] [PubMed]
- Daschner, A.; de La Osada, F.V.; Leza, B.G. Specific IgG and IgG4 in patients with IgE-antibodies against Anisakis simplex: Acute urticaria in gastro-allergic Anisakiasis versus chronic urticaria. J. Allergy Clin. Immunol. 2002, 109, S126. [Google Scholar] [CrossRef]
- Vidaček, S.; de las Heras, C.; Solas, M.T.; Mendizábal, A.; Rodriguez-Mahillo, A.I.; Tejada, M. Antigenicity and viability of Anisakis larvae infesting hake heated at different time-temperature conditions. J. Food Prot. 2010, 73, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Tejada, M.; Olivares, F.; de las Heras, C.; Careche, M.; Solas, M.T.; García, M.L.; Fernandez, A.; Mendizábal, A.; Navas, A.; Rodríguez-Mahillo, A.I.; et al. Antigenicity of Anisakis simplex s.s. L3 in parasitized fish after heating conditions used in the canning processing. J. Sci. Food Agric. 2015, 95, 922–927. [Google Scholar] [CrossRef]
- Rodríguez-Mahillo, A.I.; González-Muñoz, M.; de las Heras, C.; Tejada, M.; Moneo, I. Quantification of Anisakis simplex allergens in fresh, long-term frozen, and cooked fish muscle. Foodborne Pathog. Dis. 2010, 7, 967–973. [Google Scholar] [CrossRef] [PubMed]
- Vidaček, S.; De Las Heras, C.; Solas, M.T.; García, M.L.; Mendizabal, A.; Tejada, M. Viability and antigenicity of Anisakis simplex after conventional and microwave heating at fixed temperatures. J. Food Prot. 2011, 74, 2119–2126. [Google Scholar] [CrossRef]
- Carrera, M.; Gallardo, J.M.; Pascual, S.; González, Á.F.; Medina, I. Protein biomarker discovery and fast monitoring for the identification and detection of Anisakids by parallel reaction monitoring (PRM) mass spectrometry. J. Proteom. 2016, 142, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Ubeira, F.M. Travelling with Anisakis allergens. Int. Arch. Allergy Immunol. 2014, 163, 243–244. [Google Scholar] [CrossRef] [PubMed]
- Guarneri, F.; Guarneri, C.; Benvenga, S. Cross-reactivity of Anisakis simplex: Possible role of Ani s 2 and Ani s 3. Int. J. Dermatol. 2007, 46, 146–150. [Google Scholar] [CrossRef]
- Quiazon, K.M.A.; Zenke, K.; Yoshinaga, T. Molecular characterization and comparison of four Anisakis allergens between Anisakis simplex sensu stricto and Anisakis pegreffii from Japan. Mol. Biochem. Parasitol. 2013, 190, 23–26. [Google Scholar] [CrossRef] [PubMed]
- Hauser, M.; Roulias, A.; Ferreira, F.; Egger, M. Panallergens and their impact on the allergic patient. Allergy Asthma Clin. Immunol. 2010, 6, 1. [Google Scholar] [CrossRef] [PubMed]
- Goodman, R.E.; Ebisawa, M.; Ferreira, F.; Sampson, H.A.; van Ree, R.; Vieths, S.; Baumert, J.L.; Bohle, B.; Lalithambika, S.; Wise, J.; et al. AllergenOnline: A peer-reviewed, curated allergen database to assess novel food proteins for potential cross-reactivity. Mol. Nutr. Food Res. 2016, 60, 1183–1198. [Google Scholar] [CrossRef] [PubMed]
- Aalberse, R.C. Structural biology of allergens. J. Allergy Clin. Immunol. 2000, 106, 228–238. [Google Scholar] [CrossRef]
- Johansson, E.; Aponno, M.; Lundberg, M.; Van Hage-Hamsten, M. Allergenic cross-reactivity between the nematode Anisakis simplex and the dust mites Acarus siro, Lepidoglyphus destructor, Tyrophagus putrescentiae, and Dermatophagoides pteronyssinus. Allergy 2001, 56, 660–666. [Google Scholar] [CrossRef]
- Carballeda-Sangiao, N.; Rodríguez-Mahillo, A.; Puente, S.; Gutiérrez, M.; Moneo, I.; González-Muñoz, M. Anisakis/Ascaris IgE ratio improves specificity for the diagnosis of Anisakis simplex sensitization in travellers and immigrants. Acta Trop. 2014, 138, 1–4. [Google Scholar] [CrossRef]
- Maldonado, J.L.; Hita, L.M.; Sáez, V.D.; Almendros, I.M.; López, A.V.; Bueno, M.C. Cross-reactivity between antigens of Anisakis simplex s.l. and other ascarid nematodes. Parasite 2004, 11, 219–223. [Google Scholar] [CrossRef][Green Version]
- Sun, X.; Shan, X.; Yan, Z.; Zhang, Y.; Guan, L. Prediction and characterization of the linear IgE epitopes for the major soybean allergen β-conglycinin using immunoinformatics tools. Food Chem. Toxicol. 2013, 56, 254–260. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, Y.; Wu, Y.; Wang, H.; Ke, J. Prediction and identification of B-cell epitopes for tumor necrosis factor-α. Mol. Med. Rep. 2017, 16, 3439–3444. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-m.; Wang, H.-j.; Du, L.; Han, X.-m.; Ye, Z.-y.; Fang, Y.; Tao, H.-q.; Zhao, Z.-s.; Zhou, Y.-l. Screening and identification of novel B cell epitopes in human heparanase and their anti-invasion property for hepatocellular carcinoma. Cancer Immunol. Immunother. 2009, 58, 1387–1396. [Google Scholar] [CrossRef] [PubMed]
- Bodelón, G.; Palomino, C.; Fernández, L.Á. Immunoglobulin domains in Escherichia coli and other enterobacteria: From pathogenesis to applications in antibody technologies. FEMS Microbiol. Rev. 2013, 37, 204–250. [Google Scholar] [CrossRef] [PubMed]
- Halaby, D.; Poupon, A.; Mornon, J.P. The immunoglobulin fold family: Sequence analysis and 3D structure comparisons. Protein Eng. 1999, 12, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Lopata, A.; Kleine-Tebbe, J.; Kamath, S. Allergens and molecular diagnostics of shellfish allergy. In Molecular Allergy Diagnostics; Springer: Berlin, Germany, 2017; pp. 399–414. [Google Scholar]
- Emameh, R.Z.; Masoori, L.; Taheri, R.A.; Falak, R. Identification and characterization of parvalbumin-like protein in Trichophyton violaceum. Fungal Biol. 2020, 124, 592–600. [Google Scholar] [CrossRef] [PubMed]
- Gangl, K.; Niederberger, V.; Davies, J.; Valenta, R.; Nandy, A. Marker allergens and panallergens in tree and grass pollen allergy. In Molecular Allergy Diagnostics; Springer: Berlin, Germany, 2017; pp. 203–226. [Google Scholar]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Moseley, P.L. Heat shock proteins and heat adaptation of the whole organism. J. Appl. Physiol. 1997, 83, 1413–1417. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Bannai, H.; Sakurai, T.; Inoue, N.; Sugimoto, C.; Igarashi, I. Cloning and expression of mitochondrial heat shock protein 70 of Trypanosoma congolense and potential use as a diagnostic antigen. Clin. Diagn. Lab. Immunol. 2003, 10, 926–933. [Google Scholar] [CrossRef]
- Tazir, Y.; Steisslinger, V.; Soblik, H.; Younis, A.E.; Beckmann, S.; Grevelding, C.G.; Steen, H.; Brattig, N.W.; Erttmann, K.D. Molecular and functional characterisation of the heat shock protein 10 of Strongyloides ratti. Mol. Biochem. Parasitol. 2009, 168, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Aki, T.; Fujikawa, A.; Wada, T.; Jyo, T.; Shigeta, S.; Murooka, Y.; Oka, S.; Ono, K. Cloning and expression of cDNA coding for a new allergen from the house dust mite, Dermatophagoides farinae: Homology with human heat shock cognate proteins in the heat shock protein 70 family. J. Biochem. 1994, 115, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.G.; Su, S.N.; Chiang, B.L.; Lee, H.J.; Chow, L.P. Proteome mining for novel IgE-binding proteins from the German cockroach (Blattella germanica) and allergen profiling of patients. Proteomics 2010, 10, 3854–3867. [Google Scholar] [CrossRef]
- Rao, K.; Eswaran, M.; Ravi, V.; Gnanasekhar, B.; Narayanan, R.; Kaliraj, P.; Jayaraman, K.; Marson, A.; Raghavan, N.; Scott, A.L. The Wuchereria bancrofti orthologue of Brugia malayi SXP1 and the diagnosis of bancroftian filariasis. Mol. Biochem. Parasitol. 2000, 107, 71–80. [Google Scholar] [CrossRef]
- Chandrashekar, R.; Curtis, K.C.; Ramzy, R.M.; Liftis, F.; Li, B.W.; Weil, G.J. Molecular cloning of Brugia malayi antigens for diagnosis of lymphatic filariasis. Mol. Biochem. Parasitol. 1994, 64, 261–271. [Google Scholar] [CrossRef]
- Gallin, M.Y.; Tan, M.; Kron, M.A.; Rechnitzer, D.; Greene, B.M.; Newland, H.S.; White, A.T.; Taylor, H.R.; Unnasch, T.R. Onchocerca volvulus recombinant antigen: Physical characterization and clinical correlates with serum reactivity. J. Infect. Dis. 1989, 160, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Faber, M.; Pascal, M.; El Kharbouchi, O.; Sabato, V.; Hagendorens, M.; Decuyper, I.; Bridts, C.; Ebo, D. Shellfish allergens: Tropomyosin and beyond. Allergy 2017, 72, 842–848. [Google Scholar] [CrossRef]
- Wong, L.; Huang, C.H.; Lee, B.W. Shellfish and house dust mite allergies: Is the link tropomyosin? Allergy Asthma Immunol. Res. 2016, 8, 101–106. [Google Scholar] [CrossRef]
- Reese, G.; Ayuso, R.; Lehrer, S.B. Tropomyosin: An invertebrate pan–allergen. Int. Arch. Allergy Immunol. 1999, 119, 247–258. [Google Scholar] [CrossRef]
- Kamath, S.D.; Rahman, A.M.A.; Komoda, T.; Lopata, A.L. Impact of heat processing on the detection of the major shellfish allergen tropomyosin in crustaceans and molluscs using specific monoclonal antibodies. Food Chem. 2013, 141, 4031–4039. [Google Scholar] [CrossRef]
- Bessot, J.; Metz-Favre, C.; Rame, J.; De Blay, F.; Pauli, G. Tropomyosin or not tropomyosin, what is the relevant allergen in house dust mite and snail cross allergies? Eur. Ann. Allergy Clin. Immunol. 2010, 42, 3–10. [Google Scholar] [PubMed]
- Laclette, J.; Shoemaker, C.; Richter, D.; Arcos, L.; Pante, N.; Cohen, C.; Bing, D.; Nicholson-Weller, A. Paramyosin inhibits complement C1. J. Immunol. 1992, 148, 124–128. [Google Scholar] [PubMed]
- Kalinna, B.; McManus, D. An IgG (Fc γ,)-binding protein of Taenia crassiceps (Cestoda) exhibits sequence homology and antigenic similarity with schistosome paramyosin. Parasitology 1993, 106, 289–296. [Google Scholar] [CrossRef]
- Park, T.-J.; Kang, J.-M.; Na, B.-K.; Sohn, W.-M. Molecular cloning and characterization of a paramyosin from Clonorchis sinensis. Korean J. Parasitol. 2009, 47, 359–367. [Google Scholar] [CrossRef][Green Version]
- Pérez-Pérez, J.; Fernández-Caldas, E.; Marañón, F.; Sastre, J.; Bernal, M.L.; Rodríguez, J.; Bedate, C.A. Molecular cloning of paramyosin, a new allergen of Anisakis simplex. Int. Arch. Allergy Immunol. 2000, 123, 120–129. [Google Scholar] [CrossRef]
- Olivares, F.; González-Muñoz, M.; Carballeda-Sangiao, N.; Rodríguez-Mahillo, A.; Careche, M.; de las Heras, C.; Navas, A.; Tejada, M. Removal of Anisakis simplex allergens from infected fish during the washing step of surimi production. J. Sci. Food Agric. 2015, 95, 2626–2631. [Google Scholar] [CrossRef] [PubMed]

| No | UniProt ID | Protein Name 1 | InterPro Family 2 (ID) | InterPro Domain 2 (ID) | Mw 3 (Da) | pI 3 | Mascot Scor 4 | Coverage 4 (%) | emPAI Value 4 |
|---|---|---|---|---|---|---|---|---|---|
| 1 | A0A0M3K6N3 | Myosin, essential light chain (inferred by orthology to a Caenorhabditis elegans protein) | EF-hand domain pair (IPR011992); Myosin light chain alkali (IPR029655) | EF-hand domain (IPR002048) | 16,418 | 4.89 | 4418 | 90 | 201.76 |
| 2 | A0A0M3KCE6 | Tropomyosin (inferred by orthology to a C. elegans protein) | Tropomyosin (IPR000533) | -5 | 13,701 | 4.54 | 3023 | 63 | 109.71 |
| 3 | A0A0M3J349 | DUF4440 domain-containing protein | Nuclear transport factor 2 (NTF2)-like domain superfamily (IPR032710) | Domain of unknown function DUF4440 (IPR027843) | 13,671 | 6.29 | 785 | 72 | 81.05 |
| 4 | A0A0M3J4D2 | Uncharacterized protein | - | - | 23,930 | 8.72 | 1897 | 71 | 67.66 |
| 5 | A0A0M3K7H9 | Uncharacterized protein | - | - | 11,337 | 9.89 | 922 | 70 | 63.02 |
| 6 | Q9NAS5 | Tropomyosin (allergen Ani s 3) | Tropomyosin (IPR000533) | - | 33,203 | 4.68 | 3241 | 72 | 62.69 |
| 7 | A0A0M3KA05 | SXP/RAL-2 family protein 2 isoform 1 | - | Domain of unknown function DUF4440 (IPR027843) | 14,517 | 4.52 | 5725 | 71 | 45.10 |
| 8 | A0A0M3K8L6 | Uncharacterized protein | DVA-1 superfamily (IPR038289) | Polyprotein allergen, nematode (IPR032487) | 42,337 | 7.07 | 3504 | 62 | 42.49 |
| 9 | A0A0M3KF53 | Uncharacterized protein | - | - | 26,477 | 4.98 | 2630 | 70 | 38.24 |
| 10 | A0A0M3KIS5 | IF rod domain-containing protein | Intermediate filament, rod domain, coil 1B (IPR042180) | Intermediate filament, rod domain (IPR039008) | 27,871 | 5.17 | 1999 | 73 | 33.77 |
| 11 | A0A0M3JCV3 | KASH_CCD domain-containing protein | - | KASH5-like coiled-coil domain (IPR028168) | 17,230 | 5.4 | 1885 | 68 | 32.84 |
| 12 | A0A0M3J6C9 | Uncharacterized protein | - | - | 26,754 | 7.78 | 1755 | 54 | 30.87 |
| 13 | A0A0M3JGD2 | Uncharacterized protein | - | - | 10,889 | 4.86 | 771 | 52 | 23.42 |
| 14 | A0A158PP35 | Paramyosin (inferred by orthology to a C. elegans protein) | - | Myosin tail (IPR002928) | 101,686 | 5.34 | 11203 | 67 | 22.63 |
| 15 | A0A0M3JX08 | Protein lethal(2)essential for life (inferred by orthology to a Drosophila melanogaster protein) | HSP20-like chaperone (IPR008978); Alpha-crystallin/Heat shock protein (IPR001436) | Alpha-crystallin/Hsp20 domain (IPR002068) | 21,063 | 5.98 | 1284 | 80 | 21.20 |
| 16 | A0A0M3JAH1 | Intermediate filament protein ifb-1 (inferred by orthology to a C. elegans protein) | Lamin tail domain superfamily (IPR036415) | Intermediate filament, rod domain (IPR039008); Lamin tail domain (IPR001322) | 24,324 | 5.33 | 991 | 65 | 20.01 |
| 17 | A0A0M3JTQ4 | Myosin regulatory light chain 2 (inferred by orthology to a D. melanogaster protein) | EF-hand domain pair (IPR011992) | EF-hand domain (IPR002048) | 18,715 | 5.05 | 2597 | 81 | 19.51 |
| 18 | A0A0M3KKR0 | Uncharacterized protein | - | - | 5009 | 4.79 | 1077 | 56 | 18.10 |
| 19 | A0A0M3J766 | Uncharacterized protein | - | - | 13,471 | 5.82 | 522 | 48 | 16.84 |
| 20 | Q9NJA9 | Paramyosin (allergen Ani s 2) | - | Myosin tail (IPR002928) | 100,461 | 5.21 | 9369 | 69 | 16.04 |
| 21 | A1IKL2 | SXP/RAL-2 family protein Ani s 5 (allergen Ani s 5) | - | Domain of unknown function DUF4440 (IPR027843) | 14,738 | 4.85 | 1765 | 47 | 14.65 |
| 22 | A0A0M3JID9 | IF rod domain-containing protein | Intermediate filament, rod domain, coil 1B (IPR042180) | Intermediate filament, rod domain (IPR039008) | 10,555 | 6.29 | 649 | 43 | 14.45 |
| 23 | A0A0M3JGV0 | Uncharacterized protein | - | - | 13,766 | 4.73 | 844 | 42 | 14.31 |
| 24 | A0A0M3IYQ5 | Glutamate dehydrogenase, mitochondrial (inferred by orthology to a D. melanogaster protein) | NAD(P)-binding domain superfamily (IPR036291) | - | 11,634 | 8.89 | 650 | 61 | 14.09 |
| 25 | A0A0M3J159 | Small heat shock protein (SHSP) domain-containing protein | HSP20-like chaperone (IPR008978); Alpha-crystallin/Heat shock protein (IPR001436) | Alpha-crystallin/Hsp20 domain (IPR002068) | 13,230 | 6.29 | 1036 | 54 | 13.78 |
| No | UniProt ID | Allergen Name 1 | Protein Name | InterPro Family (ID) | InterPro Domain (ID) | AllFam 2 (ID) | Mw (Da) | pI | Mascot Score | Coverage (%) | emPAI Value |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Q7Z1K3 | Ani s 1 | Major allergen Ani s 1 (21 kDa allergen) | Pancreatic trypsin inhibitor Kunitz domain superfamily (IPR036880) | Pancreatic trypsin inhibitor Kunitz domain (IPR002223) | Animal Kunitz serine protease inhibitor (AF003) | 18,869 | 7.48 | 481 | 21 | 2.53 |
| 2 | Q9NJA9 | Ani s 2 | Paramyosin | - | Myosin tail (IPR002928) | Myosin heavy chain (AF100) | 100,461 | 5.21 | 9369 | 69 | 16.04 |
| 3 | Q9NAS5 | Ani s 3 | Tropomyosin | Tropomyosin (IPR000533) | - | Tropomyosin (AF054) | 33,203 | 4.68 | 3241 | 72 | 62.69 |
| 4 | Q14QT4 | Ani s 4 | Ani s 4 allergen | - | Cystatin domain (IPR000010) | Cystatin (AF005) | 10,291 | 5.54 | 196 | 27 | 3.67 |
| 5 | A1IKL2 | Ani s 5 | SXP/RAL-2 family protein Ani s 5 | - | Domain of unknown function DUF148 (IPR003677) | SXP/RAL-2 family (AF137) | 14,738 | 4.85 | 1765 | 47 | 14.65 |
| Possible Allergens 1 | Homologous Allergens 1 | AllergenOnline.org Result 1 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | UniProt ID | Protein Name | InterPro Family (ID) | InterPro Domain (ID) | Mw (Da) | pI | emPAI Value | Genbank ID | ALLERGEN name | Protein Name (Taxonomy) | AllFam (ID) | Identity 1 | Similarity 1 |
| 1 | A0A0M3KCE6 | Tropomyosin (inferred by orthology to a C. elegans protein) | Tropomyosin (IPR000533) | - | 13,701 | 4.54 | 109.71 | ACN32322.1 | Asc l 3 | Tropomyosin (Ascaris lumbricoides) | Tropomyosin (AF054) | 0.946 | 0.973 |
| 2 | A0A0M3KA05 | SXP/RAL-2 family protein 2 isoform 1 | - | Domain of unknown function DUF148 (IPR003677) | 14,517 | 4.52 | 45.10 | BAF75681.1 | Ani s 8 | SXP/RAL-2 family protein 2 isoform 1 (Anisakis simplex) | SXP/RAL-2 family (AF137) | 0.987 | 1 |
| 3 | A0A158PP35 | Paramyosin (inferred by orthology to a C. elegans protein) | - | Myosin tail (IPR002928) | 101,686 | 5.34 | 22.63 | Q9NJA9.1 | Ani s 2 | Paramyosin (Anisakis simplex) | Myosin heavy chain (AF100) | 0.854 | 0.958 |
| 4 | A0A0M3J4T5 | DUF148 domain-containing protein | - | Domain of unknown function DUF148 (IPR003677) | 16,646 | 5.56 | 7.10 | BAF43534.1 | Ani s 5 | SXP/RAL-2 family protein (Anisakis simplex) | SXP/RAL-2 family (AF137) | 0.971 | 0.993 |
| 5 | A0A0M3J693 | Tropomyosin (inferred by orthology to a C. elegans protein) | Tropomyosin (IPR000533) | - | 11,304 | 4.64 | 7.79 | ACN32322.1 | Asc l 3 | Tropomyosin (Ascaris lumbricoides) | Tropomyosin (AF054) | 0.824 | 0.953 |
| 6 | A0A0M3K9V2 | Heat shock 70 kDa protein cognate 1 (inferred by orthology to a D. melanogaster protein) | Heat shock protein 70 kD, peptide-binding domain superfamily (IPR029047); Heat shock protein 70 family (IPR013126); Heat shock protein 70 kD, C-terminal domain superfamily (IPR029048) | - | 67,585 | 5.45 | 4.84 | AOD75395.1 | Tyr p 28 | Heat shock-like protein (Tyrophagus putrescentiae) | Heat shock protein (Hsp70) (AF002) | 0.737 | 0.86 |
| 7 | A0A0M3KIW1 | Uncharacterized protein | Heat shock protein 70 kD, peptide-binding domain superfamily (IPR029047); Heat shock protein 70 family (IPR013126) | - | 24,599 | 8.49 | 2.31 | AOD75395.1 | Tyr p 28 | Heat shock-like protein (Tyrophagus putrescentiae) | Heat shock protein (Hsp70) (AF002) | 0.851 | 0.95 |
| 8 | A0A0M3J6G4 | Peptidyl-prolyl cis-trans isomerase | Cyclophilin-like domain superfamily (IPR029000) | Cyclophilin-type peptidyl-prolyl cis-trans isomerase domain (IPR002130) | 12,614 | 9.04 | 1.99 | AVV30163.1 | Ole e 15 | Cyclophilin 0101 (Olea europaea) | - | 0.789 | 0.916 |
| 9 | A0A0M3JU57 | Troponin-like protein | EF-hand domain pair (IPR011992) | EF-hand domain (IPR002048) | 19,444 | 4.15 | 1.39 | CAB58171.1 | Ani s troponin C | Troponin-like protein (Anisakis simplex) | EF hand family (AF007) | 0.994 | 1 |
| 10 | A0A0M3K5H6 | 78 kDa glucose-regulated protein | Heat shock protein 70 kD, peptide-binding domain superfamily (IPR029047); Heat shock protein 70 family (IPR013126); Heat shock protein 70 kD, C-terminal domain superfamily (IPR029048) | Endoplasmic reticulum chaperone BIP, nucleotide-binding domain (IPR042050) | 75,409 | 5.07 | 0.77 | ABF18258.1 | Aed a 8 | Heat shock cognate 70 (Aedes aegypti) | Heat shock protein (Hsp70) (AF002) | 0.777 | 0.93 |
| 11 | A0A0M3J8H0 | Uncharacterized protein | - | - | 9756 | 5.52 | 0.51 | ABV55106.1 | Ani s 9 | Ani s 9 allergen precursor (Anisakis simplex) | SXP/RAL-2 family (AF137) | 1 | 1 |
| 12 | A0A0M3K821 | Tubulin alpha chain | - | - | 47,784 | 4.83 | 0.42 | AIO08861.1 | Der f 33 | Der f 33 allergen (Dermatophagoides farinae) | Tubulin/FtsZ family (AF025) | 0.735 | 0.866 |
| 13 | A0A0M3KAH2 | Tubulin alpha chain | - | - | 48,222 | 5 | 0.51 | AUX14773.1 | Der f 33-like | Der f 33-like protein (Dermatophagoides pteronyssinus) | - | 0.743 | 0.891 |
| Peptide 1 | Epitope 3 | |||||
|---|---|---|---|---|---|---|
| Sequence 1 | Position1 | IEDB Epitope ID 3 | Sequence 3 | Position3 | B-Cell/T-Cell 3 | Reference 3 |
| Q7Z1K3 (Ani s 1) 4 | ||||||
| LFANCCK 2 | 188‒194 | 137143 | EDAKCERGKLFANCCK5 | 179‒194 | B-cell | [48] |
| 421049 | GKLFANCCK | 186‒194 | T-cell | [49] | ||
| EEELFAR | 137‒143 | 421391 | MGLCCPTKEEELFA | 129‒142 | B-cell | [49] |
| 421053 | GLCCPTKEEELFA | 130‒142 | B-cell | [49] | ||
| 420891 | EELFAREYEGVC | 138‒149 | T-cell | [49] | ||
| 420774 | CCPTKEEELFAR | 132‒143 | T-cell | [49] | ||
| GSGWMMTILGK | 160‒170 | 137278 | RGSGWMMTILGKSCD | 159‒173 | B-cell | [48] |
| 421086 | GWMMTILGKSCD | 162‒173 | T-cell | [49] | ||
| 421087 | GWMMTILGKSCDDQFCPEDA | 162‒181 | B-cell | [49] | ||
| 421233 | KMDRGSGWMMTI | 156‒167 | T-cell | [49] | ||
| 421406 | MTILGKSCDDQFCPEDA | 165‒181 | B-cell | [49] | ||
| 421613 | RGSGWMMTILG | 159‒169 | B-cell | [49] | ||
| MMAFMGLCCPTK | 125‒136 | 420792 | CPNGYQCKMMAFMG | 117‒130 | B-cell | [49] |
| 421053 | GLCCPTKEEELFA | 130‒142 | B-cell | [49] | ||
| 421091 | GYQCKMMAFMGL | 120‒131 | T-cell | [49] | ||
| 421092 | GYQCKMMAFMGLCCPTK | 120‒136 | B-cell | [49] | ||
| 421384 | MAFMGLCCPTKE | 126‒137 | T-cell | [49] | ||
| 421391 | MGLCCPTKEEELFA | 129‒142 | B-cell | [49] | ||
| MDRGSGWMMTILGK | 157‒170 | 137278 | RGSGWMMTILGKSCD | 159‒173 | B-cell | [48] |
| 421086 | GWMMTILGKSCD | 162‒173 | T-cell | [49] | ||
| 421087 | GWMMTILGKSCDDQFCPEDA | 162‒181 | B-cell | [49] | ||
| 421233 | KMDRGSGWMMTI | 156‒167 | T-cell | [49] | ||
| 421406 | MTILGKSCDDQFCPEDA | 165‒181 | B-cell | [49] | ||
| 421613 | RGSGWMMTILG | 159‒169 | B-cell | [49] | ||
| SGICLSFK | 56‒63 | 420822 | DKSGICLSFKYT | 54‒65 | T-cell | [49] |
| 421875 | WHDDKSGICLSFKY | 51‒64 | B-cell | [49] | ||
| A1IKL2 (Ani s 5) | ||||||
| ADAELSK | 105‒111 | 230144 | KKADAELSKIA | 103‒113 | B-cell | [50] |
| FETFKK | 73‒78 | - | - | - | - | - |
| TDPEIEK | 50‒56 | - | - | - | - | - |
| AKEAELAK | 82‒89 | - | - | - | - | - |
| KADAELSK | 104‒111 | 230144 | KKADAELSKIA | 103‒113 | B-cell | [50] |
| AFFELLK | 38‒44 | - | - | - | - | - |
| DETKTDPEIEK | 46‒56 | 230147 | KTDPEI | 49‒54 | B-cell | [50] |
| DELEKGIGPAVPQ | 140‒152 | 230136 | IQAIYKTLPQSVKDELEKGI | 127‒146 | B-cell | [50] |
| TLPQSVKDELEK | 133‒144 | 230136 | IQAIYKTLPQSVKDELEKGI | 127‒146 | B-cell | [50] |
| 230137 | IQKAQKIQAIYKTLPQSVKD | 121‒140 | B-cell | [50] | ||
| EAELAKAHEEAVAK | 84‒97 | - | - | - | - | - |
| DLDAWVDTLGGDYK | 57‒70 | 230149 | LDAWVDTLG | 58‒66 | B-cell | [50] |
| 230150 | LDAWVDTLGGDYKAKFETFK | 58‒77 | B-cell | [50] | ||
| DLDAWVDTLGGDYKAK | 57‒72 | 230149 | LDAWVDTLG | 58‒66 | B-cell | [50] |
| 230150 | LDAWVDTLGGDYKAKFETFK | 58‒77 | B-cell | [50] | ||
| TDPEIEKDLDAWVDTLGGDYK | 50‒70 | 230149 | LDAWVDTLG | 58‒66 | B-cell | [50] |
| 230150 | LDAWVDTLGGDYKAKFETFK | 58‒77 | B-cell | [50] | ||
| Peptide 1 | Epitope MHC II 3 | Epitope T-Cell 3 | Epitope B-Cell 3 | ||||
|---|---|---|---|---|---|---|---|
| Sequence 1 | Position 1 | Sequence 3 | Position 3 | Sequence 3 | Position 3 | Sequence 3 | Position 3 |
| Q9NJA9 (Ani s 2) 4 | |||||||
| ANLEAQK2 | 562‒568 | - | - | - | - | - | - |
| DLQVALK | 465‒471 | - | - | - | - | - | - |
| SLEEQVK | 712‒718 | - | - | - | - | - | - |
| AALAELQK | 402‒409 | - | - | LAELQKMKQLYEKAVE 5 | 404‒419 | - | - |
| TALDNAIR | 617‒624 | - | - | CKTALDN | 615‒621 | - | - |
| ELEDAEGR | 829‒836 | - | - | - | - | RELEDAEGRA | 828‒837 |
| ELHAADER | 674‒681 | - | - | LDEVTKELHAAD | 668‒679 | - | - |
| ALAELQQVR | 487‒495 | - | - | RAQRALAEL | 483‒491 | - | - |
| VALDEESAAR | 271‒280 | - | - | - | - | - | - |
| HKETQSALR | 760‒768 | - | - | - | - | RIRDLEVALDEETRRHKETQSALRKKDRRI | 745‒774 |
| LTAALADAEAR | 523‒533 | LTAALA | 523‒528 | - | - | - | - |
| AQNTIAILER | 361‒370 | - | - | - | - | - | - |
| EEEMEALRK | 506‒514 | - | - | - | - | - | - |
| VQLDNLQHVK | 222‒231 | - | - | - | - | - | - |
| LHELDLENAR | 449‒458 | - | - | - | - | - | - |
| LLQDDFESER | 43‒52 | - | - | - | - | QDDFESERELRNRIERERA | 45‒63 |
| MFVMAQDTADR | 787‒797 | - | - | DTADRMLE | 793‒800 | - | - |
| ADQAESSLNLIR | 837‒848 | - | - | - | - | - | - |
| FEQQTIELSNK | 178‒188 | - | - | - | - | - | - |
| KLHELDLENAR | 448‒458 | - | - | - | - | - | - |
| ISALSAELEECK | 605‒616 | ISALSA | 605‒610 | - | - | - | - |
| YQLAQQLEESR | 232‒242 | - | - | - | - | - | - |
| QAEADLEEAHVR | 628‒639 | AHVRISDLTS | 635‒645 | - | - | - | - |
| EVQMQIDEEHK | 776‒786 | - | - | - | - | EVQMQI | 776‒781 |
| LSLANTEITQWK | 287‒298 | - | - | - | - | - | - |
| ADLSVQLIALTDR | 63‒75 | - | - | - | - | - | - |
| DLQVALKESEAAR | 465‒477 | - | - | - | - | - | - |
| KISALSAELEECK | 604‒616 | ISALSA | 605‒610 | - | - | - | - |
| KQAEADLEEAHVR | 627‒639 | - | - | - | - | - | - |
| IRDLEVALDEETR | 746‒758 | - | - | - | - | RIRDLEVALDEETRRHKETQSALRKKDRRI | 745‒774 |
| ISDLTSINSNLTAIK | 640‒654 | AHVRISDLTS | 635‒645 | ISDLTSI | 640‒646 | - | - |
| LQSEVEVLIVDLEK | 347‒360 | - | - | - | - | - | - |
| IKEVQMQIDEEHK | 774‒786 | - | - | - | - | EVQMQI | 776-781 |
| QLQQTLDQYALAQR | 590‒603 | - | - | LEDTQRQLQQT | 584‒594 | - | - |
| ERADLSVQLIALTDR | 61‒75 | - | - | - | - | - | - |
| AVQELHEEQEHSMK | 692‒705 | - | - | ANRALADAARAVQELHE | 682‒698 | - | - |
| QLGEAEAMTMQNLQR | 808‒822 | - | - | AEAMTMQNLQRVRRYQRELEDA | 812‒833 | - | - |
| SRIDELLVELEAAQR | 384‒398 | - | - | IDELLVE | 386‒392 | - | - |
| QAEYEEQIEIMLQK | 323‒336 | - | - | - | - | - | - |
| VEDLNKHVNDLAQQR | 189‒203 | - | - | SNKVEDLNKHVNDLAQ | 186‒201 | LNKHVN | 192-197 |
| MAQKFEQQTIELSNK | 174‒188 | - | - | HVAEKMAQKFE | 169‒179 | - | - |
| QLQQTLDQYALAQRK | 590‒604 | - | - | LEDTQRQLQQT | 584‒594 | - | - |
| LQAENSDLLAEIHDQK | 206‒221 | - | - | DLLAEIH | 212‒218 | - | - |
| ISDLTSINSNLTAIKNK | 640‒656 | AHVRISDLTS | 635‒645 | ISDLTSI | 640‒646 | - | - |
| ARLQSEVEVLIVDLEK | 345‒360 | - | - | - | - | - | - |
| LETELSTAQADLDEVTK | 657‒673 | LSTAQA | 661‒666 | LDEVTKELHAAD | 668‒679 | - | - |
| LQHEVIELTATIDQLQK | 150‒166 | IELTAT | 155‒160 | EVIELTATIDQLQK | 153‒166 | - | - |
| LLEESQLENEDAMNVLR | 103‒119 | - | - | EDAMN | 112‒116 | - | - |
| YQAEIAELEMTVDNLNR | 545‒561 | - | - | EMTVDNLN | 553‒560 | - | - |
| QLQVQIQEAEAAALLGGKR | 719‒737 | - | - | - | - | - | - |
| FEQQTIELSNKVEDLNK | 178‒194 | - | - | SNKVEDLNKHVNDLAQ | 186‒201 | - | - |
| VEAEHKLSLANTEITQWK | 281‒298 | - | - | - | - | - | - |
| NKLETELSTAQADLDEVTK | 655‒673 | LSTAQA | 661‒666 | LDEVTKELHAAD | 668‒679 | - | - |
| SMQFEIDRLTAALADAEAR | 515‒533 | LTAALA | 523‒528 | - | - | - | - |
| QRLQAENSDLLAEIHDQK | 204‒221 | - | - | DLLAEIH | 212‒218 | - | - |
| SQMQAQLHQVQLELDSVR | 253‒270 | LHQVQL | 259‒264 | - | - | - | - |
| SKFDAEVALHHEEVEDLR | 299‒316 | - | - | EVEDLRKKMMQ | 311‒321 | - | - |
| SQMQAQLHQVQLELDSVR | 253‒270 | LHQVQL | 259‒264 | - | - | - | - |
| LLEESQLENEDAMNVLRK | 103‒120 | - | - | EDAMN | 112‒116 | - | - |
| KLLEESQLENEDAMNVLR | 102‒119 | - | - | EDAMN | 112‒116 | - | - |
| QRLQHEVIELTATIDQLQK | 148‒166 | - | - | EVIELTATIDQLQK | 153‒166 | - | - |
| QAEYEEQIEIMLQKVSQLEK | 323‒342 | - | - | - | - | - | - |
| MKAEIAR | 534‒540 | - | - | - | - | - | - |
| MKQLYEK | 410‒416 | - | - | - | - | - | - |
| EVELSKLR | 94‒101 | - | - | - | - | ESNRKREVELS | 88‒98 |
| DLEVALDEETRR | 748‒759 | - | - | - | - | RIRDLEVALDEETRRHKETQSALRKKDRRI | 745‒774 |
| LQDAECATDSQIESNR | 76‒91 | - | - | - | - | ESNRKREVELS | 88‒98 |
| KQSEQIIQLQANLEDTQR | 572‒589 | IIQLQA | 577‒582 | LEDTQRQLQQT | 584‒594 | - | - |
| SSTADMGALTSMSVADLGSLTR | 15‒36 | LTSMSVA | 23‒29 | MGALTSMSVADLGSLTRL | 20‒37 | - | - |
| ISALSAELEECKTALDNAIR | 605‒624 | ISALSA | 605‒610 | CKTALDN | 615‒621 | - | - |
| VSQLEK | 337‒342 | - | - | - | - | - | - |
| LNIQKR | 802‒807 | - | - | - | - | - | - |
| KSLEEQVK | 711‒718 | - | - | DALRKSLEEQV | 707‒717 | - | - |
| KYQAEIAELEMTVDNLNR | 544‒561 | - | - | EMTVDNLN | 553‒560 | - | - |
| Q9NAS5 (Ani s 3) | |||||||
| AEFAER | 239‒244 | - | - | - | - | - | - |
| ADAAEEK | 22‒28 | - | - | DAAEEKVRQMTDKLERIEEELRDTQKKMMQTENDLDKAQEDL | 23‒64 | - | - |
| KVMENR | 128‒133 | - | - | - | - | RKVMENRSFQDEE | 127‒139 |
| IEEELR | 39‒44 | - | - | - | - | - | - |
| LKEAETR | 232‒238 | - | - | - | - | - | - |
| SFQDEER | 134‒140 | - | - | - | - | RKVMENRSFQDEE | 127‒139 |
| SLEVSEEK | 206‒213 | - | - | - | - | - | - |
| ANTVESQLK | 141‒149 | - | - | - | - | - | - |
| QMTDKLER | 31‒38 | - | - | - | - | TDKLERIEEELRDT | 33‒46 |
| IEKDNALDR | 13‒21 | - | - | - | - | - | - |
| LEDELVHEK | 256‒264 | - | - | EFAERSVQKLQKEVDRLEDELVH | 240‒262 | DRLEDELV | 254‒261 |
| IVELEEELR | 190‒198 | - | - | KIVELEE | 189‒195 | LEEEL | 193‒197 |
| AEFAERSVQK | 239‒248 | - | - | EFAERSVQKLQKEVDRLEDELVH | 240‒262 | - | - |
| EDSYEEQIR | 218‒226 | - | - | EQIRTVS | 223‒229 | LQREDSYEEQIR | 215‒226 |
| MMQTENDLDK | 50‒59 | - | - | - | - | - | - |
| IEEELRDTQK | 39‒48 | - | - | - | - | TDKLERIEEELRDT | 33‒46 |
| EAQMLAEEADR | 150‒160 | - | - | - | - | - | - |
| MTLLEEELER | 92‒101 | - | - | - | - | - | - |
| SLEVSEEKALQR | 206‒217 | - | - | - | - | - | - |
| EAQMLAEEADRK | 150‒161 | - | - | - | - | - | - |
| VQEAEAEVAALNR | 78‒90 | - | - | - | - | - | - |
| DNALDRADAAEEK | 16‒28 | - | - | DAAEEKVRQMTDKLERIEEELRDTQKKMMQTENDLDKAQEDL | 23‒64 | - | - |
| RMTLLEEELER | 91‒101 | - | - | - | - | - | - |
| LEEATHTADESER | 113‒125 | - | - | LEEATHTADESERVRKVME | 113‒131 | - | - |
| KVQEAEAEVAALNR | 77‒90 | - | - | - | - | - | - |
| VQEAEAEVAALNRR | 78‒91 | - | - | - | - | - | - |
| EVDRLEDELVHEK | 252‒264 | - | - | EFAERSVQKLQKEVDRLEDELVH | 240‒262 | DRLEDELV | 254‒261 |
| ALQREDSYEEQIR | 214‒226 | - | - | EQIRTVS | 223‒229 | LQREDSYEEQIR | 215‒226 |
| AQEDLSTANSNLEEK | 60‒74 | - | - | - | - | - | - |
| SISEELDQTFQELSGY | 269-284 | - | - | - | - | - | - |
| YKSISEELDQTFQELSGY | 267‒284 | - | - | - | - | - | - |
| ANTVESQLKEAQMLAEEADR | 141‒160 | - | - | - | - | - | - |
| KMMQTENDLDK | 49‒59 | - | - | - | - | - | - |
| MMQTENDLDKAQEDLSTANSNLEEK | 50‒74 | - | - | DAAEEKVRQMTDKLERIEEELRDTQKKMMQTENDLDKAQEDL | 23‒64 | - | - |
| Q14QT4 (Ani s 4) | |||||||
| KQVVAGDK | 69‒76 | - | - | - | - | - | - |
| KWENFEEVK | 100‒108 | - | - | WQKKWENFEEVKVLKCDH | 97‒114 | - | - |
| ISAMINDGKPHELVK | 49‒63 | - | - | ELAGKSIAKISAMI | 40‒53 | - | - |
| - | - | GKPHELVKVVS | 56‒66 | - | - | ||
| ELAGKSIAK | 40‒48 | - | - | ELAGKSIAKISAMI | 40‒53 | - | - |
| WENFEEVK | 101‒108 | - | - | WQKKWENFEEVKVLKCDH | 97‒114 | - | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kochanowski, M.; Różycki, M.; Dąbrowska, J.; Bełcik, A.; Karamon, J.; Sroka, J.; Cencek, T. Proteomic and Bioinformatic Investigations of Heat-Treated Anisakis simplex Third-Stage Larvae. Biomolecules 2020, 10, 1066. https://doi.org/10.3390/biom10071066
Kochanowski M, Różycki M, Dąbrowska J, Bełcik A, Karamon J, Sroka J, Cencek T. Proteomic and Bioinformatic Investigations of Heat-Treated Anisakis simplex Third-Stage Larvae. Biomolecules. 2020; 10(7):1066. https://doi.org/10.3390/biom10071066
Chicago/Turabian StyleKochanowski, Maciej, Mirosław Różycki, Joanna Dąbrowska, Aneta Bełcik, Jacek Karamon, Jacek Sroka, and Tomasz Cencek. 2020. "Proteomic and Bioinformatic Investigations of Heat-Treated Anisakis simplex Third-Stage Larvae" Biomolecules 10, no. 7: 1066. https://doi.org/10.3390/biom10071066
APA StyleKochanowski, M., Różycki, M., Dąbrowska, J., Bełcik, A., Karamon, J., Sroka, J., & Cencek, T. (2020). Proteomic and Bioinformatic Investigations of Heat-Treated Anisakis simplex Third-Stage Larvae. Biomolecules, 10(7), 1066. https://doi.org/10.3390/biom10071066






