Rhizophagus irregularis and Rhizoctonia solani Differentially Elicit Systemic Transcriptional Expression of Polyphenol Biosynthetic Pathways Genes in Sunflower
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolates and Sunflower Cultivar
2.2. The greenhouse Experiment
Time-Course Analysis of Gene Expression Changes
RNA Extraction and cDNA Synthesis
Quantitative Real-Time PCR (qRT-PCR)
2.3. Growth Parameters Evaluation
2.4. Disease Assessment
2.5. Estimation of Mycorrhizal Colonization
2.6. Biochemical Estimations
2.7. Statistical Analyses
3. Results
3.1. Time-Course Changes in the Systemic Transcript Levels of the Polyphenol Biosynthesis-Related Genes
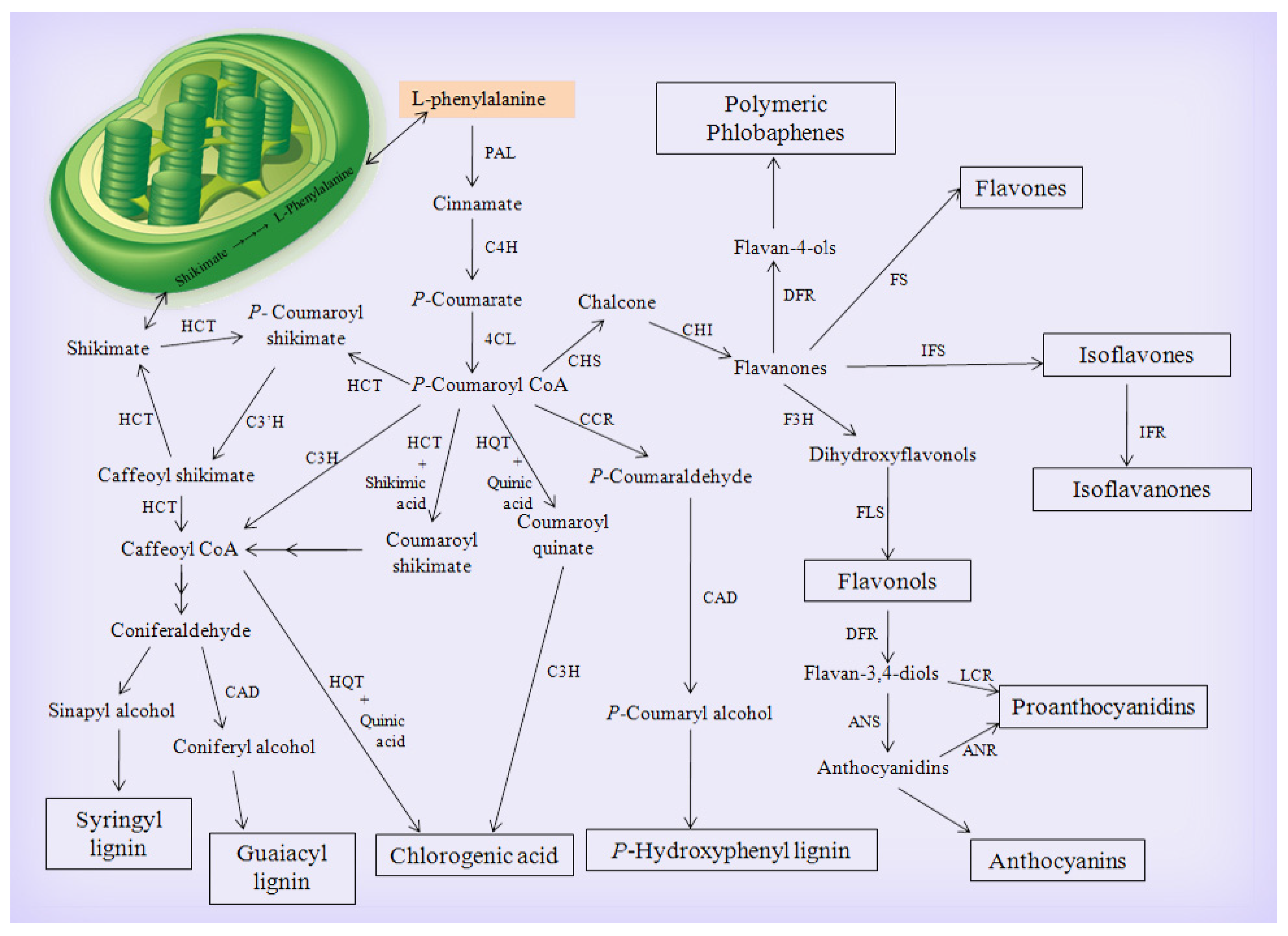
3.1.1. The Main Phenylpropanoid Biosynthetic Pathway
3.1.2. The Flavonoid Biosynthetic Pathway
3.1.3. The Chlorogenic Acid Biosynthetic Pathway
3.2. Hierarchical Clustering Analysis
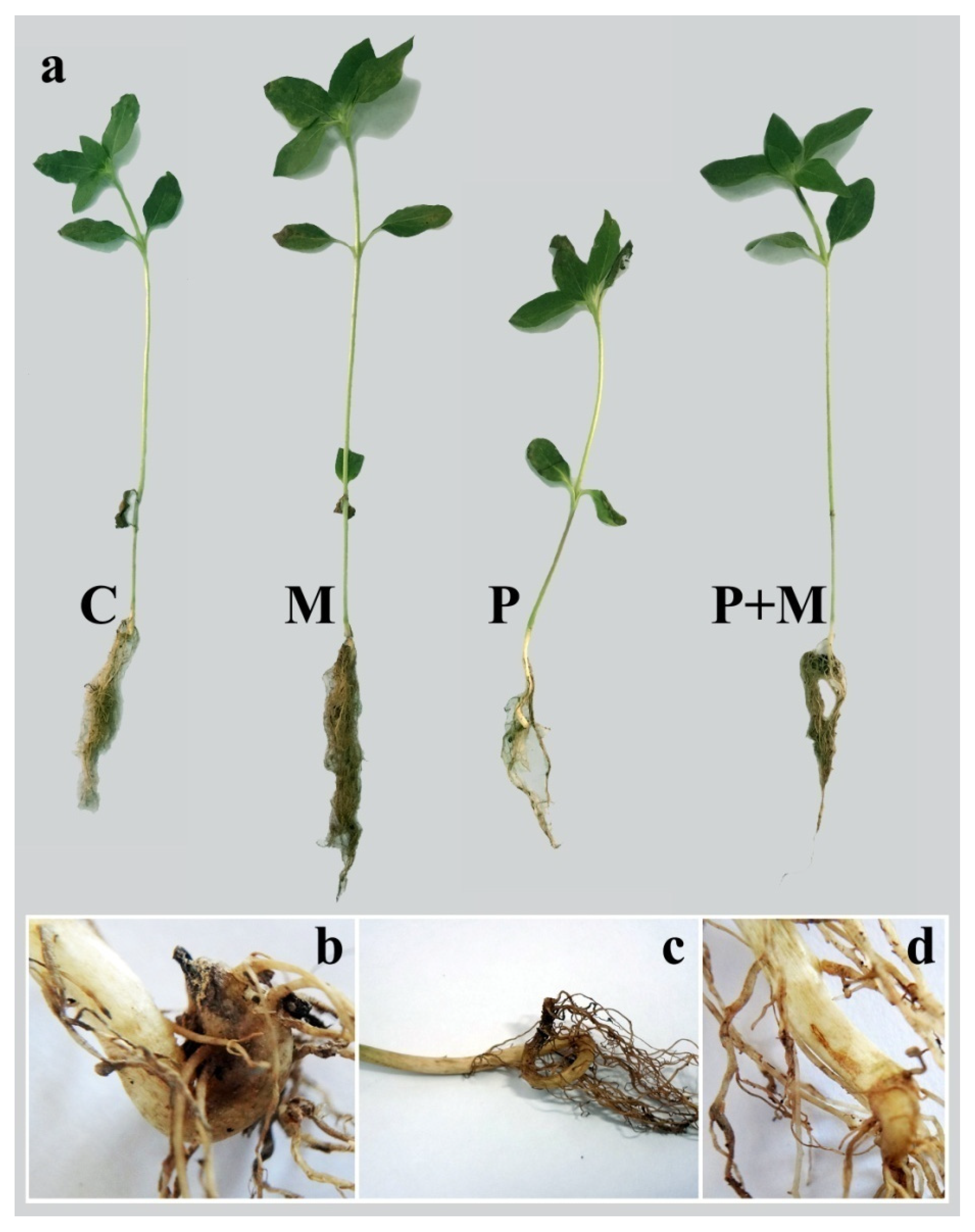
3.3. Plant Growth Evaluation
3.4. Disease Assessment
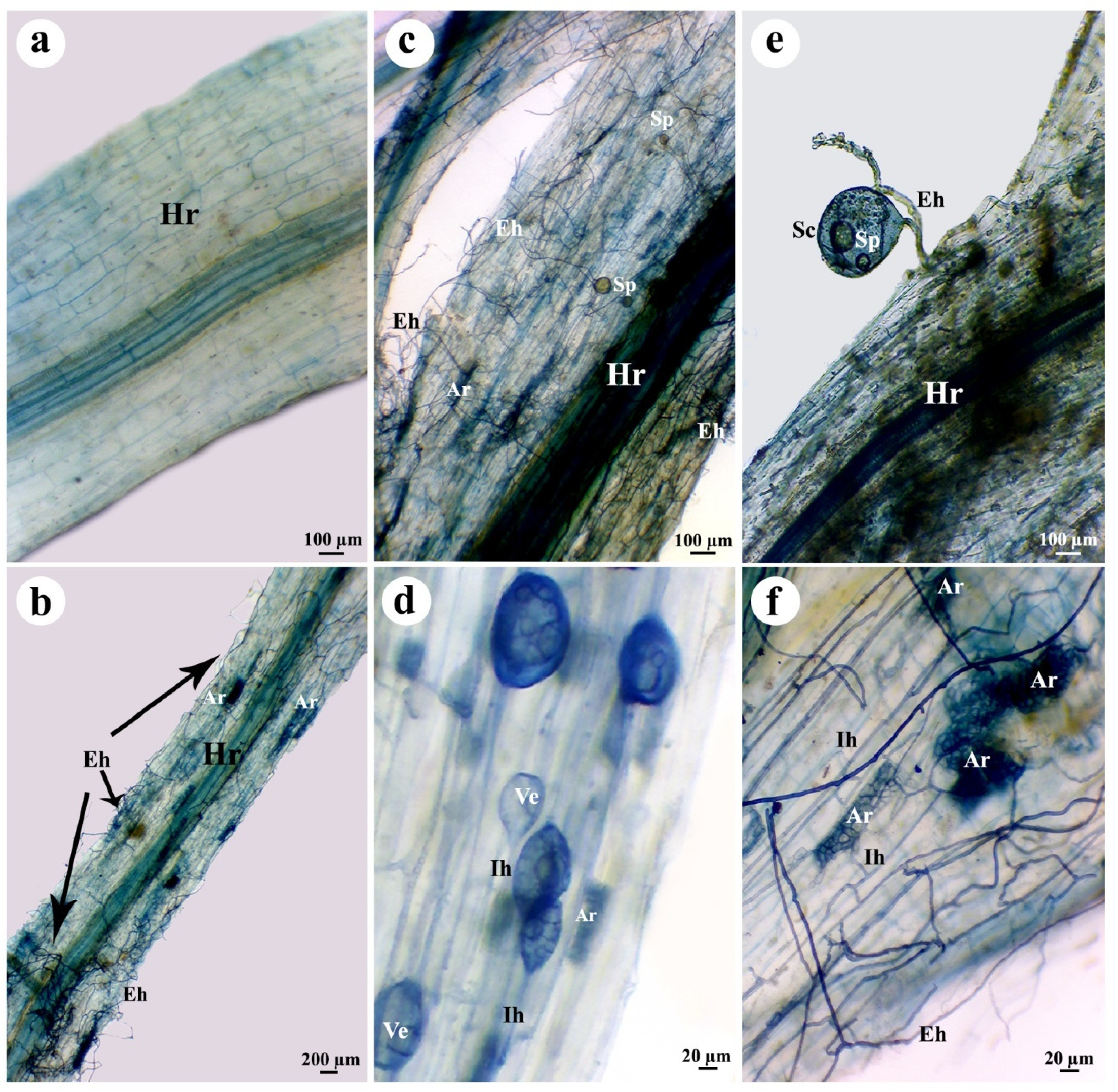
3.5. Estimation of Colonization Level
3.6. Total Phenol Content and Activities of Defense-Related Enzymes
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- FAOSTAT © FAO. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 29 February 2020).
- Alsahli, A.A.; Alaraidh, I.A.; Rashad, Y.M.; Abdel Razik, E.S. Extract from Curcuma longa L. triggers the sunflower immune system and induces defence-related genes against Fusarium root rot. Phytopathol. Mediterr. 2018, 57, 26–36. [Google Scholar]
- Gulya, T.J.; Mathew, F.; Harveson, R.; Markell, S.; Block, C. Diseases of sunflower. In Handbook of Florists’ Crops Diseases; McGovern, R.J., Elmer, W.H., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 787–837. [Google Scholar]
- Lakshmidevi, N.; Sudisha, J.; Mahadevamurthy, S.; Prakash, H.S.; Shetty, H.S. First report of the seed-borne nature of root and collar rot disease caused by Rhizoctonia solani in sunflower from India. Australas. Plant. Dis. Notes 2010, 5, 11. [Google Scholar] [CrossRef] [Green Version]
- Al-Askar, A.-A.A.; Ghoneem, K.M.; Rashad, Y.M. Seed-borne mycoflora of alfalfa (Medicago sativa L.) in the Riyadh Region of Saudi Arabia. Ann. Microbiol. 2012, 62, 273–281. [Google Scholar] [CrossRef]
- Srinivasan, K.; Visalakchi, S. First report of Rhizoctonia solani causing a disease of sunflower in India. Plant. Dis. 2010, 94, 488. [Google Scholar] [CrossRef] [PubMed]
- Tu, C.-C.; Hsieh, T.-F.; Chang, Y.-C. Vegetable diseases incited by Rhizoctonia spp. In Rhizoctonia species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control; Sneh, B., Jabaji-Hare, S., Neate, S., Dijst, G., Eds.; Springer: Dordrecht, The Netherlands, 1996; pp. 369–377. [Google Scholar]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mewalal, R.; Yin, H.; Hu, R.; Jawdy, S.; Vion, P.; Tuskan, G.A.; Le Tacon, F.; Labbé, J.L.; Yang, X. Identification of populus small RNAs responsive to mutualistic interactions with mycorrhizal fungi, Laccaria bicolor and Rhizophagus irregularis. Front. Microbiol. 2019, 10, 515. [Google Scholar] [CrossRef] [Green Version]
- Calonne-Salmon, M.; Plouznikoff, K.; Declerck, S. The arbuscular mycorrhizal fungus Rhizophagus irregularis MUCL 41833 increases the phosphorus uptake and biomass of Medicago truncatula, a benzo[a]pyrene-tolerant plant species. Mycorrhiza 2018, 28, 761–771. [Google Scholar] [CrossRef]
- Calvo-Polanco, M.; Molina, S.; Zamarreño, A.M.; García-Mina, J.M.; Aroca, R. The symbiosis with the arbuscular mycorrhizal fungus Rhizophagus irregularis drives root water transport in flooded tomato plants. Plant. Cell Physiol. 2014, 55, 1017–1029. [Google Scholar] [CrossRef]
- Bui, V.C.; Franken, P. Acclimatization of Rhizophagus irregularis enhances Zn tolerance of the fungus and the mycorrhizal plant partner. Front. Microbiol. 2018, 9, 3156. [Google Scholar] [CrossRef] [Green Version]
- Oye Anda, C.C.; Dupré de Boulois, H.; Declerck, S. The arbuscular mycorrhiza fungus Rhizophagus irregularis MUCL 41833 decreases disease severity of Black Sigatoka on banana c.v. Grande naine, under in vitro culture conditions. Fruits 2015, 70, 37–46. [Google Scholar] [CrossRef] [Green Version]
- El-Sharkawy, H.H.A.; Rashad, Y.M.; Ibrahim, S.A. Biocontrol of stem rust disease of wheat using arbuscular mycorrhizal fungi and Trichoderma spp. Physiol. Mol. Plant. Pathol. 2018, 103, 84–91. [Google Scholar] [CrossRef]
- Abdel-Fattah, G.M.; El-Haddad, S.A.; Hafez, E.E.; Rashad, Y.M. Induction of defense responses in common bean plants by arbuscular mycorrhizal fungi. Microbiol. Res. 2011, 166, 268–281. [Google Scholar] [CrossRef] [PubMed]
- Kanwal, Q.; Hussain, I.; Latif Siddiqui, H.; Javaid, A. Antifungal activity of flavonoids isolated from mango (Mangifera indica L.) leaves. Nat. Prod. Res. 2010, 24, 1907–1914. [Google Scholar] [CrossRef] [PubMed]
- Martínez, G.; Regente, M.; Jacobi, S.; Del Rio, M.; Pinedo, M.; de la Canal, L. Chlorogenic acid is a fungicide active against phytopathogenic fungi. Pestic. Biochem. Physiol. 2017, 140, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Cheevarungnapakul, K.; Khaksar, G.; Panpetch, P.; Boonjing, P.; Sirikantaramas, S. Identification and functional characterization of genes involved in the biosynthesis of caffeoylquinic acids in sunflower (Helianthus annuus L.). Front. Plant. Sci. 2019, 10, 968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- André, C.M.; Schafleitner, R.; Legay, S.; Lefèvre, I.; Aliaga, C.A.A.; Nomberto, G.; Hoffmann, L.; Hausman, J.-F.; Larondelle, Y.; Evers, D. Gene expression changes related to the production of phenolic compounds in potato tubers grown under drought stress. Phytochemistry 2009, 70, 1107–1116. [Google Scholar] [CrossRef]
- Mahesh, V.; Million-Rousseau, R.; Ullmann, P.; Chabrillange, N.; Bustamante, J.; Mondolot, L.; Morant, M.; Noirot, M.; Hamon, S.; de Kochko, A.; et al. Functional characterization of two p-coumaroyl ester 3′-hydroxylase genes from coffee tree: Evidence of a candidate for chlorogenic acid biosynthesis. Plant. Mol. Biol. 2007, 64, 145–159. [Google Scholar] [CrossRef]
- Albert, N.W.; Lewis, D.H.; Zhang, H.; Irving, L.J.; Jameson, P.E.; Davies, K.M. Light-induced vegetative anthocyanin pigmentation in Petunia. J. Exp. Bot. 2009, 60, 2191–2202. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Carling, D.E.; Pope, E.J.; Brainard, K.A.; Carter, D.A. Characterization of mycorrhizal isolates of Rhizoctonia solani from an orchid, including AG-12, a new anastomosis group. Phytopathology 1999, 89, 942–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, J.M.; Hayman, D.S. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 1970, 55, 158-IN18. [Google Scholar] [CrossRef]
- Trouvelot, A.; Kough, J.L.; Gianinazzi-Pearson, V. Estimation of vesicular arbuscular mycorrhizal infection levels. Research for methods having a functional significance. In Physiological and Genetical Aspects of Mycorrhizae; Gianinazzi-Pearson, V., Gianinazzi, S., Eds.; INRA Publ: Paris, France, 1986; pp. 217–221. [Google Scholar]
- Malik, C.P.; Singh, M.B. Plant Enzymology and Histo-enzymology; Kalyani Publishers: Delhi, India, 1980; p. 286. [Google Scholar]
- Galeazzi, M.A.M.; Sgarbieri, V.C.; Constantinides, S.M. Isolation, Purification and physicochemical characterization of polyphenoloxidases (PPO) from a dwarf variety of banana (Musa cavendishii L). J. Food Sci. 1981, 46, 150–155. [Google Scholar] [CrossRef]
- Maxwell, D.P.; Bateman, D.F. Changes in the activities of some oxidases in extracts of Rhizoctonia-infected bean hypocotyls in relation to lesion maturation. Phytopathol. 1967, 57, 132–136. [Google Scholar]
- Cohort Software. Available online: https://www.cohort.com/costat.html (accessed on 29 February 2020).
- Tanase, C.; Coșarcă, S.; Muntean, D.-L. A critical review of phenolic compounds extracted from the bark of woody vascular plants and their potential biological activity. Molecules 2019, 24, 1182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulbat, K. The role of phenolic compounds in plant resistance. Biotechnol. Food Sci. 2016, 80, 97–108. [Google Scholar]
- Tao, S.; Wang, D.; Jin, C.; Sun, W.; Liu, X.; Zhang, S.; Gao, F.; Khanizadeh, S. Cinnamate-4-hydroxylase gene is involved in the step of lignin biosynthesis in chinese white pear. J. Am. Soc. Hortic. Sci. 2015, 140, 573–579. [Google Scholar] [CrossRef] [Green Version]
- Yan, Q.; Si, J.; Cui, X.; Peng, H.; Chen, X.; Xing, H.; Dou, D. The soybean cinnamate 4-hydroxylase gene GmC4H1 contributes positively to plant defense via increasing lignin content. Plant. Growth Regul. 2019, 88, 139–149. [Google Scholar] [CrossRef]
- Fraser, C.M.; Chapple, C. The phenylpropanoid pathway in arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.P.; Matthews, M.L.; Williams, C.M.; Shi, R.; Yang, C.; Tunlaya-Anukit, S.; Chen, H.-C.; Li, Q.; Liu, J.; Lin, C.-Y.; et al. Improving wood properties for wood utilization through multi-omics integration in lignin biosynthesis. Nat. Commun. 2018, 9, 1579. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinya, T.; Iwata, E.; Nakahama, K.; Fukuda, Y.; Hayashi, K.; Nanto, K.; Rosa, A.C.; Kawaoka, A. Transcriptional profiles of hybrid eucalyptus genotypes with contrasting lignin content reveal that monolignol biosynthesis-related genes regulate wood composition. Front. Plant. Sci. 2016, 7, 443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Zhu, L.; Tu, L.; Liu, L.; Yuan, D.; Jin, L.; Long, L.; Zhang, X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011, 62, 5607–5621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eynck, C.; Séguin-Swartz, G.; Clarke, W.E.; Parkin, I.A.P. Monolignol biosynthesis is associated with resistance to Sclerotinia sclerotiorum in Camelina sativa: Monolignol biosynthesis and stem rot resistance. Mol. Plant. Pathol. 2012, 13, 887–899. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant. Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, P.; Ning, G.; Wang, Z.; Shen, Y.; Jin, H.; Li, P.; Huang, S.; Zhao, J.; Bao, M. Disequilibrium of flavonol synthase and dihydroflavonol-4-reductase expression associated tightly to white vs. red color flower formation in plants. Front. Plant. Sci. 2016, 6, 1257. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Bai, Y.; Li, S.; Chen, H.; Han, X.; Zhao, H.; Shao, J.; Park, S.; Wu, Q. Cloning, characterization, and activity analysis of a flavonol synthase gene FtFLS1 and its association with flavonoid content in Tartary buckwheat. J. Agric. Food Chem. 2012, 60, 5161–5168. [Google Scholar] [CrossRef]
- Tian, J.; Chen, M.; Zhang, J.; Li, K.; Song, T.; Zhang, X.; Yao, Y. Characteristics of dihydroflavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hortic. Res. 2017, 4, 17070. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, B.M.; Geisler, M.; Bigler, L.; Ringli, C. Flavonols accumulate asymmetrically and affect auxin transport in arabidopsis. Plant. Physiol. 2011, 156, 585–595. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin biosynthesis and degradation mechanisms in solanaceous vegetables: A review. Front. Chem. 2018, 6, 52. [Google Scholar] [CrossRef]
- Quattrocchio, F.; Wing, J.F.; Leppen, H.; Mol, J.; Koes, R.E. Regulatory genes controlling anthocyanin pigmentation are functionally conserved among plant species and have distinct sets of target genes. Plant. Cell 1993, 5, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, H.M.; Rentzsch, M.; Breuer, M. Anthocyanins reduce fungal growth in fruits. Nat. Prod. Commun. 2008, 3, 1267–1272. [Google Scholar] [CrossRef] [Green Version]
- Lev-Yadun, S.; Gould, K.S. Role of anthocyanins in plant defence. In Anthocyanins; Winefield, C., Davies, K., Gould, K., Eds.; Springer: New York, NY, USA, 2008; pp. 22–28. [Google Scholar]
- Wen, H.; Kang, J.; Li, D.; Wen, W.; Yang, F.; Hu, H.; Liu, C. Antifungal activities of anthocyanins from purple sweet potato in the presence of food preservatives. Food Sci. Biotechnol. 2016, 25, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Tellez, P.; Rojas, E.; Van Bael, S. Red coloration in young tropical leaves associated with reduced fungal pathogen damage. Biotropica 2016, 48, 150–153. [Google Scholar] [CrossRef]
- Bovy, A.; Schijlen, E.; Hall, R.D. Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): The potential for metabolomics. Metabolomics 2007, 3, 399–412. [Google Scholar] [CrossRef] [Green Version]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef] [Green Version]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant. Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef]
- Piasecka, A.; Jedrzejczak-Rey, N.; Bednarek, P. Secondary metabolites in plant innate immunity: Conserved function of divergent chemicals. New Phytol. 2015, 206, 948–964. [Google Scholar] [CrossRef]
- Gianinazzi-Pearson, V.; Dumas-Gaudot, E.; Gollotte, A.; Alaoui, A.T.; Gianinazzi, S. Cellular and molecular defence-related root responses to invasion by arbuscular mycorrhizal fungi. New Phytol. 1996, 133, 45–57. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, M.; Li, W.; Bai, G. Regulation of chlorogenic acid biosynthesis by hydroxycinnamoyl CoA quinate hydroxycinnamoyl transferase in Lonicera japonica. Plant. Physiol. Biochem. 2017, 121, 74–79. [Google Scholar] [CrossRef]
- Payyavula, R.S.; Shakya, R.; Sengoda, V.G.; Munyaneza, J.E.; Swamy, P.; Navarre, D.A. Synthesis and regulation of chlorogenic acid in potato: Rerouting phenylpropanoid flux in HQT -silenced lines. Plant. Biotechnol. J. 2015, 13, 551–564. [Google Scholar] [CrossRef] [PubMed]
- Hammerschmidt, R. Chlorogenic acid: A versatile defense compound. Physiol. Mol. Plant. Pathol. 2014, 88, iii–iv. [Google Scholar] [CrossRef]
- Lee, M.-H.; Bostock, R.M. Fruit exocarp phenols in relation to quiescence and development of Monilinia fructicola infections in Prunus spp.: A role for cellular redox? Phytopathol. 2007, 97, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Vangelisti, A.; Natali, L.; Bernardi, R.; Sbrana, C.; Turrini, A.; Hassani-Pak, K.; Hughes, D.; Cavallini, A.; Giovannetti, M.; Giordani, T. Transcriptome changes induced by arbuscular mycorrhizal fungi in sunflower (Helianthus annuus L.) roots. Sci. Rep. 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, S.C.; Martinez-Medina, A.; Lopez-Raez, J.A.; Pozo, M.J. Mycorrhiza-induced resistance and priming of plant defenses. J. Chem. Ecol. 2012, 38, 651–664. [Google Scholar] [CrossRef]
- Rivero, J.; Gamir, J.; Aroca, R.; Pozo, M.J.; Flors, V. Metabolic transition in mycorrhizal tomato roots. Front. Microbiol. 2015, 6, 598. [Google Scholar] [CrossRef]
- Okada, K.; Abe, H.; Arimura, G. Jasmonates induce both defense responses and communication in monocotyledonous and dicotyledonous plants. Plant. Cell Physiol. 2015, 56, 16–27. [Google Scholar] [CrossRef]
- Fernández, I.; Merlos, M.; López-Ráez, J.A.; Martínez-Medina, A.; Ferrol, N.; Azcón, C.; Bonfante, P.; Flors, V.; Pozo, M.J. Defense related phytohormones regulation in arbuscular mycorrhizal symbioses depends on the partner genotypes. J. Chem. Ecol. 2014, 40, 791–803. [Google Scholar] [CrossRef]
- Reynolds, G.J.; Gordon, T.R.; McRoberts, N. Using game theory to understand systemic acquired resistance as a bet-hedging option for increasing fitness when disease is uncertain. Plants 2019, 8, 219. [Google Scholar] [CrossRef] [Green Version]
- Aseel, D.G.; Rashad, Y.M.; Hammad, S.M. Arbuscular mycorrhizal fungi trigger transcriptional expression of flavonoid and chlorogenic acid biosynthetic pathways genes in tomato against Tomato Mosaic Virus. Sci. Rep. 2019, 9, 9692. [Google Scholar] [CrossRef] [Green Version]
- Bowles, T.M.; Jackson, L.E.; Cavagnaro, T.R. Mycorrhizal fungi enhance plant nutrient acquisition and modulate nitrogen loss with variable water regimes. Glob. Chang. Biol. 2018, 24, e171–e182. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Ezawa, T.; Cheng, W.; Tawaraya, K. Release of acid phosphatase from extraradical hyphae of arbuscular mycorrhizal fungus Rhizophagus clarus. Soil Sci. Plant. Nutr. 2015, 61, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Parvizi, K.; Dashti, F. The effect of in vitro mycorrhization on growth characteristics, changes in endogenous hormones and performance of microplants in potato (Solanum tuberosum L.). J. Cent. Eur. Agric. 2015, 16, 445–462. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are not available from the authors. |




| Primer Name | Abbreviation | Sequence (5’-3’) | |
|---|---|---|---|
| β-actine | β-actine | F | GTGGGCCGCTCTAGGCACCAA |
| R | CTCTTTGATGTCACGCACGATTTC | ||
| Phenylalanine ammonia-lyase 1 | PAL1 | F | ACGGGTTGCCATCTAATCTGACA |
| R | CGAGCAATAAGAAGCCATCGCAAT | ||
| Cinnamic acid 4-hydroxylase | C4H | F | CCCAGTTTTTGGAAATTGGCTTCA |
| R | GCCCCATTCTAAGCAAGAGAACATC | ||
| Chalcone synthase | CHS | F | CACCGTGGAGGAGTATCGTAAGGC |
| R | TGATCAACACAGTTGGAAGGCG | ||
| Chalcone isomerase 2 | CHI2 | F | GGCAGGCCATTGAAAAGTTCC |
| R | CTAATCGTCAATGATCCAAGCGG | ||
| Flavanone 3-hydroxylase | F3H | F | CCAAGGCATGTGTGGATATGGACC |
| R | CCTGGATCAGTATGTCGTTCAGCC | ||
| Flavonol synthase 1 | FLS1 | F | CCTCCTTCCTACAGGGAAGCAAA |
| R | CAAGCCCAAGTGACAAGCTCCTAA | ||
| Dihydroflavonol 4-reductase | DFR | F | TCACAGGAGCAGCTGGATTTATCG |
| R | TCAGGATCACGAACAGTAGCATGG | ||
| Flavonoid 3′ hydroxylase | F3′H | F | TGGGTATACCCAAACTCATTCCG |
| R | AAAAGCCCAAAGTTGATGTGAAAGG | ||
| Anthocyanin 1 transcription factor | AN1 | F | CCTCAACCTCAGAAATTCAGAAGC |
| R | TCGTTGTTGTTGTCGTTCGATGC | ||
| Anthocyanin 2 transcription factor | AN2 | F | ACAAGATGCCACTTTCCTTCACC |
| R | TGTGCATCGTTGGGAGTTAGG | ||
| Hydroxycinnamoyl Co A shikimate hydroxycinnamoyl transferase | HCT | F | TCTCCAACCCCTTTTAACGAACC |
| R | CAACTTGTCCTTCTACCACAGGGAA | ||
| Hydroxycinnamoyl Co A quinate hydroxycinnamoyl transferase | HQT | F | CCCAATGGCTGGAAGATTAGCTA |
| R | CATGAATCACTTTCAGCCTCAACAA | ||
| p-coumarate 3-hydroxylase | C3H | F | TTGGTGGCTACGACATTCCTAAGG |
| R | GGTCTGAACTCCAATGGGTTATTCC |
| Treatment * | Shoot Length (cm) | Root Length (cm) | Shoot Dry Weight (g) | Root Dry Weight (g) | No. of Leaves | Leaf Area (cm2) |
|---|---|---|---|---|---|---|
| C | 26.3 ± 0.6 b | 16.7 ± 0.5 b | 0.42 ± 0.03 b | 0.26 ± 0.02 b | 7.0 ± 0.2 a | 12.81 ± 0.3 b |
| M | 33.0 ± 1.1 a | 19.3 ± 0.7 a | 0.52 ± 0.09 a | 0.34 ± 0.06 a | 7.6 ± 0.4 a | 17.07 ± 0.2 a |
| P | 21.7 ± 1.0 c | 11.3 ± 0.9 c | 0.32 ± 0.02 c | 0.15 ± 0.09 c | 6.0 ± 0.3 a | 9.24 ± 0.2 c |
| M+P | 35.3 ± 1.0 a | 18.7 ± 0.3 a | 0.55 ± 0.05 a | 0.31 ± 0.08 a | 7.6 ± 0.5 a | 16.93 ± 0.1 a |
| Treatment * | Disease Incidence (%) | Disease Severity (%) ** |
|---|---|---|
| C | 0 c | 0 c |
| M | 0 c | 0 c |
| P | 96.7 ± 3.7 a | 58.3 ± 2.8 a |
| M+P | 53.3 ± 3.5 b | 28.3 ± 2.7 b |
| Treatment * | F (%) | I (%) | A (%) |
|---|---|---|---|
| C | 0 c | 0 c | 0 c |
| M | 96.7 ± 1.1 a | 45.25 ± 1.0 a | 23.4 ± 0.9 a |
| P | 0 c | 0 c | 0 c |
| M+P | 90.5 ± 0.9 b | 14.33 ± 0.8 b | 7.5 ± 1.0 b |
| Treatment * | Total Phenol Content (mg g−1 Fresh Weight) | PPO (U mL−1 min−1) | POD (U mL−1 min−1) |
|---|---|---|---|
| C | 1.813 ± 0.095 d | 0.213 ± 0.033 d | 0.153 ± 0.086 c |
| M | 2.129 ± 0.046 c | 0.334 ± 0.045 c | 0.210 ± 0.061 b |
| P | 2.499 ± 0.064 b | 0.455 ± 0.015 b | 0.221 ± 0.048 b |
| M+P | 2.873 ± 0.191 a | 0.554 ± 0.012 a | 0.307 ± 0.027 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashad, Y.; Aseel, D.; Hammad, S.; Elkelish, A. Rhizophagus irregularis and Rhizoctonia solani Differentially Elicit Systemic Transcriptional Expression of Polyphenol Biosynthetic Pathways Genes in Sunflower. Biomolecules 2020, 10, 379. https://doi.org/10.3390/biom10030379
Rashad Y, Aseel D, Hammad S, Elkelish A. Rhizophagus irregularis and Rhizoctonia solani Differentially Elicit Systemic Transcriptional Expression of Polyphenol Biosynthetic Pathways Genes in Sunflower. Biomolecules. 2020; 10(3):379. https://doi.org/10.3390/biom10030379
Chicago/Turabian StyleRashad, Younes, Dalia Aseel, Saad Hammad, and Amr Elkelish. 2020. "Rhizophagus irregularis and Rhizoctonia solani Differentially Elicit Systemic Transcriptional Expression of Polyphenol Biosynthetic Pathways Genes in Sunflower" Biomolecules 10, no. 3: 379. https://doi.org/10.3390/biom10030379
APA StyleRashad, Y., Aseel, D., Hammad, S., & Elkelish, A. (2020). Rhizophagus irregularis and Rhizoctonia solani Differentially Elicit Systemic Transcriptional Expression of Polyphenol Biosynthetic Pathways Genes in Sunflower. Biomolecules, 10(3), 379. https://doi.org/10.3390/biom10030379







