Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1
Abstract
1. Introduction
2. Materials and Methods
2.1. MLLE Protein Expression, Purification and Complex Crystallization
2.2. Crystallographic Methods
2.3. Cloning, Mutagenesis and Antibodies
2.4. Cell Culture and Luciferase Assay
2.5. Immunological Procedures and In Vitro Binding Studies
3. Results
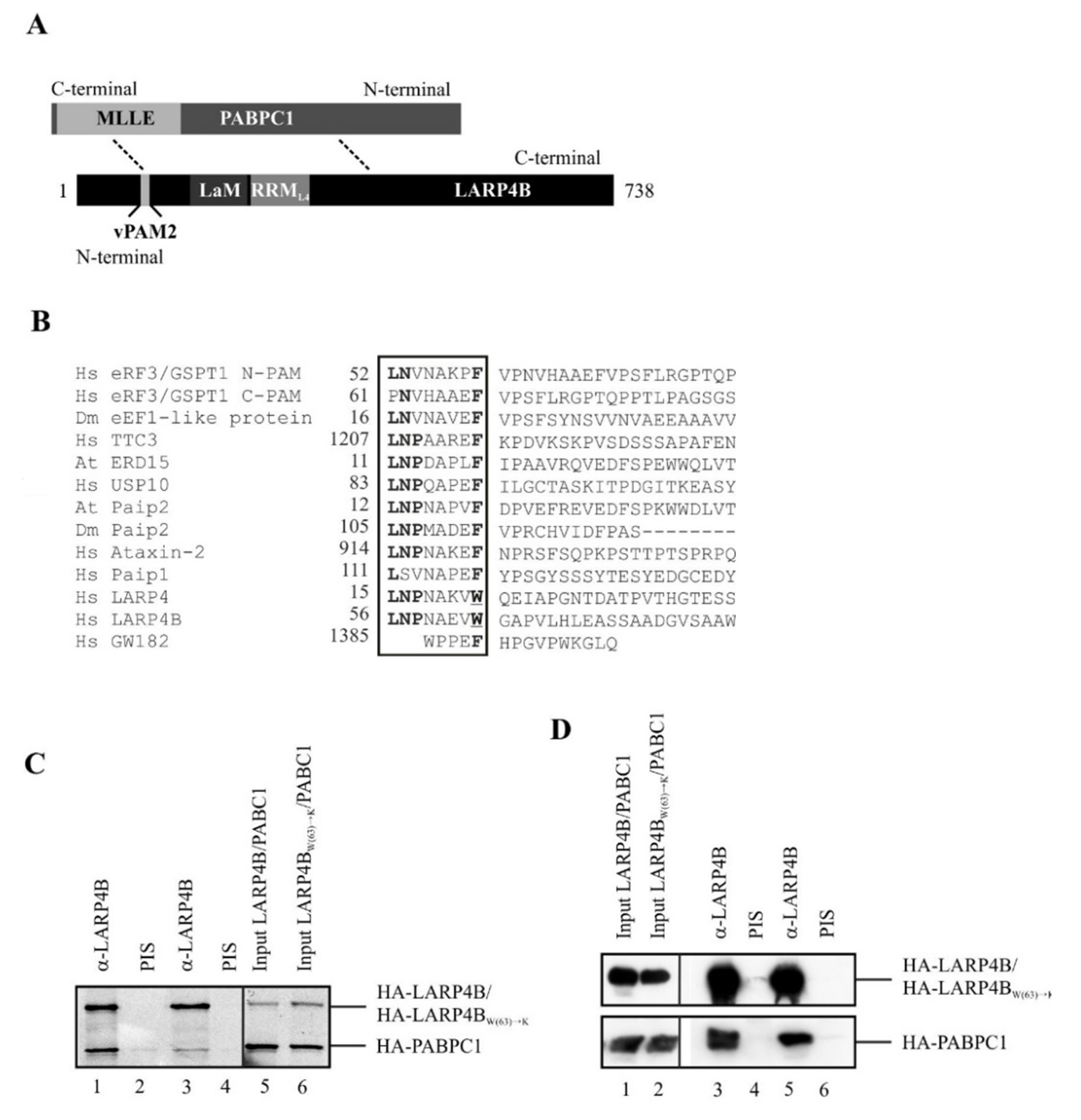
3.1. LARP4B Contains a Variant PAM2 Motif Necessary for Efficient PABC1 Binding
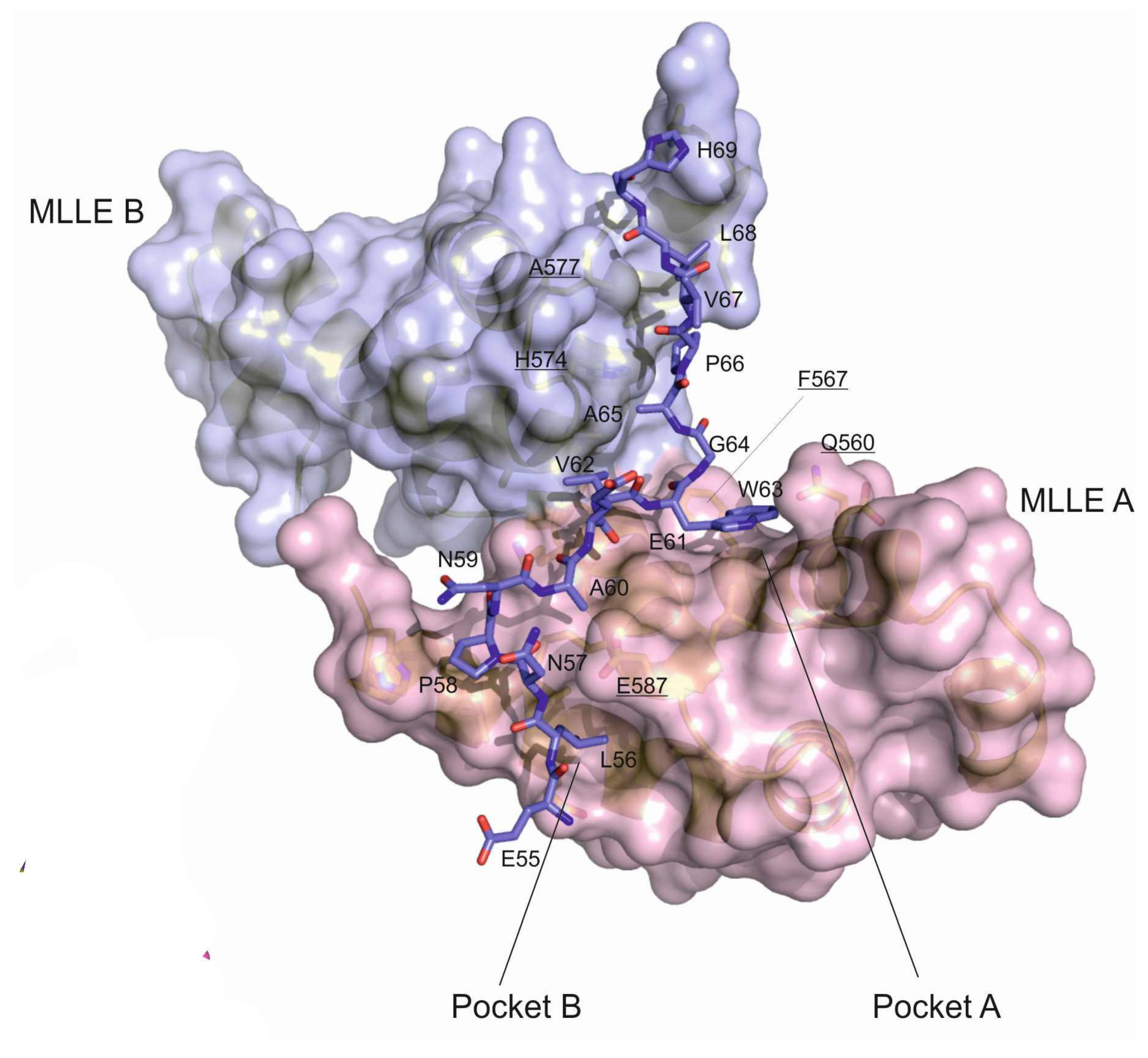
3.2. Crystal Structure of the LARP4B PAM2w/MLLE Complex
3.3. PAM2w/PAM2 Sequences Can Be Divided into a Canonical and a Variable Part
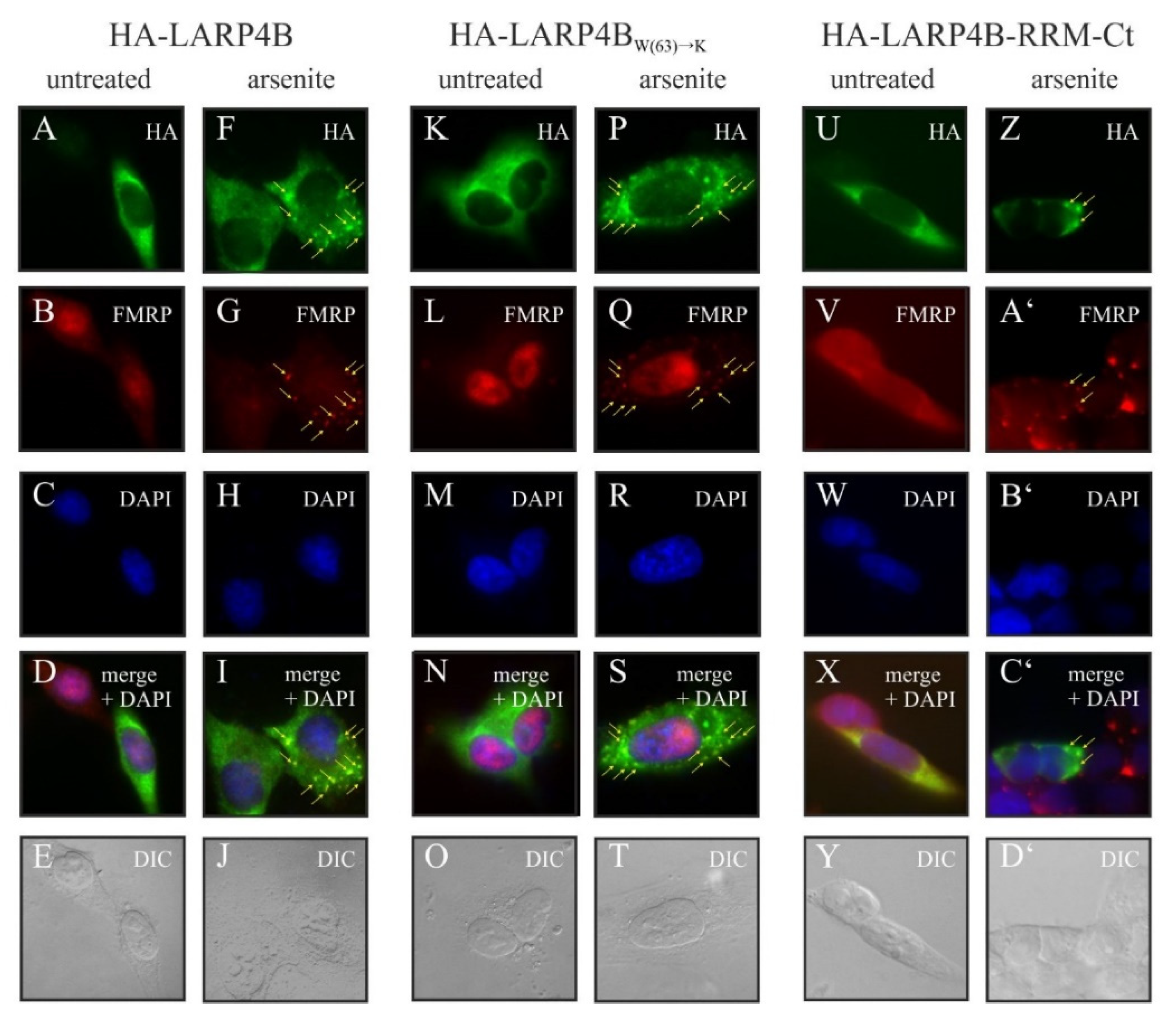
3.4. Disruption of the PAM2w/MLLE Interaction Does Not Affect Recruitment of LARP4B to Stress Granules upon Arsenite Stress
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Mangus, D.A.; Amrani, N.; Jacobson, A. Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation. Mol. Cell. Biol. 1998, 18, 7383–7396. [Google Scholar] [CrossRef] [PubMed]
- Mangus, D.A.; Evans, M.C.; Jacobson, A. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003, 4, 223. [Google Scholar] [CrossRef]
- Burd, C.G.; Dreyfuss, G. Conserved structures and diversity of functions of RNA-binding proteins. Science 1994, 265, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.G.; Matunis, E.L.; Dreyfuss, G. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol. Cell. Biol. 1991, 11, 3419–3424. [Google Scholar] [CrossRef]
- Khanam, T.; Muddashetty, R.S.; Kahvejian, A.; Sonenberg, N.; Brosius, J. Poly(A)-binding protein binds to A-rich sequences via RNA-binding domains 1+2 and 3+4. RNA Biol. 2006, 3, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Schafer, I.B.; Yamashita, M.; Schuller, J.M.; Schussler, S.; Reichelt, P.; Strauss, M.; Conti, E. Molecular Basis for poly(A) RNP Architecture and Recognition by the Pan2-Pan3 Deadenylase. Cell 2019, 177, 1619–1631.e1621. [Google Scholar] [CrossRef]
- Kuhn, U.; Pieler, T. Xenopus poly(A) binding protein: Functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 1996, 256, 20–30. [Google Scholar] [CrossRef]
- Gray, N.K.; Coller, J.M.; Dickson, K.S.; Wickens, M. Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000, 19, 4723–4733. [Google Scholar] [CrossRef]
- Collier, B.; Gorgoni, B.; Loveridge, C.; Cooke, H.J.; Gray, N.K. The DAZL family proteins are PABP-binding proteins that regulate translation in germ cells. EMBO J. 2005, 24, 2656–2666. [Google Scholar] [CrossRef]
- Mangus, D.A.; Evans, M.C.; Agrin, N.S.; Smith, M.; Gongidi, P.; Jacobson, A. Positive and negative regulation of poly(A) nuclease. Mol. Cell. Biol. 2004, 24, 5521–5533. [Google Scholar] [CrossRef]
- Patel, G.P.; Bag, J. IMP1 interacts with poly(A)-binding protein (PABP) and the autoregulatory translational control element of PABP-mRNA through the KH III-IV domain. FEBS J. 2006, 273, 5678–5690. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Safaee, N.; Rosenauer, A.; Gehring, K. Structural basis of binding of P-body-associated proteins GW182 and ataxin-2 by the Mlle domain of poly(A)-binding protein. J. Biol. Chem. 2010, 285, 13599–13606. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Gallie, D.R. eIF4G, eIFiso4G, and eIF4B bind the poly(A)-binding protein through overlapping sites within the RNA recognition motif domains. J. Biol. Chem. 2007, 282, 25247–25258. [Google Scholar] [CrossRef]
- Roy, G.; De Crescenzo, G.; Khaleghpour, K.; Kahvejian, A.; O’Connor-McCourt, M.; Sonenberg, N. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol. Cell. Biol. 2002, 22, 3769–3782. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Menade, M.; Rosenauer, A.; Nguyen, L.; Gehring, K. Molecular determinants of PAM2 recognition by the MLLE domain of poly(A)-binding protein. J. Mol. Biol. 2010, 397, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Lim, N.S.; Kozlov, G.; Chang, T.C.; Groover, O.; Siddiqui, N.; Volpon, L.; De Crescenzo, G.; Shyu, A.B.; Gehring, K. Comparative peptide binding studies of the PABC domains from the ubiquitin-protein isopeptide ligase HYD and poly(A)-binding protein. Implications for HYD function. J. Biol. Chem. 2006, 281, 14376–14382. [Google Scholar] [CrossRef]
- Xie, J.; Kozlov, G.; Gehring, K. The “tale” of poly(A) binding protein: The MLLE domain and PAM2-containing proteins. Biochim. Biophys. Acta 2014, 1839, 1062–1068. [Google Scholar] [CrossRef]
- Kozlov, G.; Siddiqui, N.; Coillet-Matillon, S.; Trempe, J.F.; Ekiel, I.; Sprules, T.; Gehring, K. Solution structure of the orphan PABC domain from Saccharomyces cerevisiae poly(A)-binding protein. J. Biol. Chem. 2002, 277, 22822–22828. [Google Scholar] [CrossRef]
- Deo, R.C.; Sonenberg, N.; Burley, S.K. X-ray structure of the human hyperplastic discs protein: An ortholog of the C-terminal domain of poly(A)-binding protein. Proc. Natl. Acad. Sci. USA 2001, 98, 4414–4419. [Google Scholar] [CrossRef]
- Kozlov, G.; De Crescenzo, G.; Lim, N.S.; Siddiqui, N.; Fantus, D.; Kahvejian, A.; Trempe, J.F.; Elias, D.; Ekiel, I.; Sonenberg, N.; et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J. 2004, 23, 272–281. [Google Scholar] [CrossRef]
- Tritschler, F.; Huntzinger, E.; Izaurralde, E. Role of GW182 proteins and PABPC1 in the miRNA pathway: A sense of deja vu. Nat. Rev. Mol. Cell. Biol. 2010, 11, 379–384. [Google Scholar] [CrossRef]
- Dos Santos Rodrigues, F.H.; Firczuk, H.; Breeze, A.L.; Cameron, A.D.; Walko, M.; Wilson, A.J.; Zanchin, N.I.T.; McCarthy, J.E.G. The Leishmania PABP1-eIF4E4 interface: A novel 5′-3′ interaction architecture for trans-spliced mRNAs. Nucleic Acids Res. 2019, 47, 1493–1504. [Google Scholar] [CrossRef] [PubMed]
- Bousquet-Antonelli, C.; Deragon, J.M. A comprehensive analysis of the La-motif protein superfamily. RNA 2009, 15, 750–764. [Google Scholar] [CrossRef]
- Bayfield, M.A.; Yang, R.; Maraia, R.J. Conserved and divergent features of the structure and function of La and La-related proteins (LARPs). Biochim. Biophys. Acta 2010, 1799, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Maraia, R.J.; Mattijssen, S.; Cruz-Gallardo, I.; Conte, M.R. The La and related RNA-binding proteins (LARPs): Structures, functions, and evolving perspectives. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Alfano, C.; Babon, J.; Kelly, G.; Curry, S.; Conte, M.R. Resonance assignment and secondary structure of an N-terminal fragment of the human La protein. J. Biomol. NMR 2003, 27, 93–94. [Google Scholar] [CrossRef] [PubMed]
- Alfano, C.; Sanfelice, D.; Babon, J.; Kelly, G.; Jacks, A.; Curry, S.; Conte, M.R. Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat. Struct. Mol. Biol. 2004, 11, 323–329. [Google Scholar] [CrossRef]
- Dong, G.; Chakshusmathi, G.; Wolin, S.L.; Reinisch, K.M. Structure of the La motif: A winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004, 23, 1000–1007. [Google Scholar] [CrossRef]
- Stefano, J.E. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 1984, 36, 145–154. [Google Scholar] [CrossRef]
- Wolin, S.L.; Cedervall, T. The La protein. Annu. Rev. Biochem. 2002, 71, 375–403. [Google Scholar] [CrossRef]
- Maraia, R.J.; Lamichhane, T.N. 3′ processing of eukaryotic precursor tRNAs. Wiley Interdiscip. Rev. RNA 2011, 2, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Muniz, L.; Egloff, S.; Kiss, T. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res. 2013, 41, 4686–4698. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, C.D.; Yang, Y.; Repeta, L.; Feigon, J. Structural basis for recognition of human 7SK long noncoding RNA by the La-related protein Larp7. Proc. Natl. Acad. Sci. USA 2018, 115, E6457–E6466. [Google Scholar] [CrossRef]
- Prathapam, R.; Witkin, K.L.; O’Connor, C.M.; Collins, K. A telomerase holoenzyme protein enhances telomerase RNA assembly with telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2005, 12, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Hasler, D.; Meduri, R.; Bak, M.; Lehmann, G.; Heizinger, L.; Wang, X.; Li, Z.T.; Sement, F.M.; Bruckmann, A.; Dock-Bregeon, A.C.; et al. The Alazami Syndrome-Associated Protein LARP7 Guides U6 Small Nuclear RNA Modification and Contributes to Splicing Robustness. Mol. Cell 2020, 77, 1014–1031 e1016. [Google Scholar] [CrossRef]
- Wang, X.; Li, Z.T.; Yan, Y.; Lin, P.; Tang, W.; Hasler, D.; Meduri, R.; Li, Y.; Hua, M.M.; Qi, H.T.; et al. LARP7-Mediated U6 snRNA Modification Ensures Splicing Fidelity and Spermatogenesis in Mice. Mol. Cell 2020, 77, 999–1013 e1016. [Google Scholar] [CrossRef]
- Markert, A.; Grimm, M.; Martinez, J.; Wiesner, J.; Meyerhans, A.; Meyuhas, O.; Sickmann, A.; Fischer, U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008, 9, 569–575. [Google Scholar] [CrossRef]
- Aigner, S.; Postberg, J.; Lipps, H.J.; Cech, T.R. The Euplotes La motif protein p43 has properties of a telomerase-specific subunit. Biochemistry 2003, 42, 5736–5747. [Google Scholar] [CrossRef]
- Stone, M.D.; Mihalusova, M.; O’Connor C, M.; Prathapam, R.; Collins, K.; Zhuang, X. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 2007, 446, 458–461. [Google Scholar] [CrossRef]
- Mattijssen, S.; Arimbasseri, A.G.; Iben, J.R.; Gaidamakov, S.; Lee, J.; Hafner, M.; Maraia, R.J. LARP4 mRNA codon-tRNA match contributes to LARP4 activity for ribosomal protein mRNA poly(A) tail length protection. Elife 2017, 6. [Google Scholar] [CrossRef]
- Fonseca, B.D.; Lahr, R.M.; Damgaard, C.K.; Alain, T.; Berman, A.J. LARP1 on TOP of ribosome production. Wiley Interdiscip. Rev. RNA 2018, e1480. [Google Scholar] [CrossRef]
- Zhang, Y.; Stefanovic, B. LARP6 Meets Collagen mRNA: Specific Regulation of Type I Collagen Expression. Int. J. Mol. Sci. 2016, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Kuspert, M.; Murakawa, Y.; Schaffler, K.; Vanselow, J.T.; Wolf, E.; Juranek, S.; Schlosser, A.; Landthaler, M.; Fischer, U. LARP4B is an AU-rich sequence associated factor that promotes mRNA accumulation and translation. RNA 2015, 21, 1294–1305. [Google Scholar] [CrossRef] [PubMed]
- Merret, R.; Martino, L.; Bousquet-Antonelli, C.; Fneich, S.; Descombin, J.; Billey, E.; Conte, M.R.; Deragon, J.M. The association of a La module with the PABP-interacting motif PAM2 is a recurrent evolutionary process that led to the neofunctionalization of La-related proteins. RNA 2013, 19, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, B.D.; Zakaria, C.; Jia, J.J.; Graber, T.E.; Svitkin, Y.; Tahmasebi, S.; Healy, D.; Hoang, H.D.; Jensen, J.M.; Diao, I.T.; et al. La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1). J. Biol. Chem. 2015, 290, 15996–16020. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Gallardo, I.; Martino, L.; Kelly, G.; Atkinson, R.A.; Trotta, R.; De Tito, S.; Coleman, P.; Ahdash, Z.; Gu, Y.; Bui, T.T.T.; et al. LARP4A recognizes polyA RNA via a novel binding mechanism mediated by disordered regions and involving the PAM2w motif, revealing interplay between PABP, LARP4A and mRNA. Nucleic Acids Res. 2019, 47, 4272–4291. [Google Scholar] [CrossRef]
- Schaffler, K.; Schulz, K.; Hirmer, A.; Wiesner, J.; Grimm, M.; Sickmann, A.; Fischer, U. A stimulatory role for the La-related protein 4B in translation. RNA 2010, 16, 1488–1499. [Google Scholar] [CrossRef]
- Dock-Bregeon, A.C.; Lewis, K.A.; Conte, M.R. The La-related proteins: Structures and interactions of a versatile superfamily of RNA-binding proteins. RNA Biol. 2019, 1–16. [Google Scholar] [CrossRef]
- Yang, R.; Gaidamakov, S.A.; Xie, J.; Lee, J.; Martino, L.; Kozlov, G.; Crawford, A.K.; Russo, A.N.; Conte, M.R.; Gehring, K.; et al. La-related protein 4 binds poly(A), interacts with the poly(A)-binding protein MLLE domain via a variant PAM2w motif, and can promote mRNA stability. Mol. Cell. Biol. 2011, 31, 542–556. [Google Scholar] [CrossRef]
- Kabsch, W. Xds. Acta Crystallogr D Biol. Crystallogr 2010, 66, 125–132. [Google Scholar] [CrossRef]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol. Crystallogr 1997, 53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Nakano, S.; Shinde, A.; Ito, H.; Ito, H.; Kusaka, H. Messenger RNA degradation may be inhibited in sporadic inclusion body myositis. Neurology 2005, 65, 420–425. [Google Scholar] [CrossRef]
- Otter, S.; Grimmler, M.; Neuenkirchen, N.; Chari, A.; Sickmann, A.; Fischer, U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007, 282, 5825–5833. [Google Scholar] [CrossRef]
- Thoma, C.; Ostareck-Lederer, A.; Hentze, M.W. A poly(A) tail-responsive in vitro system for cap- or IRES-driven translation from HeLa cells. Methods Mol. Biol. 2004, 257, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Plottner, O.; Kroiss, M.; Hartmann, E.; Laggerbauer, B.; Meister, G.; Keidel, E.; Fischer, U. Tdrd3 is a novel stress granule-associated protein interacting with the Fragile-X syndrome protein FMRP. Hum. Mol. Genet. 2008, 17, 3236–3246. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. Stress granules: The Tao of RNA triage. Trends Biochem. Sci 2008, 33, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Arimoto, K.; Fukuda, H.; Imajoh-Ohmi, S.; Saito, H.; Takekawa, M. Formation of stress granules inhibits apoptosis by suppressing stress-responsive MAPK pathways. Nat. Cell Biol. 2008, 10, 1324–1332. [Google Scholar] [CrossRef] [PubMed]
- Deragon, J.M. Distribution, organization an evolutionary history of La and LARPs in eukaryotes. RNA Biol. 2020, 1–9. [Google Scholar] [CrossRef]

| Data Collection | |
|---|---|
| Space group | P 21 21 21 |
| Cell dimensions a, b, c [Å] | 29.15, 57.19, 59.51 |
| Resolution [Å] (outer shell) | 41.2–1.7 (1.8–1.7) |
| I/sigma(I) (outer shell) | 14.4 (2.4) |
| Completeness [%] (outer shell) | 96.9 (86.8) |
| Redundancy (outer shell) | 3.2 (2.4) |
| Wilson B-factor [Å2] | 26.1 |
| Refinement | |
| Resolution [Å] | 41.2–1.7 |
| No. of reflections | 10805 |
| Rwork/ Rfree | 17.2/21.8 |
| No. atoms | 769 |
| Water molecules | 82 |
| B-factors [Å2] | |
| overall | 22.2 |
| MLLE (Chain A) | 19.2 |
| Peptide (chain B) | 24.0 |
| Solvent (chain Z) | 40.3 |
| RMS (root-mean-square deviation) deviations from ideal | |
| Bond lengths [Å] | 0.017 |
| Bond angles (°) | 1.6 |
| Ramachandran statistics, residues in [%] | |
| Most favoured regions | 96.9 |
| Additionally allowed regions | 3.1 |
| Generously allowed regions | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Grimm, C.; Pelz, J.-P.; Schneider, C.; Schäffler, K.; Fischer, U. Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1. Biomolecules 2020, 10, 872. https://doi.org/10.3390/biom10060872
Grimm C, Pelz J-P, Schneider C, Schäffler K, Fischer U. Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1. Biomolecules. 2020; 10(6):872. https://doi.org/10.3390/biom10060872
Chicago/Turabian StyleGrimm, Clemens, Jann-Patrick Pelz, Cornelius Schneider, Katrin Schäffler, and Utz Fischer. 2020. "Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1" Biomolecules 10, no. 6: 872. https://doi.org/10.3390/biom10060872
APA StyleGrimm, C., Pelz, J.-P., Schneider, C., Schäffler, K., & Fischer, U. (2020). Crystal Structure of a Variant PAM2 Motif of LARP4B Bound to the MLLE Domain of PABPC1. Biomolecules, 10(6), 872. https://doi.org/10.3390/biom10060872




