Genome-Wide Identification of RNA Silencing-Related Genes and Their Expressional Analysis in Response to Heat Stress in Barley (Hordeum vulgare L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of HvDCL, HvAGO and HvRDR Homologues
2.2. Phylogenetic Analysis, Chromosomal Localisation and Identification of Conserved Motifs
2.3. Plant Material and Abiotic Stress Treatments
2.4. RNA Isolation, RT-PCR, Semiquantitative RT-PCR and RT-qPCR Analysis
2.5. RNA-Seq Analysis
3. Results
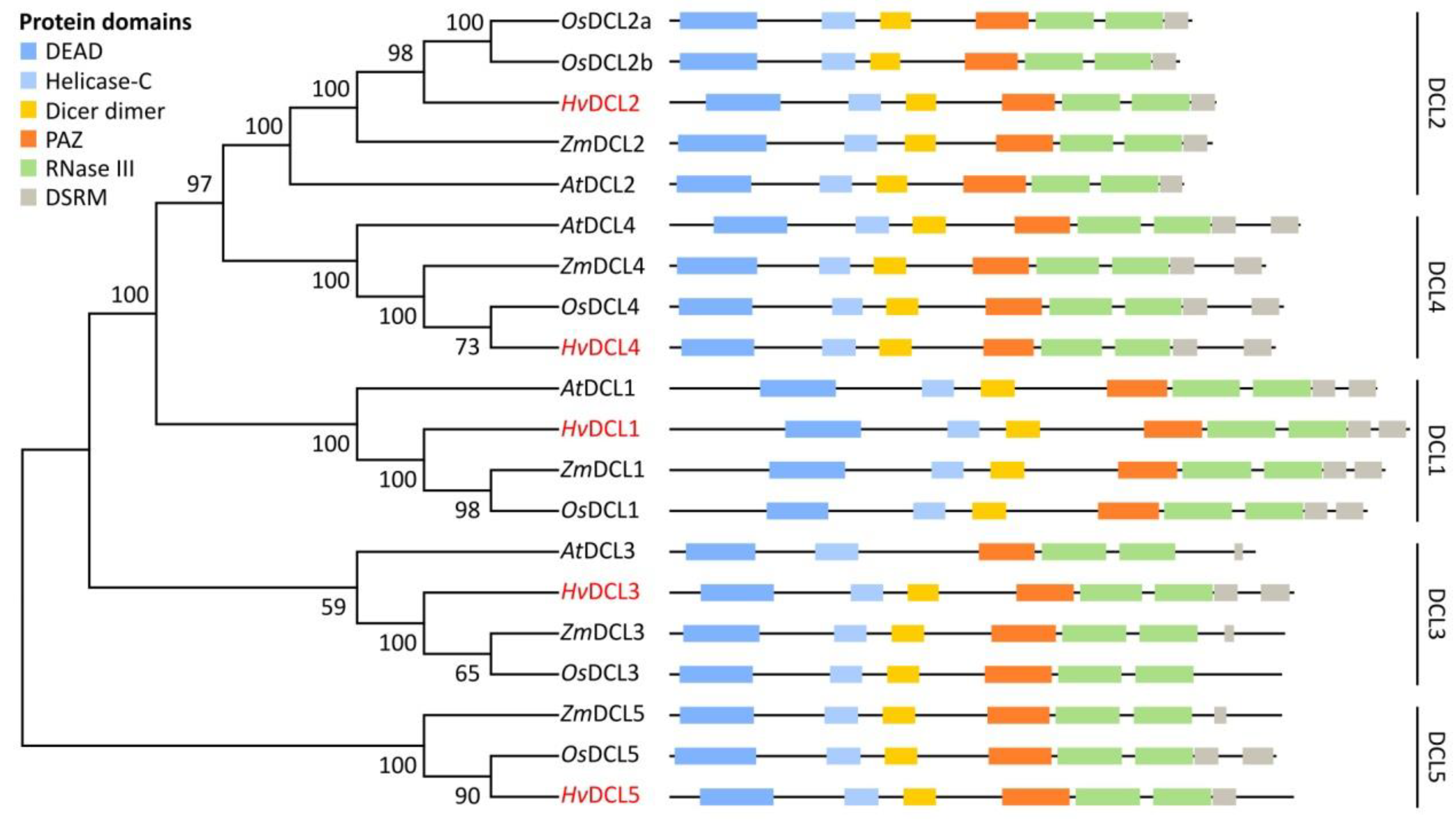
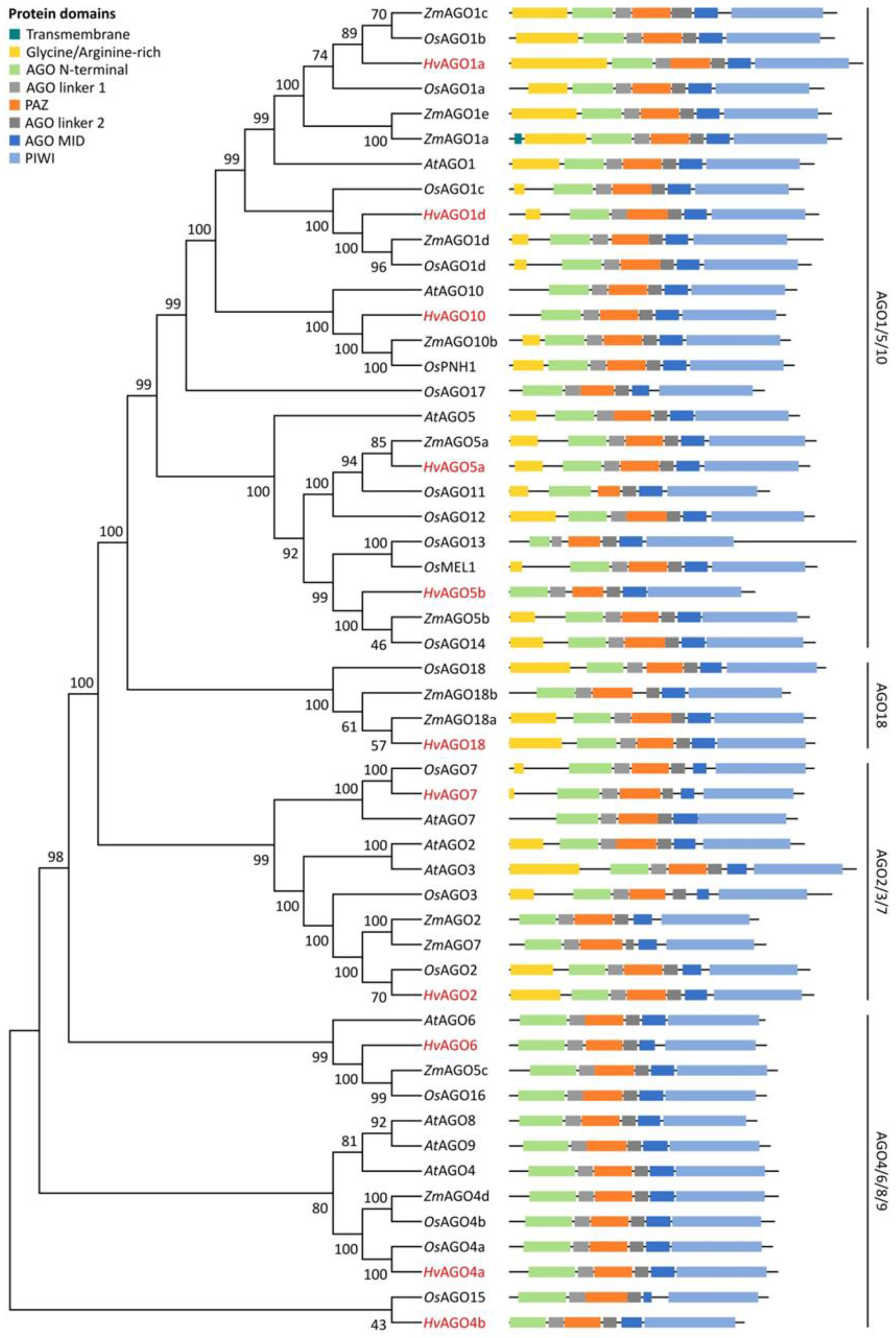
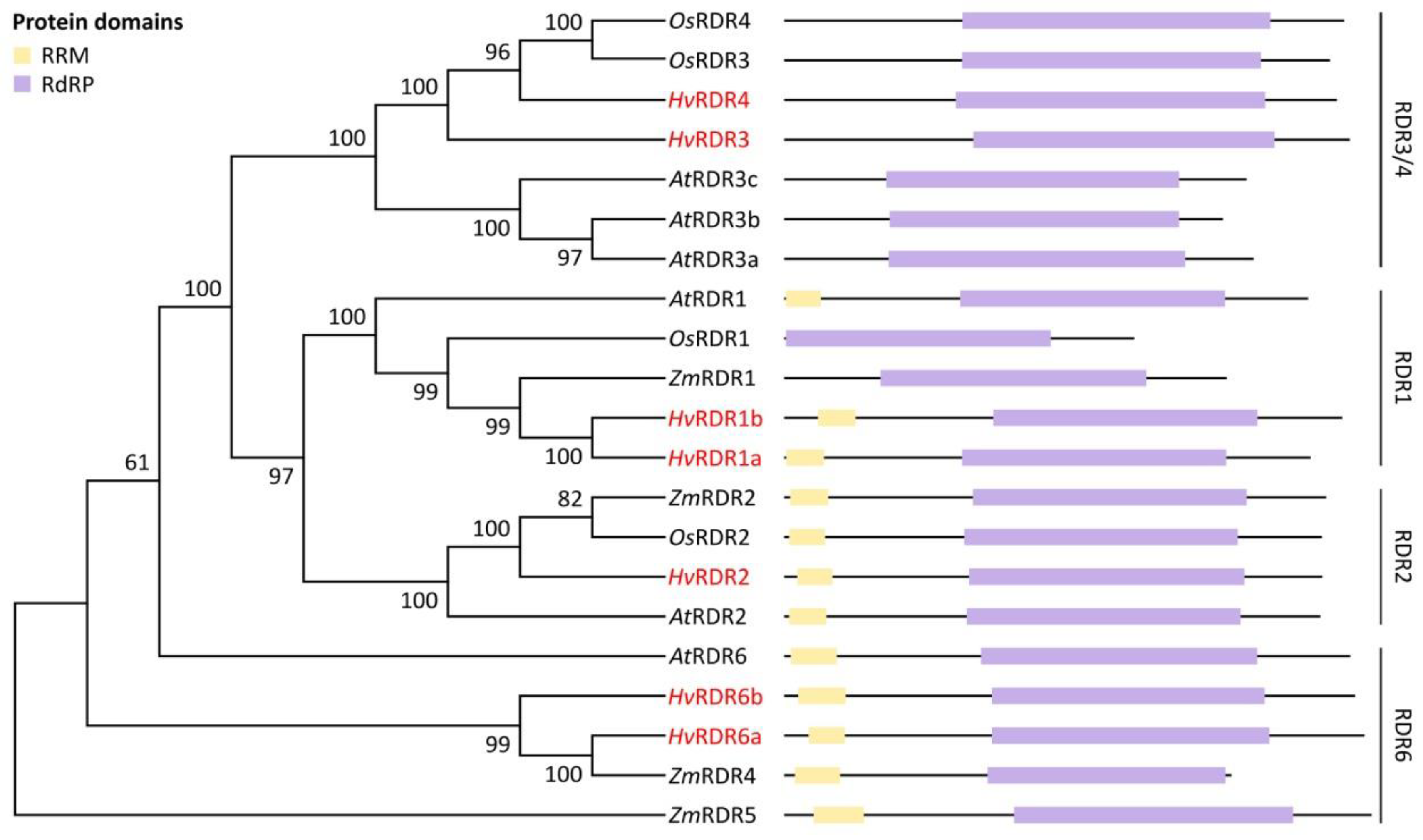
3.1. Identification and Structural Analysis of DCL, AGO and RDR Genes in Barley
3.2. Domain Analysis and Phylogenetic Relationship of Identified Barley DCL, AGO and RDR Proteins
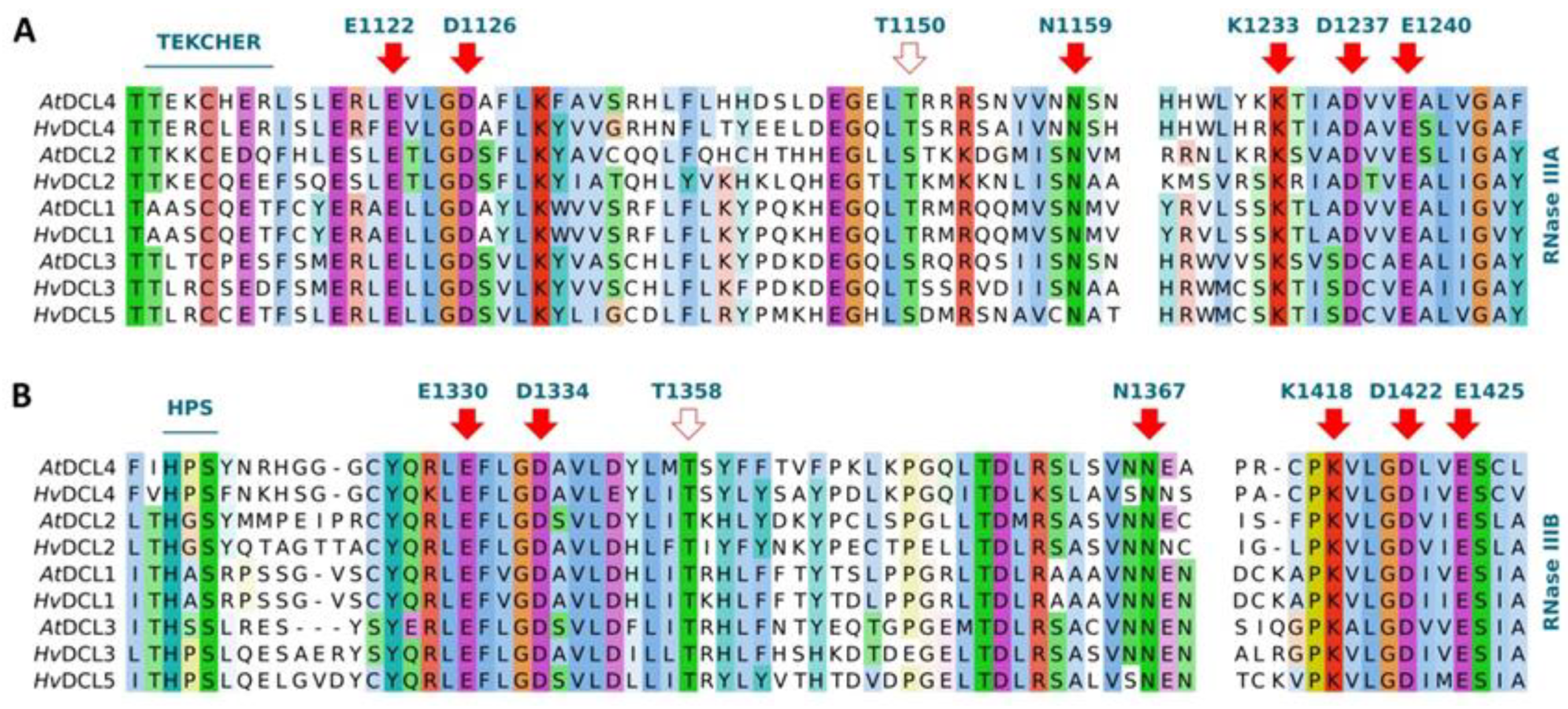
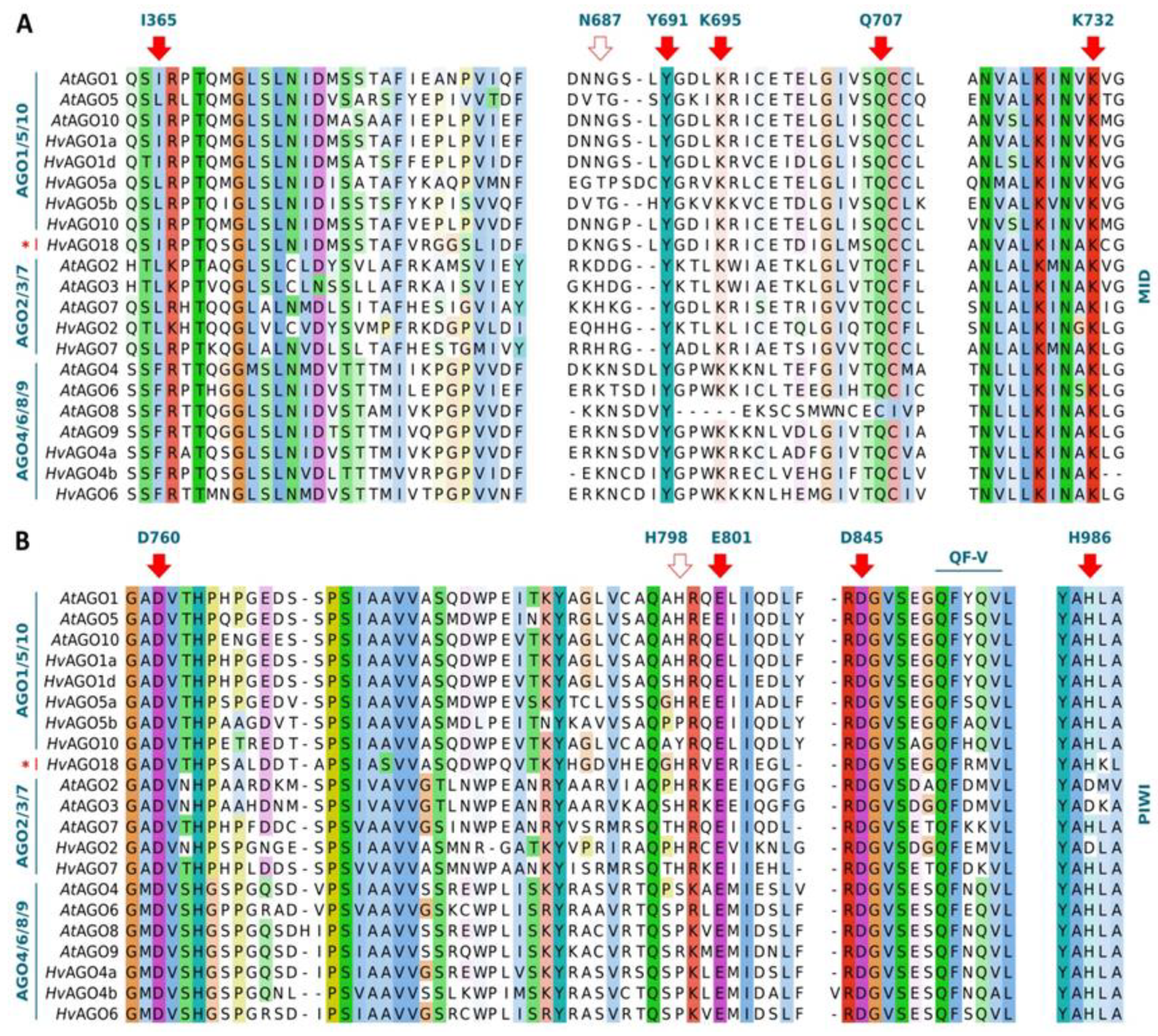
3.3. Functionally Conserved Amino Acid Residues in Domains of the Identified RNA Silencing Factors
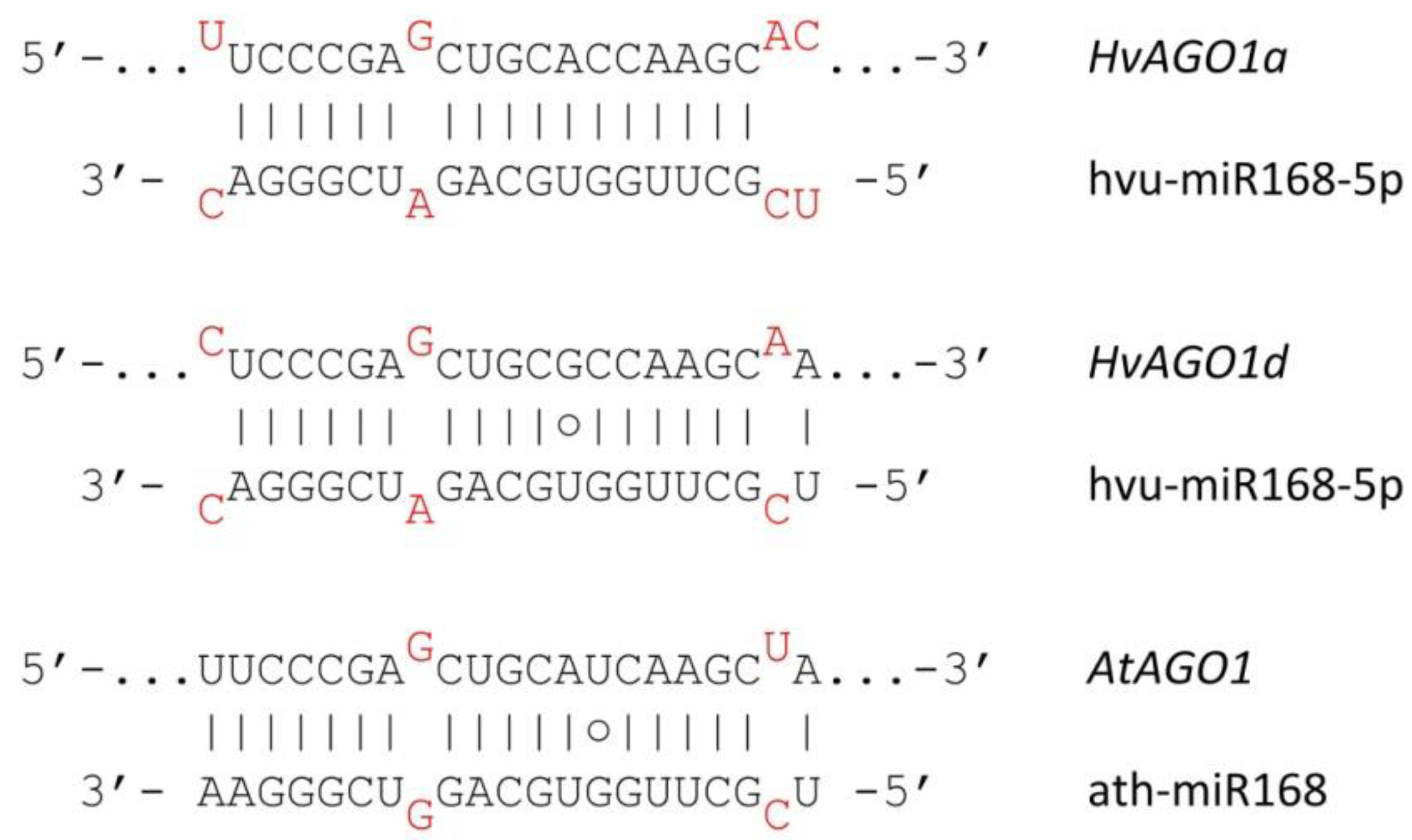
3.4. Autoregulation of RNA Silencing
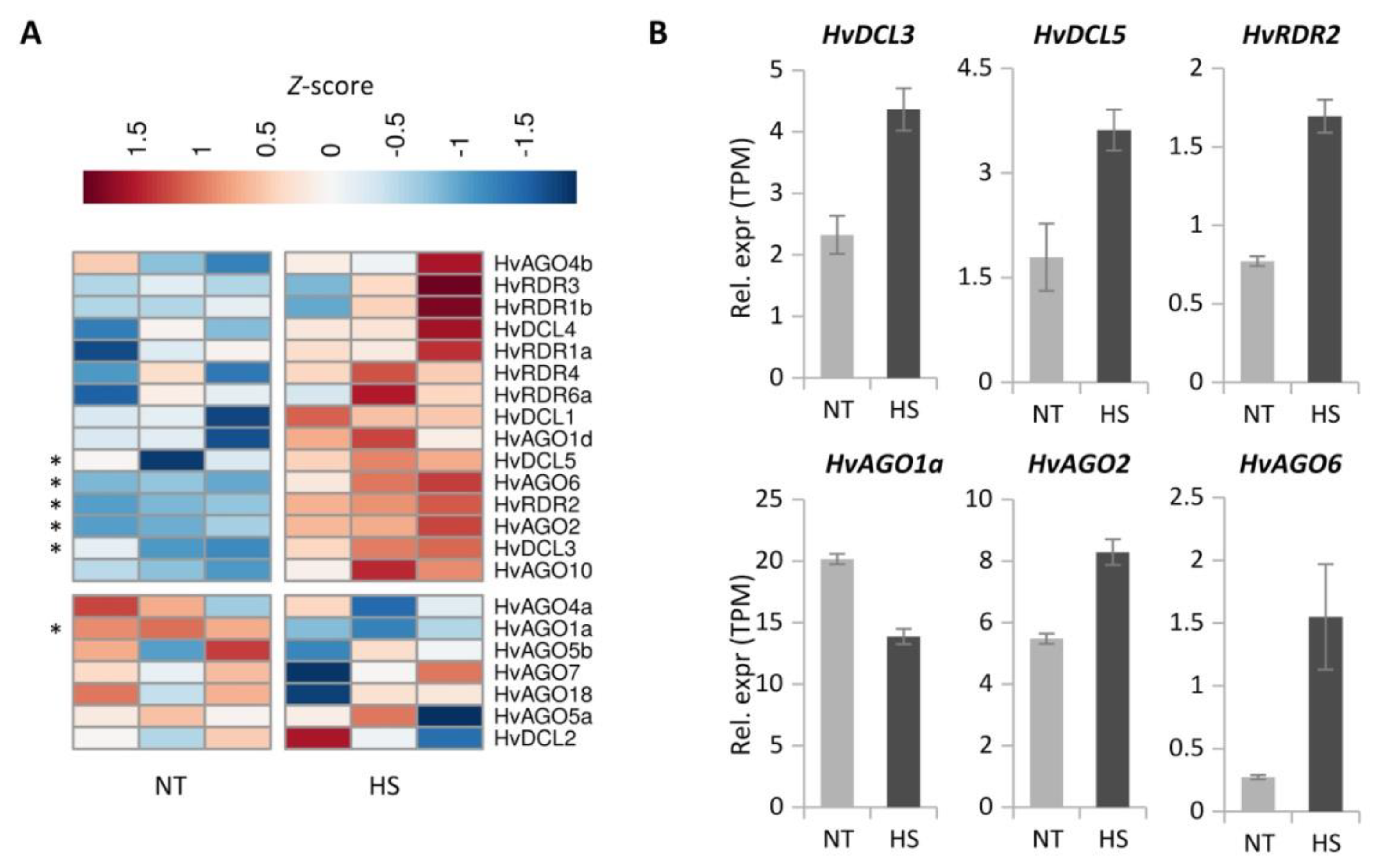
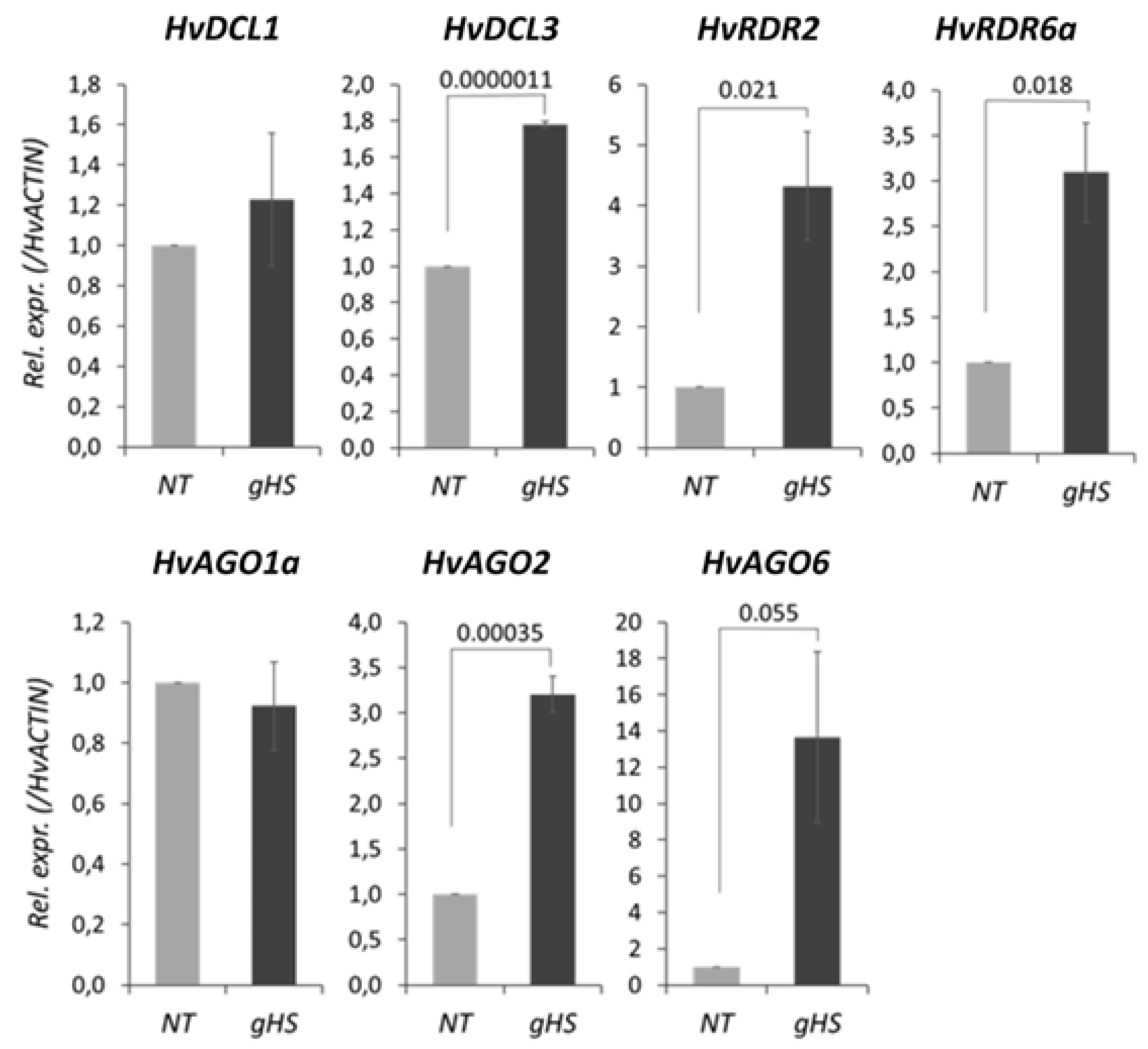
3.5. Heat Stress Significantly Alters the Expression of RNA Silencing Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: http://www.fao.org/faostat/en/#home (accessed on 21 March 2020).
- Barley Grain | Feedipedia. Available online: https://feedipedia.org/node/227 (accessed on 20 March 2020).
- Xie, W.; Xiong, W.; Pan, J.; Ali, T.; Cui, Q.; Guan, D.; Meng, J.; Mueller, N.D.; Lin, E.; Davis, S.J. Decreases in global beer supply due to extreme drought and heat. Nat. Plants 2018, 4, 964–973. [Google Scholar] [CrossRef]
- Mullen, C.A.; Boateng, A.A.; Hicks, K.B.; Goldberg, N.M.; Moreau, R.A. Analysis and Comparison of Bio-Oil Produced by Fast Pyrolysis from Three Barley Biomass/Byproduct Streams. Energy Fuels 2010, 24, 699–706. [Google Scholar] [CrossRef]
- Dawson, I.K.; Russell, J.; Powell, W.; Steffenson, B.; Thomas, W.T.B.; Waugh, R. Barley: A translational model for adaptation to climate change. New Phytol. 2015, 206, 913–931. [Google Scholar] [CrossRef] [PubMed]
- Savin, R.; Nicolas, M.E. Effects of timing of heat stress and drought on growth and quality of barley grains. Aust. J. Agric. Res. 1999, 50, 357–364. [Google Scholar] [CrossRef]
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; von Korff, M. Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef]
- Knüpffer, H. Triticeae Genetic Resources in ex situ Genebank Collections. In Genetics and Genomics of the Triticeae; Muehlbauer, G.J., Feuillet, C., Eds.; Springer US: New York, NY, USA, 2009; Volume 7, pp. 31–79. [Google Scholar]
- Voinnet, O. Origin, Biogenesis, and Activity of Plant MicroRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef]
- Shabalina, S.A.; Koonin, E.V. Origins and evolution of eukaryotic RNA interference. Trends Ecol. Evol. 2008, 23, 578–587. [Google Scholar] [CrossRef]
- Wilson, R.C.; Doudna, J.A. Molecular mechanisms of RNA interference. Annu. Rev. Biophys. 2013, 42, 217–239. [Google Scholar] [CrossRef]
- Castel, S.E.; Martienssen, R.A. RNA interference (RNAi) in the Nucleus: Roles for small RNA in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef]
- Napoli, C.; Lemieux, C.; Jorgensen, R. Introduction of a Chimeric Chalcone Synthase Gene into Petunia Results in Reversible Co-Suppression of Homologous Genes in trans. Plant Cell 1990, 2, 279–289. [Google Scholar] [CrossRef]
- Sen, G.L.; Blau, H.M. A brief history of RNAi: The silence of the genes. FASEB J. 2006, 20, 1293–1299. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, A.J.; Baulcombe, D.C. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999, 286, 950–952. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Bologna, N.G.; Mateos, J.L.; Bresso, E.G.; Palatnik, J.F. A loop-to-base processing mechanism underlies the biogenesis of plant microRNAs miR319 and miR159. EMBO J. 2009, 28, 3646–3656. [Google Scholar] [CrossRef]
- Bologna, N.G.; Schapire, A.L.; Zhai, J.; Chorostecki, U.; Boisbouvier, J.; Meyers, B.C.; Palatnik, J.F. Multiple RNA recognition patterns during microRNA biogenesis in plants. Genome Res. 2013, 23, 1675–1689. [Google Scholar] [CrossRef]
- Holoch, D.; Moazed, D. RNA-mediated epigenetic regulation of gene expression. Nat. Rev. Genet. 2015, 16, 71–84. [Google Scholar] [CrossRef]
- Fei, Q.; Xia, R.; Meyers, B.C. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell 2013, 25, 2400–2415. [Google Scholar] [CrossRef]
- Qi, T.; Guo, J.; Peng, H.; Liu, P.; Kang, Z.; Guo, J. Host-Induced Gene Silencing: A Powerful Strategy to Control Diseases of Wheat and Barley. Int. J. Mol. Sci. 2019, 20, 206. [Google Scholar] [CrossRef]
- Csorba, T.; Kontra, L.; Burgyán, J. Viral silencing suppressors: Tools forged to fine-tune host-pathogen coexistence. Virology 2015, 479–480. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, M.; Arora, R.; Lama, T.; Nijhawan, A.; Khurana, J.P.; Tyagi, A.K.; Kapoor, S. Genome-wide identification, organization and phylogenetic analysis of Dicer-like, Argonaute and RNA-dependent RNA Polymerase gene families and their expression analysis during reproductive development and stress in rice. BMC Genom. 2008, 9, 451. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Cheng, Y.; Cheng, X.; Jiang, H.; Zhu, S.; Cheng, B. Identification and characterization of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families in maize. Plant Cell Rep. 2011, 30, 1347. [Google Scholar] [CrossRef] [PubMed]
- Šečić, E.; Zanini, S.; Kogel, K.H. Further Elucidation of the Argonaute and Dicer Protein Families in the Model Grass Species Brachypodium distachyon. Front. Plant Sci. 2019, 10, 1332. [Google Scholar] [CrossRef] [PubMed]
- Margis, R.; Fusaro, A.F.; Smith, N.A.; Curtin, S.J.; Watson, J.M.; Finnegan, E.J.; Waterhouse, P.M. The evolution and diversification of Dicers in plants. FEBS Lett. 2006, 580, 2442–2450. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Chen, C.; Pokhrel, S.; Ma, W.; Huang, K.; Patel, P.; Wang, F.; Xu, J.; Liu, Z.; Li, J.; et al. 24-nt reproductive phasiRNAs are broadly present in angiosperms. Nat. Commun. 2019, 10, 1–8. [Google Scholar] [CrossRef]
- Song, X.; Li, P.; Zhai, J.; Zhou, M.; Ma, L.; Liu, B.; Jeong, D.H.; Nakano, M.; Cao, S.; Liu, C.; et al. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012, 69, 462–474. [Google Scholar] [CrossRef]
- Qin, H.; Chen, F.; Huan, X.; Machida, S.; Song, J.; Yuan, Y.A. Structure of the Arabidopsis thaliana DCL4 DUF283 domain reveals a noncanonical double-stranded RNA-binding fold for protein–protein interaction. RNA 2010, 16, 474–481. [Google Scholar] [CrossRef]
- Meister, G. Argonaute proteins: Functional insights and emerging roles. Nat. Rev. Genet. 2013, 14, 447–459. [Google Scholar] [CrossRef]
- Fang, X.; Qi, Y. RNAi in Plants: An Argonaute-Centered View. Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef]
- Kwak, P.B.; Tomari, Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol. 2012, 19, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Baumberger, N.; Baulcombe, D.C. Arabidopsis ARGONAUTE1 is an RNA Slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 2005, 102, 11928–11933. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Fahlgren, N.; Garcia-Ruiz, H.; Gilbert, K.B.; Montgomery, T.A.; Nguyen, T.; Cuperus, J.T.; Carrington, J.C. Functional Analysis of Three Arabidopsis ARGONAUTES Using Slicer-Defective Mutants. Plant Cell 2012, 24, 3613–3629. [Google Scholar] [CrossRef] [PubMed]
- Arribas-Hernández, L.; Marchais, A.; Poulsen, C.; Haase, B.; Hauptmann, J.; Benes, V.; Meister, G.; Brodersen, P. The Slicer Activity of ARGONAUTE1 Is Required Specifically for the Phasing, Not Production, of Trans-Acting Short Interfering RNAs in Arabidopsis. Plant Cell 2016, 28, 1563–1580. [Google Scholar] [CrossRef]
- Swarts, D.C.; Makarova, K.; Wang, Y.; Nakanishi, K.; Ketting, R.F.; Koonin, E.V.; Patel, D.J.; van der Oost, J. The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol. 2014, 21, 743–753. [Google Scholar] [CrossRef] [PubMed]
- Mallory, A.; Vaucheret, H. Form, function, and regulation of ARGONAUTE proteins. Plant Cell 2010, 22, 3879–3889. [Google Scholar] [CrossRef]
- Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009, 457, 413–420. [Google Scholar] [CrossRef]
- Wassenegger, M.; Krczal, G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006, 11, 142–151. [Google Scholar] [CrossRef]
- Komiya, R. Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing. J. Plant Res. 2017, 130, 17–23. [Google Scholar] [CrossRef]
- Willmann, M.R.; Endres, M.W.; Cook, R.T.; Gregory, B.D. The Functions of RNA-Dependent RNA Polymerases in Arabidopsis. Arabidopsis Book 2011, 9, e0146. [Google Scholar] [CrossRef]
- Branscheid, A.; Marchais, A.; Schott, G.; Lange, H.; Gagliardi, D.; Andersen, S.U.; Voinnet, O.; Brodersen, P. SKI2 mediates degradation of RISC 5′-cleavage fragments and prevents secondary siRNA production from miRNA targets in Arabidopsis. Nucleic Acids Res. 2015, 43, 10975–10988. [Google Scholar] [CrossRef] [PubMed]
- Parent, J.S.; Jauvion, V.; Bouché, N.; Béclin, C.; Hachet, M.; Zytnicki, M.; Vaucheret, H. Post-transcriptional gene silencing triggered by sense transgenes involves uncapped antisense RNA and differs from silencing intentionally triggered by antisense transgenes. Nucleic Acids Res. 2015, 43, 8464–8475. [Google Scholar] [CrossRef] [PubMed]
- Martínez de Alba, A.E.; Moreno, A.B.; Gabriel, M.; Mallory, A.C.; Christ, A.; Bounon, R.; Balzergue, S.; Aubourg, S.; Gautheret, D.; Crespi, M.D.; et al. In plants, decapping prevents RDR6-dependent production of small interfering RNAs from endogenous mRNAs. Nucleic Acids Res. 2015, 43, 2902–2913. [Google Scholar] [CrossRef] [PubMed]
- Carbonell, A.; Carrington, J.C. Antiviral Roles of Plant ARGONAUTES. Curr. Opin. Plant Biol. 2015, 27, 111–117. [Google Scholar] [CrossRef]
- Ding, S.W. RNA-based antiviral immunity. Nat. Rev. Immunol. 2010, 10, 632–644. [Google Scholar] [CrossRef]
- Voinnet, O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 2008, 13, 317–328. [Google Scholar] [CrossRef]
- Barnabás, B.; Jäger, K.; Fehér, A. The effect of drought and heat stress on reproductive processes in cereals. Plant Cell Environ. 2008, 31, 11–38. [Google Scholar] [CrossRef]
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564. [Google Scholar] [CrossRef]
- Ohama, N.; Sato, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Heat Stress Response. Trends Plant Sci. 2017, 22, 53–65. [Google Scholar] [CrossRef]
- Gyula, P.; Baksa, I.; Tóth, T.; Mohorianu, I.; Dalmay, T.; Szittya, G. Ambient temperature regulates the expression of a small set of sRNAs influencing plant development through NF-YA2 and YUC2. Plant Cell Environ. 2018, 41, 2404–2417. [Google Scholar] [CrossRef]
- Szaker, H.M.; Gyula, P.; Szittya, G.; Csorba, T. Regulation of High-Temperature Stress Response by Small RNAs. In Plant microRNAs: Shaping Development and Environmental Responses; Miguel, C., Dalmay, T., Chaves, I., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 171–197. [Google Scholar]
- Szaker, H.M.; Darkó, É.; Medzihradszky, A.; Janda, T.; Liu, H.; Charng, Y.; Csorba, T. miR824/AGAMOUS-LIKE16 Module Integrates Recurring Environmental Heat Stress Changes to Fine-Tune Poststress Development. Front. Plant Sci. 2019, 10, 1454. [Google Scholar] [CrossRef] [PubMed]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.R.; Bäurle, I. Arabidopsis miR156 Regulates Tolerance to Recurring Environmental Stress through SPL Transcription Factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Ravichandran, S.; Ragupathy, R.; Edwards, T.; Domaratzki, M.; Cloutier, S. MicroRNA-guided regulation of heat stress response in wheat. BMC Genom. 2019, 20, 488. [Google Scholar] [CrossRef] [PubMed]
- Kruszka, K.; Pacak, A.; Swida-Barteczka, A.; Nuc, P.; Alaba, S.; Wroblewska, Z.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J. Exp. Bot. 2014, 65, 6123–6135. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Schölkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” Browser for Exploring and Analyzing Large-Scale Biological Data Sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef]
- Sibout, R.; Proost, S.; Hansen, B.O.; Vaid, N.; Giorgi, F.M.; Ho-Yue-Kuang, S.; Legée, F.; Cézart, L.; Bouchabké-Coussa, O.; Soulhat, C.; et al. Expression atlas and comparative coexpression network analyses reveal important genes involved in the formation of lignified cell wall in Brachypodium distachyon. New Phytol. 2017, 215, 1009–1025. [Google Scholar] [CrossRef]
- Zhai, L.; Teng, F.; Zheng, K.; Xiao, J.; Deng, W.; Sun, W. Expression analysis of Argonaute genes in maize (Zea mays L.) in response to abiotic stress. Hereditas 2019, 156, 27. [Google Scholar] [CrossRef]
- Zhao, H.; Zhao, K.; Wang, J.; Chen, X.; Chen, Z.; Cai, R.; Xiang, Y. Comprehensive Analysis of Dicer-Like, Argonaute, and RNA-dependent RNA Polymerase Gene Families in Grapevine (Vitis Vinifera). J. Plant Growth Regul. 2015, 34, 108–121. [Google Scholar] [CrossRef]
- Qin, L.; Mo, N.; Muhammad, T.; Liang, Y. Genome-Wide Analysis of DCL, AGO, and RDR Gene Families in Pepper (Capsicum Annuum L.). Int. J. Mol. Sci. 2018, 19, 1038. [Google Scholar] [CrossRef]
- Bai, M.; Yang, G.S.; Chen, W.T.; Mao, Z.C.; Kang, H.X.; Chen, G.H.; Yang, Y.H.; Xie, B.Y. Genome-wide identification of Dicer-like, Argonaute and RNA-dependent RNA polymerase gene families and their expression analyses in response to viral infection and abiotic stresses in Solanum lycopersicum. Gene 2012, 501, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Yadav, C.B.; Muthamilarasan, M.; Pandey, G.; Prasad, M. Identification, Characterization and Expression Profiling of Dicer-Like, Argonaute and RNA-Dependent RNA Polymerase Gene Families in Foxtail Millet. Plant Mol. Biol. Report. 2015, 33, 43–55. [Google Scholar] [CrossRef]
- Gan, D.; Liang, D.; Wu, J.; Zhan, M.; Yang, F.; Xu, W.; Zhu, S.; Shi, J. Genome-Wide Identification of the Dicer-Like, Argonaute, and RNA-Dependent RNA Polymerase Gene Families in Cucumber (Cucumis sativus L.). J. Plant Growth Regul. 2016, 35, 135–150. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef]
- Hubbard, T.; Barker, D.; Birney, E.; Cameron, G.; Chen, Y.; Clark, L.; Cox, T.; Cuff, J.; Curwen, V.; Down, T.; et al. The Ensembl genome database project. Nucleic Acids Res. 2002, 30, 38–41. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME Suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Stothard, P. The Sequence Manipulation Suite: JavaScript Programs for Analyzing and Formatting Protein and DNA Sequences. BioTechniques 2000, 28, 1102–1104. [Google Scholar] [CrossRef]
- Pacak, A.; Barciszewska-Pacak, M.; Swida-Barteczka, A.; Kruszka, K.; Sega, P.; Milanowska, K.; Jakobsen, I.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Heat Stress Affects Pi-related Genes Expression and Inorganic Phosphate Deposition/Accumulation in Barley. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Pimentel, H.; Bray, N.L.; Puente, S.; Melsted, P.; Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 2017, 14, 687–690. [Google Scholar] [CrossRef] [PubMed]
- Kolde, R. Pheatmap: Pretty heatmaps. R Package Version 2012, 61, 926. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Madsen, C.T.; Stephens, J.; Hornyik, C.; Shaw, J.; Collinge, D.B.; Lacomme, C.; Albrechtsen, M. Identification and characterization of barley RNA-directed RNA polymerases. Biochim. Biophys. Acta BBA—Gene Regul. Mech. 2009, 1789, 375–385. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Höck, J.; Meister, G. The Argonaute protein family. Genome Biol. 2008, 9, 210. [Google Scholar] [CrossRef]
- Chung, B.Y.W.; Valli, A.; Deery, M.J.; Navarro, F.J.; Brown, K.; Hnatova, S.; Howard, J.; Molnar, A.; Baulcombe, D.C. Distinct roles of Argonaute in the green alga Chlamydomonas reveal evolutionary conserved mode of miRNA-mediated gene expression. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xia, R.; Meyers, B.C.; Walbot, V. Evolution, functions, and mysteries of plant ARGONAUTE proteins. Curr. Opin. Plant Biol. 2015, 27, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, D.E.; Nakanishi, K.; Patel, D.J.; Bartel, D.P. The Inside-Out Mechanism of Dicers from Budding Yeasts. Cell 2011, 146, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Burdisso, P.; Suarez, I.P.; Bologna, N.G.; Palatnik, J.F.; Bersch, B.; Rasia, R.M. Second Double-Stranded RNA Binding Domain of Dicer-like Ribonuclease 1: Structural and Biochemical Characterization. Biochemistry 2012, 51, 10159–10166. [Google Scholar] [CrossRef]
- Mickiewicz, A.; Sarzyńska, J.; Miłostan, M.; Kurzyńska-Kokorniak, A.; Rybarczyk, A.; Łukasiak, P.; Kuliński, T.; Figlerowicz, M.; Błażewicz, J. Modeling of the catalytic core of Arabidopsis thaliana Dicer-like 4 protein and its complex with double-stranded RNA. Comput. Biol. Chem. 2017, 66, 44–56. [Google Scholar] [CrossRef]
- Lingel, A.; Simon, B.; Izaurralde, E.; Sattler, M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 2003, 426, 465–469. [Google Scholar] [CrossRef]
- Yan, K.S.; Yan, S.; Farooq, A.; Han, A.; Zeng, L.; Zhou, M.M. Structure and conserved RNA binding of the PAZ domain. Nature 2003, 426, 469–474. [Google Scholar] [CrossRef]
- Ma, J.B.; Ye, K.; Patel, D.J. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 2004, 429, 318–322. [Google Scholar] [CrossRef]
- Ma, J.B.; Yuan, Y.R.; Meister, G.; Pei, Y.; Tuschl, T.; Patel, D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 2005, 434, 666–670. [Google Scholar] [CrossRef]
- Frank, F.; Hauver, J.; Sonenberg, N.; Nagar, B. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012, 31, 3588–3595. [Google Scholar] [CrossRef]
- Thieme, C.J.; Schudoma, C.; May, P.; Walther, D. Give It AGO: The Search for miRNA-Argonaute Sorting Signals in Arabidopsis thaliana Indicates a Relevance of Sequence Positions Other than the 5′-Position Alone. Front. Plant Sci. 2012, 3, 272. [Google Scholar] [CrossRef] [PubMed]
- Mi, S.; Cai, T.; Hu, Y.; Chen, Y.; Hodges, E.; Ni, F.; Wu, L.; Li, S.; Zhou, H.; Long, C.; et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5′ terminal nucleotide. Cell 2008, 133, 116–127. [Google Scholar] [CrossRef] [PubMed]
- Zha, X.; Xia, Q.; Yuan, Y.A. Structural insights into small RNA sorting and mRNA target binding by Arabidopsis Argonaute Mid domains. FEBS Lett. 2012, 586, 3200–3207. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Zhang, Q.; Zhou, H.; Ni, F.; Wu, X.; Qi, Y. Rice MicroRNA Effector Complexes and Targets. Plant Cell 2009, 21, 3421–3435. [Google Scholar] [CrossRef]
- Montgomery, T.A.; Howell, M.D.; Cuperus, J.T.; Li, D.; Hansen, J.E.; Alexander, A.L.; Chapman, E.J.; Fahlgren, N.; Allen, E.; Carrington, J.C. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 2008, 133, 128–141. [Google Scholar] [CrossRef]
- Klum, S.M.; Chandradoss, S.D.; Schirle, N.T.; Joo, C.; MacRae, I.J. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 2018, 37, 75–88. [Google Scholar] [CrossRef]
- Baker, C.M.; Grant, G.H. Role of aromatic amino acids in protein–nucleic acid recognition. Biopolymers 2007, 85, 456–470. [Google Scholar] [CrossRef]
- Nakama, M.; Kawakami, K.; Kajitani, T.; Urano, T.; Murakami, Y. DNA–RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells 2012, 17, 218–233. [Google Scholar] [CrossRef]
- Fátyol, K.; Ludman, M.; Burgyán, J. Functional dissection of a plant Argonaute. Nucleic Acids Res. 2016, 44, 1384–1397. [Google Scholar] [CrossRef]
- Zhang, X.; Niu, D.; Carbonell, A.; Wang, A.; Lee, A.; Tun, V.; Wang, Z.; Carrington, J.C.; Chang, C.E.A.; Jin, H. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat. Commun. 2014, 5, 5468. [Google Scholar] [CrossRef]
- Till, S.; Lejeune, E.; Thermann, R.; Bortfeld, M.; Hothorn, M.; Enderle, D.; Heinrich, C.; Hentze, M.W.; Ladurner, A.G. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007, 14, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Giner, A.; Lakatos, L.; García-Chapa, M.; López-Moya, J.J.; Burgyán, J. Viral Protein Inhibits RISC Activity by Argonaute Binding through Conserved WG/GW Motifs. PLOS Pathog. 2010, 6, e1000996. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, J.; Garcia, D.; Pontier, D.; Ohnesorge, S.; Yu, A.; Garcia, S.; Braun, L.; Bergdoll, M.; Hakimi, M.A.; Lagrange, T.; et al. Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 2010, 24, 904–915. [Google Scholar] [CrossRef] [PubMed]
- Vaucheret, H.; Vazquez, F.; Crété, P.; Bartel, D.P. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004, 18, 1187–1197. [Google Scholar] [CrossRef] [PubMed]
- Várallyay, É.; Válóczi, A.; Ágyi, Á.; Burgyán, J.; Havelda, Z. Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. EMBO J. 2010, 29, 3507–3519. [Google Scholar] [CrossRef]
- Lu, C.; Jeong, D.H.; Kulkarni, K.; Pillay, M.; Nobuta, K.; German, R.; Thatcher, S.R.; Maher, C.; Zhang, L.; Ware, D.; et al. Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc. Natl. Acad. Sci. USA 2008, 105, 4951–4956. [Google Scholar] [CrossRef]
- Schreiber, A.W.; Shi, B.J.; Huang, C.Y.; Langridge, P.; Baumann, U. Discovery of barley miRNAs through deep sequencing of short reads. BMC Genom. 2011, 12, 129. [Google Scholar] [CrossRef]
- Jagtap, S.; Shivaprasad, P.V. Diversity, expression and mRNA targeting abilities of Argonaute-targeting miRNAs among selected vascular plants. BMC Genom. 2014, 15, 1049. [Google Scholar] [CrossRef]
- Chávez-Hernández, E.C.; Alejandri-Ramírez, N.D.; Juárez-González, V.T.; Dinkova, T.D. Maize miRNA and target regulation in response to hormone depletion and light exposure during somatic embryogenesis. Front. Plant Sci. 2015, 6, 555. [Google Scholar] [CrossRef]
- Ozhuner, E.; Eldem, V.; Ipek, A.; Okay, S.; Sakcali, S.; Zhang, B.; Boke, H.; Unver, T. Boron Stress Responsive MicroRNAs and Their Targets in Barley. PLoS ONE 2013, 8, e59543. [Google Scholar] [CrossRef]
- Harvey, J.J.W.; Lewsey, M.G.; Patel, K.; Westwood, J.; Heimstädt, S.; Carr, J.P.; Baulcombe, D.C. An Antiviral Defense Role of AGO2 in Plants. PLoS ONE 2011, 6, e14639. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Kasschau, K.D.; Carrington, J.C. Negative Feedback Regulation of Dicer-Like1 in Arabidopsis by microRNA-Guided mRNA Degradation. Curr. Biol. 2003, 13, 784–789. [Google Scholar] [CrossRef]
- Grover, A.; Mittal, D.; Negi, M.; Lavania, D. Generating high temperature tolerant transgenic plants: Achievements and challenges. Plant Sci. 2013, 205, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Zinn, K.E.; Tunc-Ozdemir, M.; Harper, J.F. Temperature stress and plant sexual reproduction: Uncovering the weakest links. J. Exp. Bot. 2010, 61, 1959–1968. [Google Scholar] [CrossRef]
- Richter, K.; Haslbeck, M.; Buchner, J. The Heat Shock Response: Life on the Verge of Death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- Teng, C.; Zhang, H.; Hammond, R.; Huang, K.; Meyers, B.C.; Walbot, V. Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. bioRxiv 2018, 498410. [Google Scholar] [CrossRef]
- Meng, F.; Jia, H.; Ling, N.; Xue, Y.; Liu, H.; Wang, K.; Yin, J.; Li, Y. Cloning and characterization of two Argonaute genes in wheat (Triticum aestivum L.). BMC Plant Biol. 2013, 13, 18. [Google Scholar] [CrossRef]
- Waterhouse, P. RNA interference: A dark horse in the AGO stable. Nat. Plants 2016, 2, 1–2. [Google Scholar] [CrossRef]
- Liu, H.; Nonomura, K.I. A wide reprogramming of histone H3 modifications during male meiosis I in rice is dependent on the Argonaute protein MEL1. J. Cell Sci. 2016, 129, 3553–3561. [Google Scholar] [CrossRef]
- Zhu, H.; Hu, F.; Wang, R.; Zhou, X.; Sze, S.H.; Liou, L.W.; Barefoot, A.; Dickman, M.; Zhang, X. Arabidopsis Argonaute 10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell 2011, 145, 242–256. [Google Scholar] [CrossRef]
- Yu, Y.; Ji, L.; Le, B.H.; Zhai, J.; Chen, J.; Luscher, E.; Gao, L.; Liu, C.; Cao, X.; Mo, B.; et al. ARGONAUTE10 promotes the degradation of miR165/6 through the SDN1 and SDN2 exonucleases in Arabidopsis. PLOS Biol. 2017, 15, e2001272. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Yang, Z.; Wang, Y.; Zheng, L.; Ye, R.; Ji, Y.; Zhao, S.; Ji, S.; Liu, R.; Xu, L.; et al. Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA. eLife 2015, 4, e05733. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Zhang, H.; Arikit, S.; Huang, K.; Nan, G.L.; Walbot, V.; Meyers, B.C. Spatiotemporally dynamic, cell-type–dependent premeiotic and meiotic phasiRNAs in maize anthers. Proc. Natl. Acad. Sci. USA 2015, 112, 3146–3151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhao, H.; Gao, S.; Wang, W.C.; Katiyar-Agarwal, S.; Huang, H.D.; Raikhel, N.; Jin, H. Arabidopsis Argonaute 2 Regulates Innate Immunity via miRNA393∗-Mediated Silencing of a Golgi-Localized SNARE Gene, MEMB12. Mol. Cell 2011, 42, 356–366. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Rendtlew Danielsen, J.M.; Yang, Y.G.; Qi, Y. A Role for Small RNAs in DNA Double-Strand Break Repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, X.; Guo, X.; Wang, X.J.; Zhang, X. Arabidopsis AGO3 predominantly recruits 24-nt small RNAs to regulate epigenetic silencing. Nat. Plants 2016, 2, 16049. [Google Scholar] [CrossRef]
- Douglas, R.N.; Wiley, D.; Sarkar, A.; Springer, N.; Timmermans, M.C.P.; Scanlon, M.J. Ragged seedling2 Encodes an ARGONAUTE7-Like Protein Required for Mediolateral Expansion, but Not Dorsiventrality, of Maize Leaves. Plant Cell 2010, 22, 1441–1451. [Google Scholar] [CrossRef]
- Shen, D.; Wang, S.; Chen, H.; Zhu, Q.H.; Helliwell, C.; Fan, L. Molecular phylogeny of miR390-guided trans-acting siRNA genes (TAS3) in the grass family. Plant Syst. Evol. 2009, 283, 125. [Google Scholar] [CrossRef]
- Havecker, E.R.; Wallbridge, L.M.; Hardcastle, T.J.; Bush, M.S.; Kelly, K.A.; Dunn, R.M.; Schwach, F.; Doonan, J.H.; Baulcombe, D.C. The Arabidopsis RNA-Directed DNA Methylation Argonautes Functionally Diverge Based on Their Expression and Interaction with Target Loci. Plant Cell 2010, 22, 321–334. [Google Scholar] [CrossRef]
- Duan, C.G.; Zhang, H.; Tang, K.; Zhu, X.; Qian, W.; Hou, Y.J.; Wang, B.; Lang, Z.; Zhao, Y.; Wang, X.; et al. Specific but interdependent functions for Arabidopsis AGO4 and AGO6 in RNA-directed DNA methylation. EMBO J. 2015, 34, 581–592. [Google Scholar] [CrossRef]
- Olmedo-Monfil, V.; Durán-Figueroa, N.; Arteaga-Vázquez, M.; Demesa-Arévalo, E.; Autran, D.; Grimanelli, D.; Slotkin, R.K.; Martienssen, R.A.; Vielle-Calzada, J.P. Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 2010, 464, 628–632. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Mao, L.; Qi, Y. Roles of DICER-LIKE and ARGONAUTE Proteins in TAS-Derived Small Interfering RNA-Triggered DNA Methylation. Plant Physiol. 2012, 160, 990–999. [Google Scholar] [CrossRef] [PubMed]
- Matzke, M.A.; Mosher, R.A. RNA-directed DNA methylation: An epigenetic pathway of increasing complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Goel, S.; Meeley, R.B.; Dantec, C.; Parrinello, H.; Michaud, C.; Leblanc, O.; Grimanelli, D. Production of Viable Gametes without Meiosis in Maize Deficient for an ARGONAUTE Protein. Plant Cell 2011, 23, 443–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jiao, X.; Kong, X.; Hamera, S.; Wu, Y.; Chen, X.; Fang, R.; Yan, Y. A Signaling Cascade from miR444 to RDR1 in Rice Antiviral RNA Silencing Pathway. Plant Physiol. 2016, 170, 2365–2377. [Google Scholar] [CrossRef] [PubMed]
- Alleman, M.; Sidorenko, L.; McGinnis, K.; Seshadri, V.; Dorweiler, J.E.; White, J.; Sikkink, K.; Chandler, V.L. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature 2006, 442, 295–298. [Google Scholar] [CrossRef]
- Nagasaki, H.; Itoh, J.; Hayashi, K.; Hibara, K.; Satoh-Nagasawa, N.; Nosaka, M.; Mukouhata, M.; Ashikari, M.; Kitano, H.; Matsuoka, M.; et al. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proc. Natl. Acad. Sci. USA 2007, 104, 14867–14871. [Google Scholar] [CrossRef]
- Nakasugi, K.; Crowhurst, R.N.; Bally, J.; Wood, C.C.; Hellens, R.P.; Waterhouse, P.M. De Novo Transcriptome Sequence Assembly and Analysis of RNA Silencing Genes of Nicotiana benthamiana. PLoS ONE 2013, 8, e59534. [Google Scholar] [CrossRef]
- Dalmadi, Á.; Gyula, P.; Bálint, J.; Szittya, G.; Havelda, Z. AGO-unbound cytosolic pool of mature miRNAs in plant cells reveals a novel regulatory step at AGO1 loading. Nucleic Acids Res. 2019, 47, 9803–9817. [Google Scholar] [CrossRef]
- Zhong, S.H.; Liu, J.Z.; Jin, H.; Lin, L.; Li, Q.; Chen, Y.; Yuan, Y.X.; Wang, Z.Y.; Huang, H.; Qi, Y.J.; et al. Warm temperatures induce transgenerational epigenetic release of RNA silencing by inhibiting siRNA biogenesis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2013, 110, 9171–9176. [Google Scholar] [CrossRef]
- Yu, X.; Wang, H.; Lu, Y.; de Ruiter, M.; Cariaso, M.; Prins, M.; van Tunen, A.; He, Y. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 2012, 63, 1025–1038. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Liu, J.; Liu, Z.; Li, X.; Wu, F.; He, Y. HEAT-INDUCED TAS1 TARGET1 Mediates Thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a–Directed Pathways in Arabidopsis. Plant Cell 2014, 26, 1764–1780. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.S.; Kuo, C.C.; Yang, I.C.; Tsai, W.A.; Shen, Y.H.; Lin, C.C.; Liang, Y.C.; Li, Y.C.; Kuo, Y.W.; King, Y.C.; et al. MicroRNA160 Modulates Plant Development and Heat Shock Protein Gene Expression to Mediate Heat Tolerance in Arabidopsis. Front. Plant Sci. 2018, 9, 68. [Google Scholar] [CrossRef] [PubMed]
- Nuthikattu, S.; McCue, A.D.; Panda, K.; Fultz, D.; DeFraia, C.; Thomas, E.N.; Slotkin, R.K. The Initiation of Epigenetic Silencing of Active Transposable Elements Is Triggered by RDR6 and 21–22 Nucleotide Small Interfering RNAs. Plant Physiol. 2013, 162, 116–131. [Google Scholar] [CrossRef] [PubMed]


| Predicted Gene Name * | Chromosomal Location (5′-3′) | Accession No. of Ensembl Transcript | ORF Length (bp) | No. Exons | No. of Amino Acids | Isoelectric Point (PI) ** | Molecular Weight (Da) ** |
|---|---|---|---|---|---|---|---|
| HvDCLs | |||||||
| HvDCL1 | chr4H:630334902–630347021 | HORVU4Hr1G084890.7 | 5994 | 19 | 1997 | 6.26 | 222,185.89 |
| HvDCL2 | chr5H:84450148–84493265 | HORVU5Hr1G019300.8 | 4428 | 20 | 1475 | 8.56 | 165,696.76 |
| HvDCL3 | chr3H:638064590–638074096 | HORVU3Hr1G092430.9 | 5058 | 26 | 1685 | 5.92 | 188,438.84 |
| HvDCL4 | chr2H:632322356–632371458 | HORVU2Hr1G088270.9 | 4908 | 25 | 1635 | 6.17 | 185,174.48 |
| HvDCL5 | chr1H:310007028–310025536 | HORVU1Hr1G042710.5 | 5052 | 26 | 1683 | 6.21 | 189,830.24 |
| HvAGOs | |||||||
| HvAGO1a | chr7H:9459266–9467181 | HORVU7Hr1G007000.14 | 3654 | 23 | 1217 | 9.56 | 133,685.87 |
| HvAGO1d | chr7H:651899946–651906855 | HORVU7Hr1G120600.3 | 3195 | 22 | 1064 | 9.05 | 117,524.50 |
| HvAGO2 | chr2H:684121508–684127760 | HORVU2Hr1G098650.1 | 3144 | 3 | 1047 | 9.34 | 113,039.59 |
| HvAGO4a | chr3H:226356734–226364526 | HORVU3Hr1G038830.1 | 2769 | 23 | 922 | 9.10 | 102,605.58 |
| HvAGO4b | chr7H:623895748–623891202 | HORVU7Hr1G107770.0 *** | 2451 | 22 | 816 | 4.88 | 201,998.97 |
| HvAGO5a | chr4H:8972435–8981437 | HORVU4Hr1G004030.1 | 3102 | 22 | 1033 | 9.47 | 114,317.97 |
| HvAGO5b | chr2H:116787397–116793928 | HORVU2Hr1G031130.2 | 2532 | 22 | 843 | 9.17 | 94,171.10 |
| HvAGO6 | chrUn:71291842–71302762 | HORVU0Hr1G012490.1 | 2652 | 23 | 883 | 9.25 | 98,208.02 |
| HvAGO7 | chrUn:29969371–29973587 | HORVU0Hr1G005350.1 | 3039 | 3 | 1012 | 9.28 | 114,045.18 |
| HvAGO10 | chr6H:183574909–183587426 | HORVU6Hr1G036840.2 | 2850 | 21 | 949 | 9.43 | 105,648.60 |
| HvAGO18 | chr3H:68596766–68603419 | HORVU3Hr1G021290.1 | 3156 | 21 | 1051 | 9.17 | 114,177.28 |
| HvRDRs | |||||||
| HvRDR1a | chr6H:512834480–512842253 | HORVU6Hr1G074180.1 | 3348 | 5 | 1115 | 7.73 | 126,707.04 |
| HvRDR1b | chr6H:513108807–513120207 | HORVU6Hr1G074220.2 | 3549 | 5 | 1182 | 8.66 | 134,118.47 |
| HvRDR2 | chr2H:589496751–589507376 | HORVU2Hr1G081260.6 | 3414 | 4 | 1137 | 6.80 | 127,637.79 |
| HvRDR3 | chr3H:116156819–116191312 | HORVU3Hr1G027290.6 | 3588 | 19 | 1195 | 6.30 | 134,565.32 |
| HvRDR4 | chr3H:116503595–116535566 | HORVU3Hr1G027340.4 | 3507 | 19 | 1168 | 6.83 | 131,898.43 |
| HvRDR6a | chr3H:673186268–673191806 | HORVU3Hr1G107690.2 | 3684 | 2 | 1227 | 7.38 | 137,814.98 |
| HvRDR6b | chr3H:17012603–17027858 | HORVU7Hr1G012280.1 | 3621 | 6 | 1206 | 8.17 | 135,686.38 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hamar, É.; Szaker, H.M.; Kis, A.; Dalmadi, Á.; Miloro, F.; Szittya, G.; Taller, J.; Gyula, P.; Csorba, T.; Havelda, Z. Genome-Wide Identification of RNA Silencing-Related Genes and Their Expressional Analysis in Response to Heat Stress in Barley (Hordeum vulgare L.). Biomolecules 2020, 10, 929. https://doi.org/10.3390/biom10060929
Hamar É, Szaker HM, Kis A, Dalmadi Á, Miloro F, Szittya G, Taller J, Gyula P, Csorba T, Havelda Z. Genome-Wide Identification of RNA Silencing-Related Genes and Their Expressional Analysis in Response to Heat Stress in Barley (Hordeum vulgare L.). Biomolecules. 2020; 10(6):929. https://doi.org/10.3390/biom10060929
Chicago/Turabian StyleHamar, Éva, Henrik Mihály Szaker, András Kis, Ágnes Dalmadi, Fabio Miloro, György Szittya, János Taller, Péter Gyula, Tibor Csorba, and Zoltán Havelda. 2020. "Genome-Wide Identification of RNA Silencing-Related Genes and Their Expressional Analysis in Response to Heat Stress in Barley (Hordeum vulgare L.)" Biomolecules 10, no. 6: 929. https://doi.org/10.3390/biom10060929
APA StyleHamar, É., Szaker, H. M., Kis, A., Dalmadi, Á., Miloro, F., Szittya, G., Taller, J., Gyula, P., Csorba, T., & Havelda, Z. (2020). Genome-Wide Identification of RNA Silencing-Related Genes and Their Expressional Analysis in Response to Heat Stress in Barley (Hordeum vulgare L.). Biomolecules, 10(6), 929. https://doi.org/10.3390/biom10060929





