Aldose Reductase Differential Inhibitors in Green Tea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Assay of Aldose Reductase
2.3. Purification of Human Recombinant AKR1B1
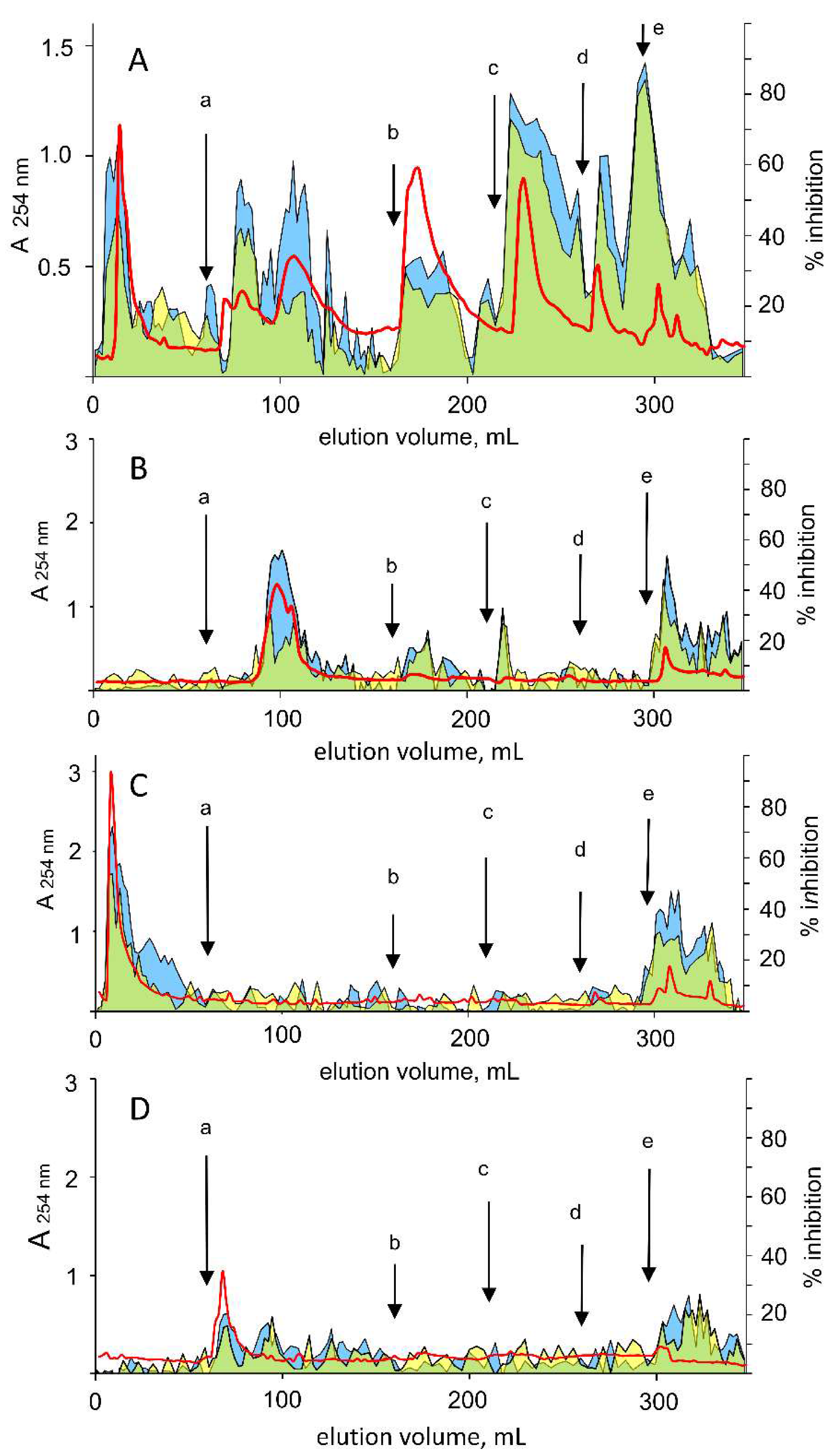
2.4. Preparation of a Green Tea “Water-Soluble Fraction”
2.5. Green Tea Extracts Fractionation
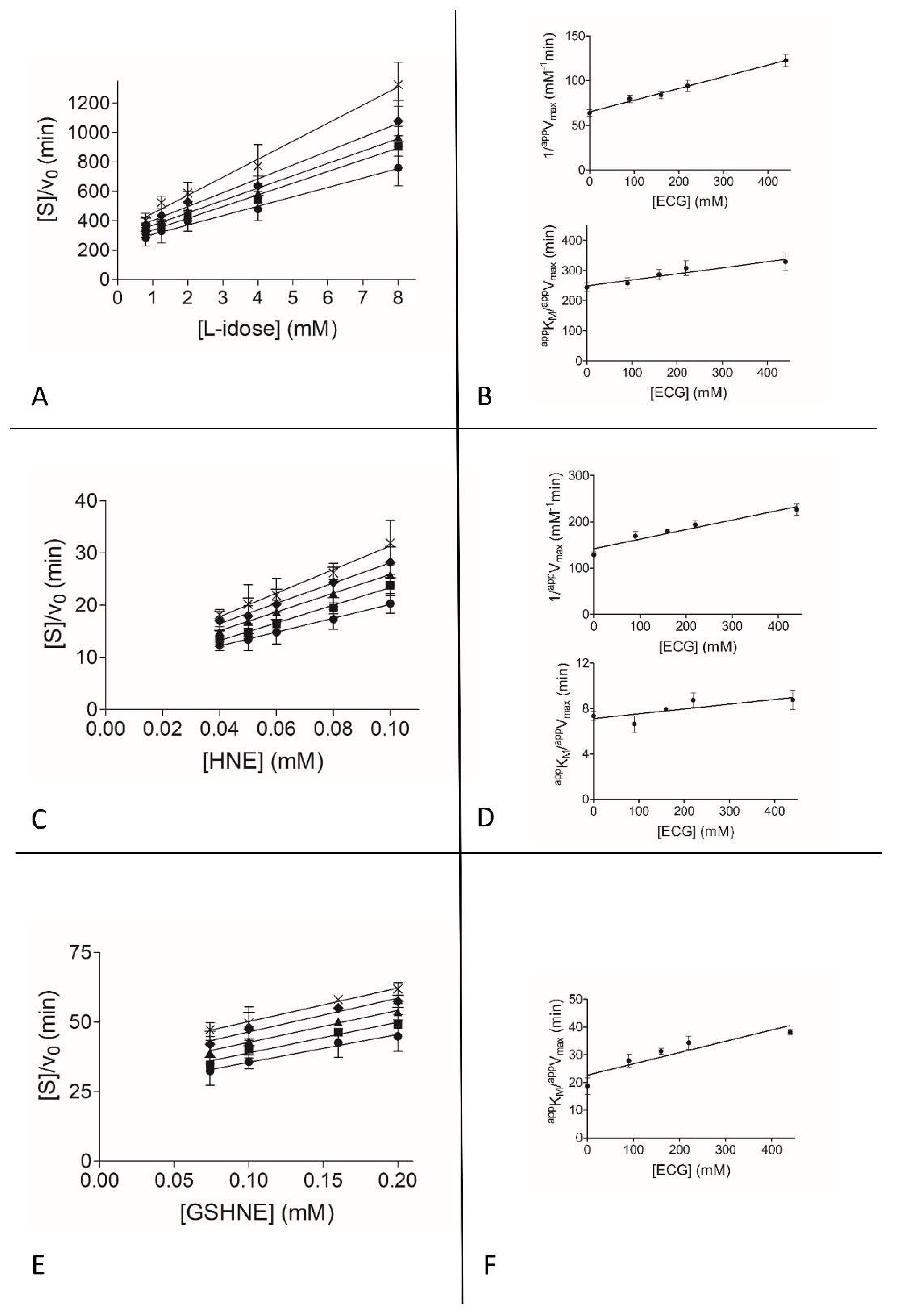
2.6. Kinetic Analysis
2.7. Molecular Docking
2.8. Molecular Dynamics Simulations
2.9. Binding Energy Evaluation
2.10. Other Methods
3. Results and Discussion
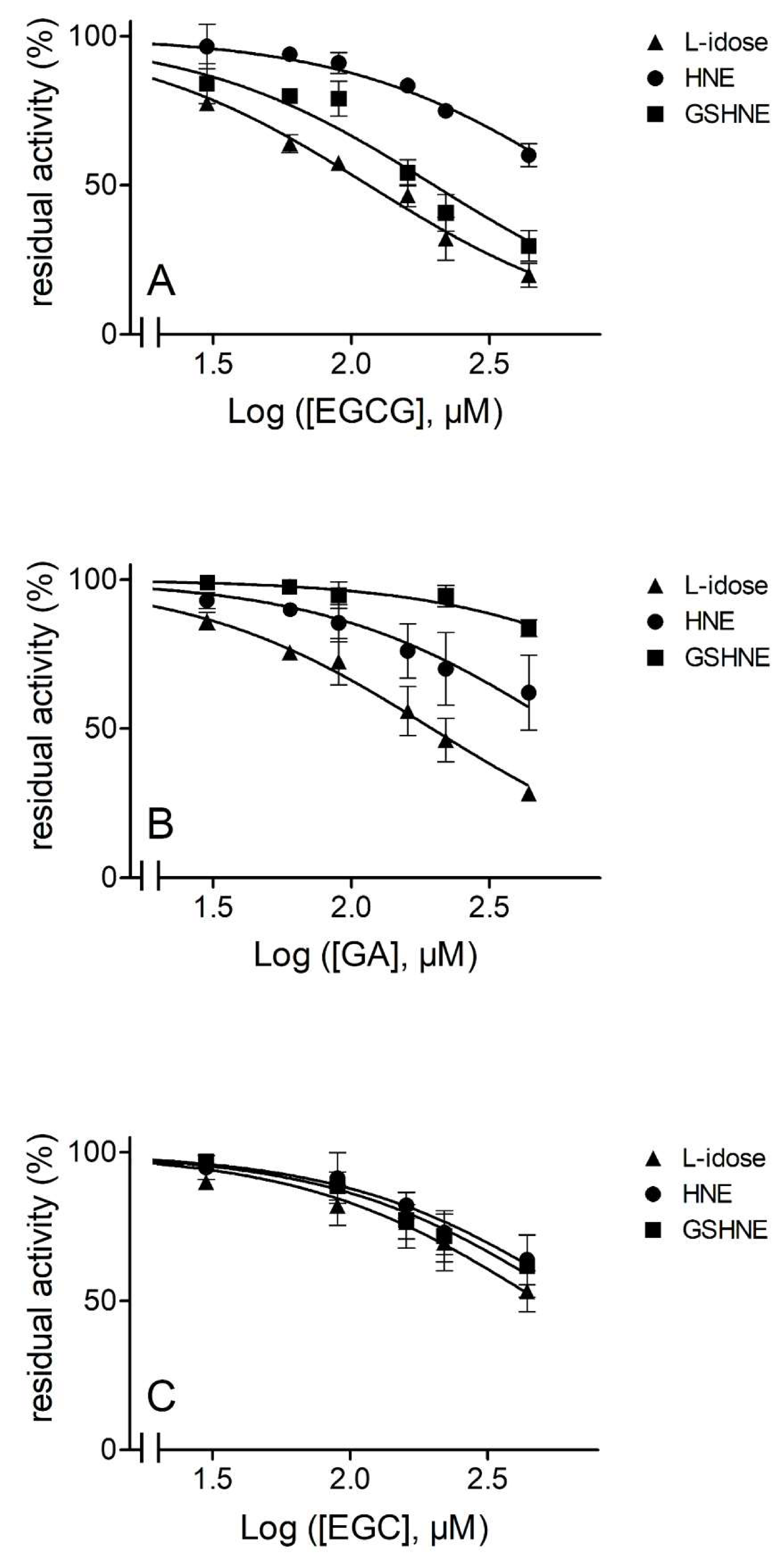
3.1. Evidence of Differential Inhibition for AKR1B1-Dependent Reactions
3.2. Inhibition Kinetic Analysis
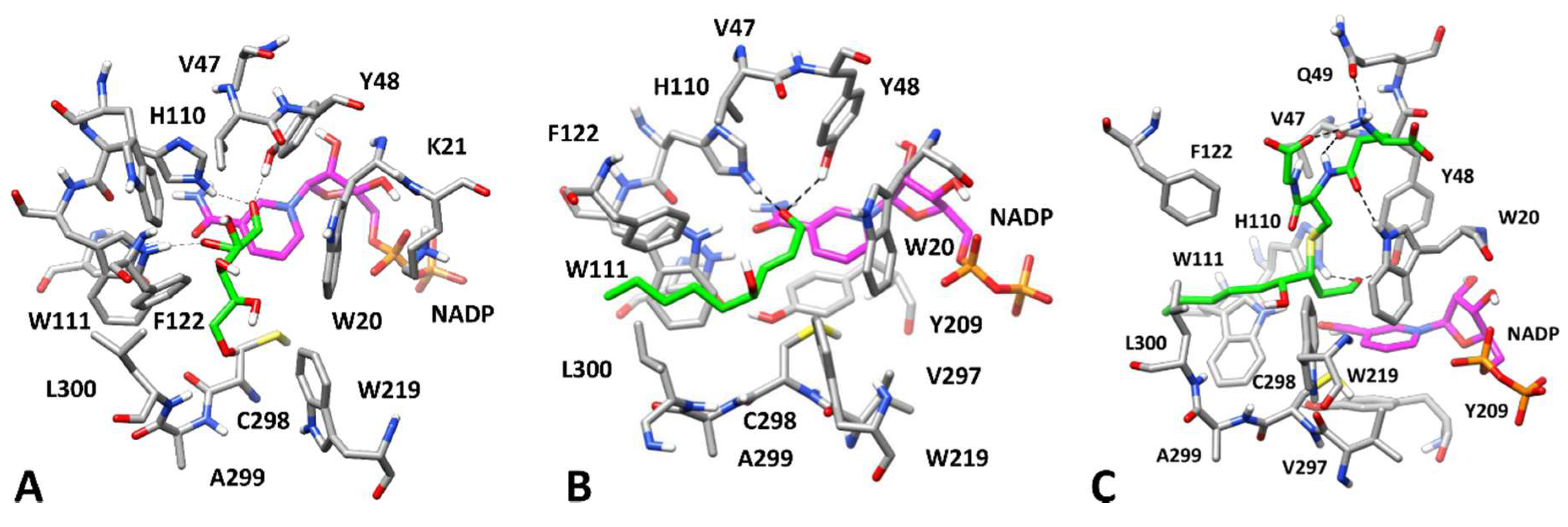
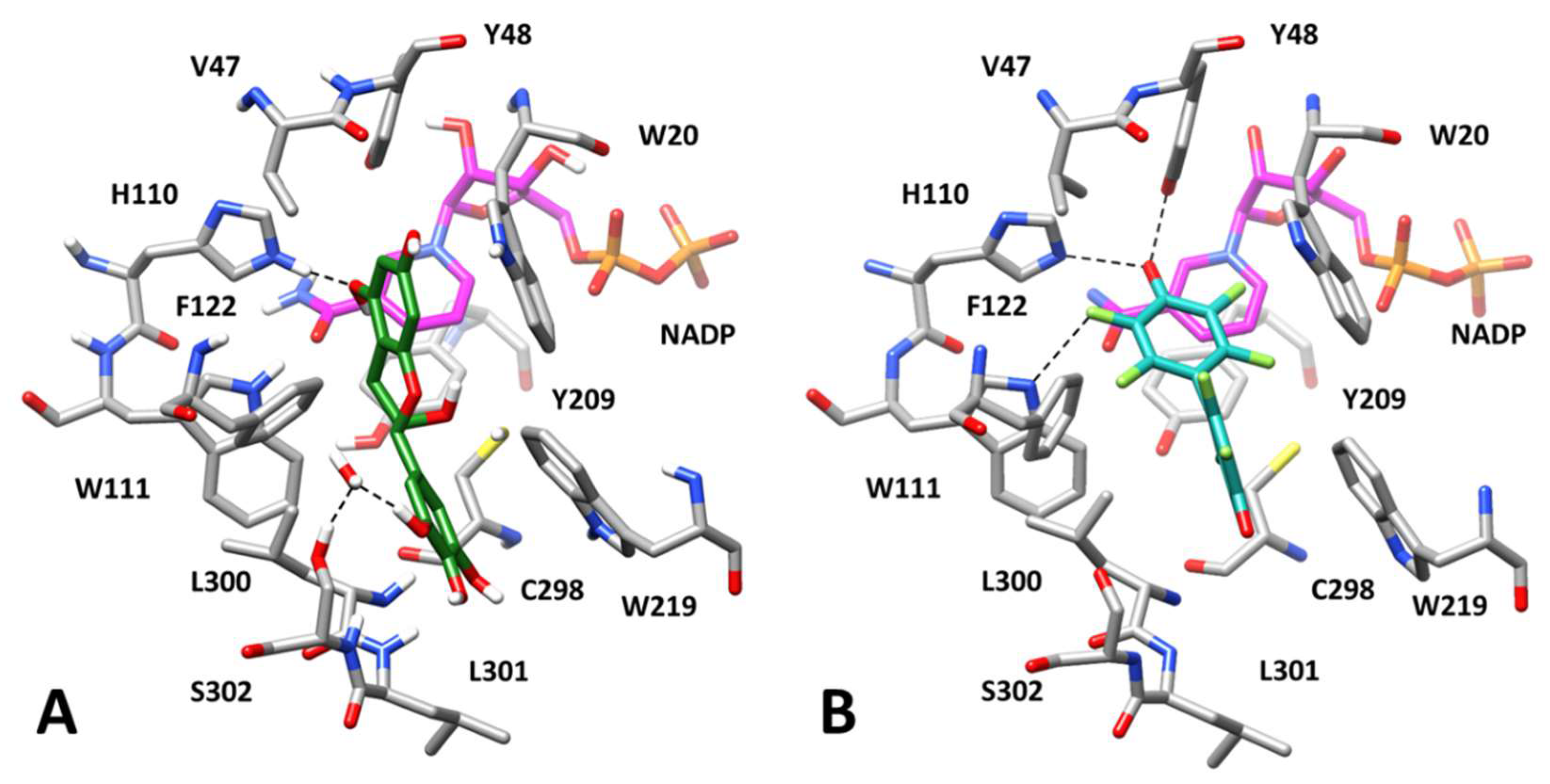
3.3. Computational Study of AKR1B1 Differential Inhibition
3.3.1. Substrates Allocation at the Enzyme Active Site
3.3.2. Inhibition by EGCG and Derivatives in the Reduction of Different Substrates
3.4. Differential Inhibition of AKR1B1 in Green Tea Extracts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Higdon, J.V.; Frei, B. Tea catechins and polyphenols: Health effects, metabolism, and antioxidant functions. Crit. Rev. Food Sci. Nutr. 2003, 43, 89–143. [Google Scholar] [CrossRef] [PubMed]
- Dufresne, C.J.; Farnworth, E.R. A review of latest research findings on the health promotion properties of tea. J. Nutr. Biochem. 2001, 12, 404–421. [Google Scholar] [CrossRef]
- Cabrera, C.; Artacho, R.; Gimenez, R. Beneficial effects of green tea-A review. J. Am. Coll. Nutr. 2006, 25, 79–99. [Google Scholar] [CrossRef] [PubMed]
- Chacko, S.M.; Thambi, P.T.; Kuttan, R.; Nishigaki, I. Beneficial effects of green tea: A literature review. Chin. Med. 2010, 5, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, H.; Peng, A. The Green Tea Polyphenol (–)-epigallocatechin-3-gallate and its beneficial roles in chronic kidney disease. J. Transl. Intern. Med. 2016, 4, 99–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipoe, G.L.; Leung, T.M.; Liong, E.C.; Lau, T.Y.H.; Fung, M.L.; Nanji, A.A. Epigallocatechin-3-gallate (EGCG) reduces liver inflammation, oxidative stress and fibrosis in carbon tetrachloride (CCl4)-induced liver injury in mice. Toxicology 2010, 273, 45–52. [Google Scholar] [CrossRef]
- Shen, K.; Feng, X.; Su, R.; Xie, H.; Zhou, L.; Zheng, S. Epigallocatechin 3-gallate ameliorates bile duct ligation induced liver injury in mice by modulation of mitochondrial oxidative stress and inflammation. PLoS ONE 2015, 10, e0126278. [Google Scholar] [CrossRef]
- Xiao, J.; Ho, C.T.; Liong, E.C.; Nanji, A.A.; Leung, T.M.; Lau, T.Y.; Fung, M.L.; Tipoe, G.L. Epigallocatechin gallate attenuates fibrosis, oxidative stress, and inflammation in nonalcoholic fatty liver disease rat model through TGF/SMAD, PI3 K/Akt/FoxO1, and NF-kappa B pathways. Eur. J. Nutr. 2014, 53, 187–199. [Google Scholar] [CrossRef]
- Oliveira, M.R.; Nabavi, S.F.; Daglia, M.; Rastrelli, L.; Nabavi, S.M. Epigallocatechin gallate and mitochondria—A story of life and death. Pharm. Res. 2016, 104, 70–85. [Google Scholar] [CrossRef]
- Morris, J.; Fang, Y.; De Mukhopdhyay, K.; Wargovich, M.J. Natural agents used in chemoprevention of aerodigestive and GI cancers. Curr. Pharm. Rep. 2016, 2, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; Xin, Y.; Xu, C.; Xiao, Y. Capecitabine combined with (-)-epigallocatechin-3-gallate inhibits angiogenesis and tumor growth in nude mice with gastric cancer xenografts. Exp. Med. 2012, 3, 650–654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Liu, F.; Zhang, W.; Liu, X.; Lin, B.; Tang, X. Epigallocatechin-3-gallate inhibits nicotine induced migration and invasion by the suppression of angiogenesis and epithelial mesenchymal transition in non-small cell lung cancer cells. Oncol. Rep. 2015, 33, 2972–2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.C.; Chung, L.C.; Chen, Y.J.; Feng, T.H.; Chen, W.T.; Juang, H.H. Upregulation of B-cell translocation gene 2 by epigallo-catechin-3-gallate via p38 and ERK signaling blocks cell proliferation in human oral squamous cell carcinoma cells. Cancer Lett. 2015, 360, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Karamese, M.; Guvendi, B.; Karamese, S.A.; Ciinar, I.; Can, S.; Erol, H.S.; Aydin, H.; Gelen, V.; Karakus, E. The protective effects of epigallocatechin gallate on lipopolysaccharide-induced hepatotoxicity: An in vitro study on Hep3B cells. Iran. J. Basic Med. Sci. 2016, 19, 483–489. [Google Scholar]
- Tak, E.; Park, C.G.; Kim, S.K.; Jun, D.Y.; Lee, J.; Hwang, S.; Song, G.W.; Lee, S.G. Epigallocatechin-3-gallate protects against hepatic ischaemia–reperfusion injury by reducing oxidative stress and apoptotic cell death. J. Int. Med. Res. 2016, 44, 1248–1262. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Meng, Z.J.; Xiong, R.B.; Guo, J.Q.; Lu, X.C.; Zheng, Z.W.; Deng, Y.P.; Luo, B.D.; Zou, F.; Li, H. Green tea polyphenol epigallocatechin-3-gallate ameliorates insulin resistance in non-alcoholic fatty liver disease mice. Acta Pharm. Sin. 2015, 36, 597–605. [Google Scholar] [CrossRef] [Green Version]
- Panpan, Y.; Kana, L.; Zhaochun, L.; Jian, L.; Ke, Y.; Wen, X. Epigallocatechin gallate regulates expression of apoptotic genes and protects cultured human lens epithelial cells under hyperglycemia. Mol. Biol. 2013, 47, 222–227. [Google Scholar]
- Takamura, Y.; Kubo, E.; Tsuzuki, S.; Akagi, Y. Apoptotic cell death in the lens epithelium of rat sugar cataract. Exp. Eye Res. 2003, 77, 51–57. [Google Scholar] [CrossRef]
- Kubo, E.; Urakami, T.; Fatma, N.; Akagi, Y.; Singh, D.P. Polyol pathway_dependent osmotic and oxidative stresses in aldose reductase_mediated apoptosis in human lens epithelial cells: Role of AOP2. Biochem. Biophys. Res. Commun. 2004, 314, 1050–1056. [Google Scholar] [CrossRef]
- Ravindranath, M.H.; Ramasamy, V.; Moon, S.; Ruiz, C.; Muthugounder, S. Differential growth suppression of human melanoma cells by tea (Camellia sinensis) epicatechins (ECG, EGC and EGCG). Evid. Based Complement. Altern. Med. 2009, 6, 523–530. [Google Scholar] [CrossRef] [Green Version]
- Vergote, D.; Cren-Olivé, C.; Chopin, V.; Toillon, R.A.; Rolando, C.; Hondermarck, H.; Le Bourhis, X. (-)-Epigallocatechin (EGC) of green tea induces apoptosis of human breast cancer cells but not of their normal counterparts. Breast Cancer Res. Treat. 2002, 76, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Suzuki, R.; Sakaguchi, N.; Li, Z.; Takeda, T.; Ogihara, Y.; Jiang, B.; Chen, Y. Selective induction of cell death in cancer cells by gallic acid. Biol. Pharm. Bull. 1995, 18, 1526–1530. [Google Scholar] [CrossRef] [PubMed]
- Chia, Y.C.; Rajbanshi, R.; Calhoun, C.; Chiu, R.H. Anti-neoplastic effects of gallic acid, a major component of Toona sinensis leaf extract, on oral squamous carcinoma cells. Molecules 2010, 15, 8377–8389. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Guan, X.; Grün, C.; Zhou, Z.; Schepers, U.; Nick, P. Gallic acid induces mitotic catastrophe and inhibits centrosomal clustering in HeLa cells. Toxicol. Vitr. 2015, 30, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Strlic, M.; Radovic, T.; Kolar, J.; Pihlar, B. Anti- and pro-oxidative properties of gallic acid in fenton-type systems. J. Agric. Food Chem. 2002, 50, 6313–6317. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Carranza, J.N.; Diaz, J.F.; Redondo-Horcaio, M.; Barasoain, I.; Alvarez, L.; Lastres, P.; Romero-Estrada, A.; Aller, P.; Gonzalez-Maya, L. Gallic acid sensitizes paclitaxel-resistant human ovarian carcinoma cells through an increase in reactive oxygen species and subsequent downregulation of ERK activation. Oncol. Rep. 2018, 39, 3007–3014. [Google Scholar]
- Ou, B.R.; Park, W.H. Gallic acid-induced lung cancer cell death is related to glutathione depletion as well as reactive oxygen species increase. Toxicol. Vitr. 2010, 24, 1356–1362. [Google Scholar]
- Son, N.H.; Ananthakrishnan, R.; Yu, S.; Khan, R.S.; Jiang, H.; Ji, R.; Akashi, H.; Li, Q.; O’Shea, K.; Homma, S.; et al. Cardiomyocyte aldose reductase causes heart failure and impairs recovery from ischemia. PLoS ONE 2012, 7, e46549. [Google Scholar] [CrossRef] [Green Version]
- Obrosova, I.G.; Kador, P.F. Aldose reductase/polyol inhibitors for diabetic retinopathy. Curr. Pharm. Biotechnol. 2011, 12, 373–385. [Google Scholar] [CrossRef]
- El-Gamal, H.; Munusamy, S. Aldose reductase as a drug target for treatment of diabetic nephropathy: Promises and challenges. Protein. Pept. Lett. 2017, 24, 71–77. [Google Scholar] [CrossRef]
- Yagihashi, S.; Yamagishi, S.I.; Wada-Ri, R.; Baba, M.; Hohman, T.C.; Yabe-Nishimura, C.; Kokai, Y. Neuropathy in diabetic mice overexpressing human aldose reductase and effects of aldose reductase inhibitor. Brain 2001, 124, 2448–2458. [Google Scholar] [CrossRef] [Green Version]
- Elimam, D.M.; Ibrahim, A.S.; Liou, G.I.; Badria, F.A. Olive and ginkgo extracts as potential cataract therapy with differential inhibitory activity on aldose reductase. Drug Discov. 2017, 11, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Shukla, K.; Pal, P.B.; Sonowal, H.; Srivastava, S.K.; Ramana, K.V. Aldose reductase inhibitor protects against hyperglycemic stress by activating NRF2-dependent antioxidant proteins. J. Diabetes Res. 2017, 2017, 6785852. [Google Scholar] [CrossRef] [PubMed]
- Tammali, R.; Reddy, A.B.M.; Saxena, A.; Rychahou, P.G.; Evers, B.M.; Qiu, S.; Awasthi, S.; Ramana, K.V.; Srivastava, S.K. Inhibition of aldose reductase prevents colon cancer metastasis. Carcinogenesis 2011, 32, 1259–1267. [Google Scholar] [CrossRef] [Green Version]
- Esterbauer, H.; Schaur, R.J.; Zollner, H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic. Biol. Med. 1991, 11, 81–128. [Google Scholar] [CrossRef]
- Srivastava, S.; Chandra, A.; Bhatnagar, A.; Srivastava, S.K.; Ansari, N.H. Lipid peroxidation product, 4-hydroxynonenal and its conjugate with GSH are excellent substrates of bovine lens aldose reductase. Biochem. Biophys. Res. Commun. 1995, 217, 741–746. [Google Scholar] [CrossRef] [PubMed]
- Vander Jagt, D.L.; Kolb, N.S.; Vander Jagt, T.J.; Chino, J.; Martinez, F.J.; Hunsaker, L.A.; Royer, R.E. Substrate specificity of human Aldose Reductase: Identification of 4-hydroxynonenal as an endogenous substrate. Biochim. Biophys. Acta 1995, 1249, 117–126. [Google Scholar] [CrossRef]
- Frohnert, B.I.; Bernlohr, D.A. Glutathionylated products of lipid peroxidation. A novel mechanism of adipocyte to macrophage signaling. Adipocyte 2014, 3, 224–229. [Google Scholar] [CrossRef] [Green Version]
- Frohnert, B.I.; Long, E.K.; Hahn, W.S.; Bernlohr, D.A. Glutathionylated lipid aldehydes are products of adipocyte oxidative stress and activators of macrophage inflammation. Diabetes 2014, 63, 89–100. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, S.; Ramana, K.V.; Bhatnagar, A.; Srivastava, S.K. Synthesis, quantification, characterization, and signaling properties of glutathionyl conjugates of enals. Meth. Enzym. 2010, 474, 297–313. [Google Scholar]
- Del Corso, A.; Cappiello, M.; Mura, U. From a dull enzyme to something else: Facts and perspectives regarding aldose reductase. Curr. Med. Chem. 2008, 15, 1452–1461. [Google Scholar] [CrossRef] [PubMed]
- Del Corso, A.; Balestri, F.; Di Bugno, E.; Moschini, R.; Cappiello, M.; Sartini, S.; La Motta, C.; Da Settimo, F.; Mura, U. A new approach to control the enigmatic activity of aldose reductase. PLoS ONE 2013, 8, e74076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappiello, M.; Moschini, R.; Balestri, F.; Mura, U.; Del Corso, A. Basic models for differential inhibition of enzymes. Biochem. Biophys. Res. Commun. 2014, 445, 556–560. [Google Scholar] [CrossRef] [PubMed]
- Balestri, F.; Sorce, C.; Moschini, R.; Cappiello, M.; Misuri, L.; Del Corso, A.; Mura, U. Edible vegetables as a source of aldose reductase differential inhibitors. Chem. Biol. Interact. 2017, 276, 155–159. [Google Scholar] [CrossRef]
- Balestri, F.; Rotondo, R.; Moschini, R.; Pellegrino, M.; Cappiello, M.; Barracco, V.; Misuri, L.; Sorce, C.; Andreucci, A.; Del-Corso, A.; et al. Zolfino landrace (Phaseolus vulgaris L.) from Pratomagno: General and specific features of a functional food. Food Nutr. Res. 2016, 60, 31792. [Google Scholar] [CrossRef] [Green Version]
- Balestri, F.; De Leo, M.; Sorce, C.; Cappiello, M.; Quattrini, L.; Moschini, R.; Pineschi, C.; Braca, A.; La Motta, C.; Da Settimo, F.; et al. Soyasaponins from Zolfino bean as aldose reductase differential inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 350–360. [Google Scholar] [CrossRef]
- Murata, M.; Irie, J.; Homma, S. Aldose reductase inhibitors from green tea. Lwt-Food Sci. Technol. 1994, 27, 401–405. [Google Scholar] [CrossRef]
- Sampath, C.; Sang, S.; Ahmedna, M. In vitro and in vivo inhibition of aldose reductase and advanced glycation end products by phloretin, epigallocatechin 3-gallate and [6]-gingerol. Biomed. Pharm. 2016, 84, 502–513. [Google Scholar] [CrossRef]
- Aslan, H.E.; Beydemir, S. Phenolic compounds: The inhibition effect on polyol pathway enzymes. Chem. Biol. Interact. 2017, 266, 47–55. [Google Scholar] [CrossRef]
- Alim, Z.; Kilinc, N.; Sengul, B.; Beydemir, S. Inhibition behaviours of some phenolic acids on rat kidney aldose reductase enzyme: An in vitro study. J. Enzym. Inhib. Med. Chem. 2017, 32, 277–284. [Google Scholar] [CrossRef] [Green Version]
- Balestri, F.; Cappiello, M.; Moschini, R.; Rotondo, R.; Abate, M.; Del Corso, A.; Mura, U. Modulation of aldose reductase activity by aldose hemiacetals. Biochim. Biophys. Acta 2015, 1850, 2329–2339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balestri, F.; Cappiello, M.; Moschini, R.; Rotondo, R.; Buggiani, I.; Pelosi, P.; Mura, U.; Del Corso, A. L-Idose: An attractive substrate alternative to D-glucose for measuring aldose reductase activity. Biochem. Biophys. Res. Commun. 2015, 456, 891–895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanes, C.S. Studies on plant amylases: The effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley. Biochem. J. 1932, 26, 1406–1421. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Zhang, L.; Chen, W.; Chen, Y.; Xie, W.; Hu, X. Partial inhibition of aldose reductase by nitazoxanide and its molecular basis. Chem. Med. Chem. 2012, 7, 1921–1923. [Google Scholar] [CrossRef] [PubMed]
- Steuber, H.; Heine, A.; Klebe, G. Structural and thermodynamic study on aldose reductase: Nitro-substituted inhibitors with strong enthalpic binding contribution. J. Mol. Biol. 2007, 368, 618–638. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Verdonk, M.L.; Cole, J.C.; Hartshorn, M.J.; Murray, C.W.; Taylor, R.D. Improved protein-ligand docking using GOLD. Proteins 2013, 52, 609–623. [Google Scholar] [CrossRef]
- Case, D.A.; Berryman, J.T.; Betz, R.M.; Cerutti, D.S.; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Homeyer, N.; et al. AMBER, Version 16; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Poli, G.; Lapillo, M.; Jha, V.; Mouawad, N.; Caligiuri, I.; Macchia, M.; Rizzolio, F.; Tuccinardi, T.; Granchi, C. Computationally driven discovery of phenyl(piperazin-1-yl)methanone derivatives as reversible monoacylglycerol lipase (MAGL) inhibitors. J. Enzym. Inhib. Med. Chem. 2019, 34, 589–596. [Google Scholar] [CrossRef] [Green Version]
- Bononi, G.; Granchi, C.; Lapillo, M.; Giannotti, M.; Nieri, D.; Fortunato, S.; Boustani, M.E.; Caligiuri, I.; Poli, G.; Carlson, K.E.; et al. Discovery of long-chain salicylketoxime derivatives as monoacylglycerol lipase (MAGL) inhibitors. Eur. J. Med. Chem. 2018, 157, 817–836. [Google Scholar] [CrossRef]
- Poli, G.; Lapillo, M.; Granchi, C.; Caciolla, J.; Mouawad, N.; Caligiuri, I.; Rizzolio, F.; Langer, T.; Minutolo, F.; Tuccinardi, T. Binding investigation and preliminary optimisation of the 3-amino-1,2,4-triazin-5(2H)-one core for the development of new Fyn inhibitors. J. Enzym. Inhib. Med. Chem. 2018, 33, 956–961. [Google Scholar] [CrossRef] [Green Version]
- Granchi, C.; Caligiuri, I.; Bertelli, E.; Poli, G.; Rizzolio, F.; Macchia, M.; Martinelli, A.; Minutolo, F.; Tuccinardi, T. Development of terphenyl-2-methyloxazol-5(4H)-one derivatives as selective reversible MAGL inhibitors. J. Enzym. Inhib. Med. Chem. 2017, 32, 1240–1252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Moschini, R.; Peroni, E.; Rotondo, R.; Renzone, G.; Melck, D.; Cappiello, M.; Srebot, M.; Napolitano, E.; Motta, A.; Scaloni, A.; et al. NADP+-dependent dehydrogenase activity of carbonyl reductase on glutathionyl hydroxynonanal as a new pathway for hydroxynonenal detoxification. Free Radic. Biol. Med. 2015, 83, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Rotondo, R.; Moschini, R.; Renzone, G.; Tuccinardi, T.; Balestri, F.; Cappiello, M.; Scaloni, A.; Mura, U.; Del Corso, A. Human carbonyl reductase 1 as efficient catalyst for the reduction of glutathionylated aldehydes derived from lipid peroxidation. Free Radic. Biol. Med. 2016, 99, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Cappiello, M.; Peroni, E.; Lepore, A.; Moschini, R.; Del Corso, A.; Balestri, F.; Mura, U. Rapid colorimetric determination of reduced and oxidized glutathione using an end point coupled enzymatic assay. Anal. Bioanal. Chem. 2013, 405, 1779–1785. [Google Scholar] [CrossRef]
- Cappiello, M.; Balestri, F.; Moschini, R.; Mura, U.; Del Corso, A. Intra-site differential inhibition of multi-specific enzyme. J. Enzym. Inhib. Med. Chem. 2020, 35, 840–846. [Google Scholar] [CrossRef]
- Urzhumtsev, A.; Tête-Favier, F.; Mitschler, A.; Barbanton, J.; Barth, P.; Urzhumtseva, L.; Biellmann, J.F.; Podjarny, A.; Moras, D. A ‘specificity’ pocket inferred from the crystal structures of the complexes of aldose reductase with the pharmaceutically important inhibitors tolrestat and sorbinil. Structure 1197, 5, 601–612. [Google Scholar] [CrossRef] [Green Version]
- Balestri, F.; Barracco, V.; Renzone, G.; Tuccinardi, T.; Pomelli, C.S.; Cappiello, M.; Lessi, M.; Rotondo, R.; Bellina, F.; Scaloni, A.; et al. Stereoselectivity of Aldose Reductase in the reduction of glutathionyl-hydroxynonanal adduct. Antioxidants 2019, 8, 502. [Google Scholar] [CrossRef] [Green Version]





| ECGC | GA | EGC | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Substrate | Ki’ | Ki | Ki/Ki’ | Ki’ | Ki | Ki/Ki’ | Ki’ | Ki | Ki/Ki’ |
| L-idose | 61 ± 9 | 425 ± 64 | 7.0 | 103 ± 7 | n.d. | n.a. | 496 ± 35 | 1239 ± 2 33 | 2.5 |
| HNE | 116 ± 11 | n.d. | n.a. | 253 ± 23 | n.d. | n.a. | 686 ± 63 | 1682 ± 289 | 2.5 |
| GSHNE | 144 ± 22 | 196 ± 17 | 1.3 | 793 ± 102 | n.d. | n.a. | n.d. | 553 ± 87 | n.a. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balestri, F.; Poli, G.; Pineschi, C.; Moschini, R.; Cappiello, M.; Mura, U.; Tuccinardi, T.; Del Corso, A. Aldose Reductase Differential Inhibitors in Green Tea. Biomolecules 2020, 10, 1003. https://doi.org/10.3390/biom10071003
Balestri F, Poli G, Pineschi C, Moschini R, Cappiello M, Mura U, Tuccinardi T, Del Corso A. Aldose Reductase Differential Inhibitors in Green Tea. Biomolecules. 2020; 10(7):1003. https://doi.org/10.3390/biom10071003
Chicago/Turabian StyleBalestri, Francesco, Giulio Poli, Carlotta Pineschi, Roberta Moschini, Mario Cappiello, Umberto Mura, Tiziano Tuccinardi, and Antonella Del Corso. 2020. "Aldose Reductase Differential Inhibitors in Green Tea" Biomolecules 10, no. 7: 1003. https://doi.org/10.3390/biom10071003
APA StyleBalestri, F., Poli, G., Pineschi, C., Moschini, R., Cappiello, M., Mura, U., Tuccinardi, T., & Del Corso, A. (2020). Aldose Reductase Differential Inhibitors in Green Tea. Biomolecules, 10(7), 1003. https://doi.org/10.3390/biom10071003












