Targeted Isolation of Lignans from Trachelospermum asiaticum Using Molecular Networking and Hierarchical Clustering Analysis
Abstract
1. Introduction
2. Results and Discussion
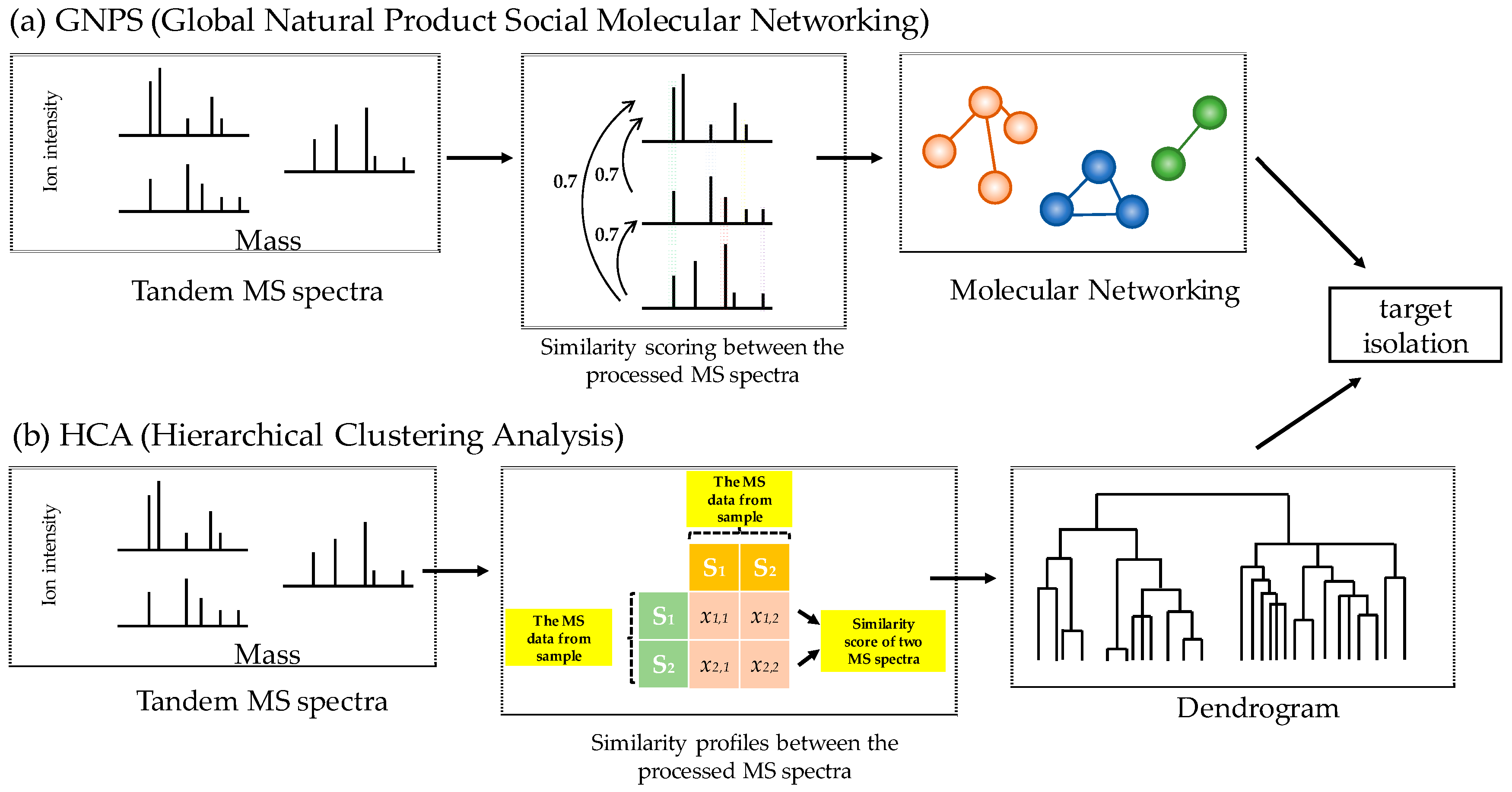
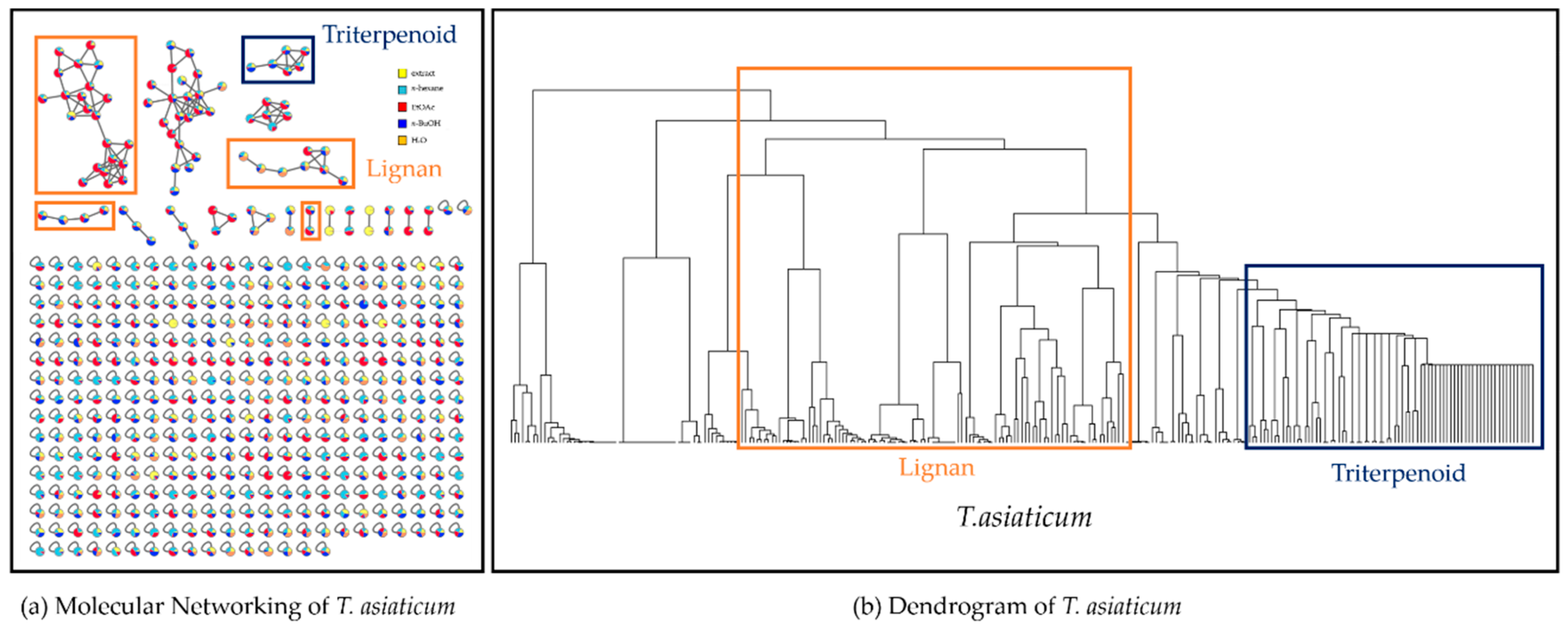
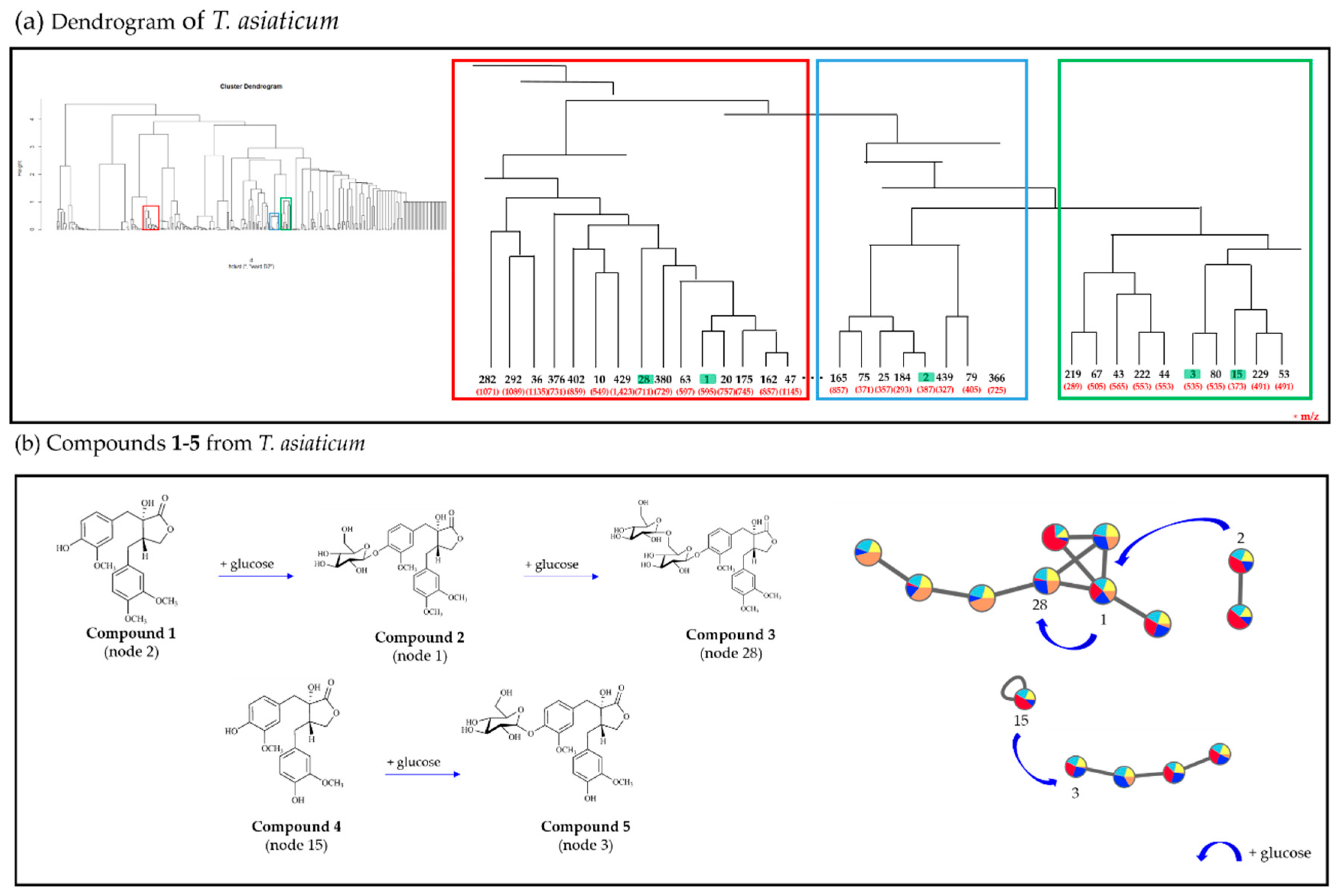
2.1. Molecular Networking and Hierarchical Clustering Analysis of Mass Spectral Data from T. asiaticum
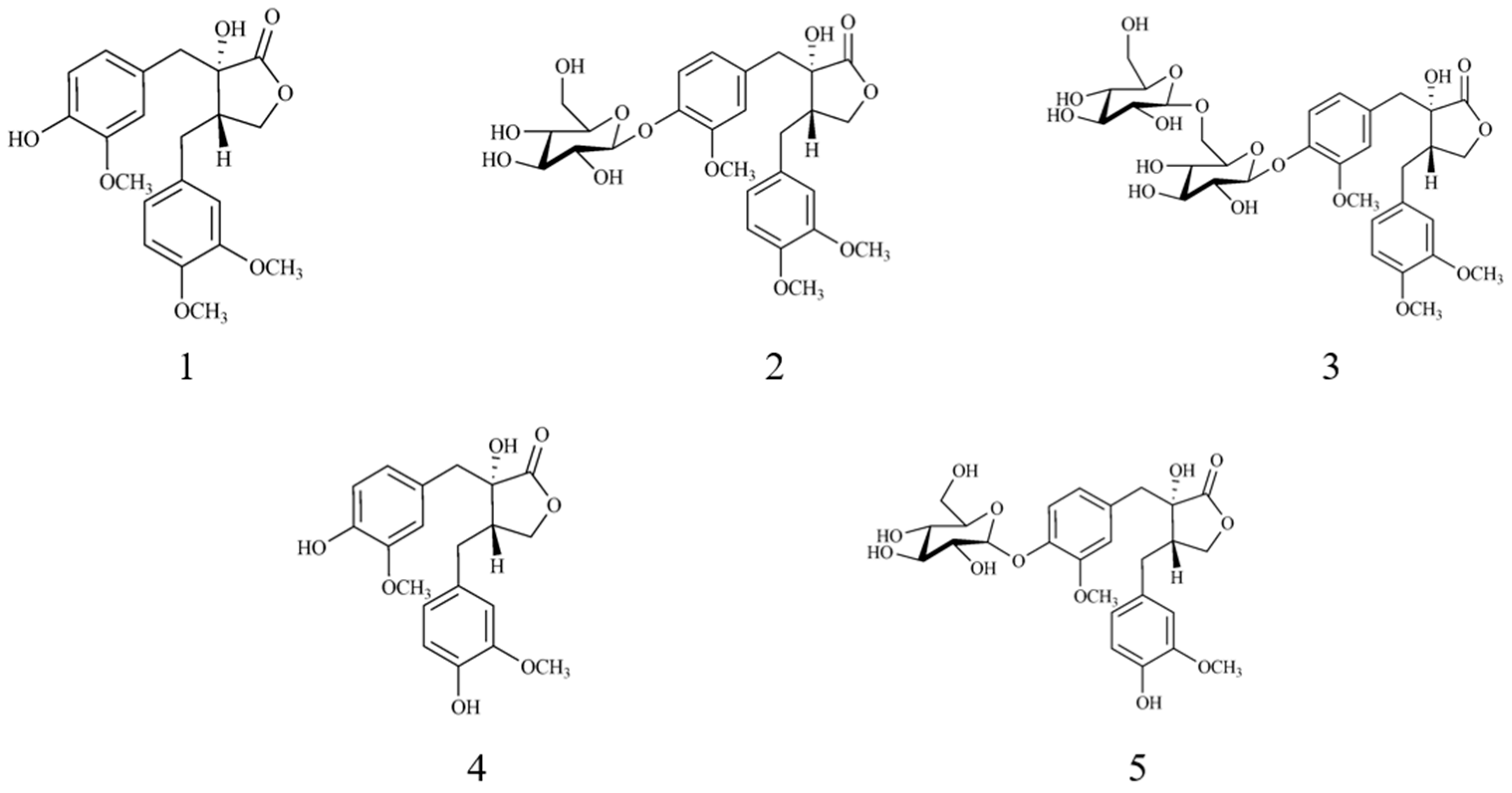
2.2. Targeted Isolation of Compounds 1–5 Using Molecular Networking and HCA Results
2.3. Cytotoxic Activity of Isolated Compounds 1–5
3. Materials and Methods
3.1. Plant Material
3.2. Apparatus and Reagents
3.3. Extraction and Isolation
3.3.1. Trachelogenin (1)
3.3.2. Tracheloside (2)
3.3.3. Trachelogenin β-Gentionbioside (3)
3.3.4. Nortrachelogenin (4)
3.3.5. Nortracheloside (5)
3.4. GNPS Analysis
3.5. Dendrogram Analysis
3.6. Cytotoxicity Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Abe, F.; Yamauchi, T. Glycosides of 19α-hydroxyoleanane-type triterpenoids from Trachelospermum asiaticum (Trachelospermum. IV). Chem. Pharm. Bull. 1987, 35, 1833–1838. [Google Scholar] [CrossRef]
- Nishibe, S.; Hisada, S.; Inagaki, I. The ether-soluble lignans of Trachelospermum asiaticum var. intermedium. Phytochemistry 1971, 10, 2231–2232. [Google Scholar] [CrossRef]
- Zhang, J.; Yin, Z.; Liang, J. A new isoflavonoid glycoside from the aerial parts of Trachelospermum jasminoides. Chin. J. Nat. Med. 2013, 11, 274–276. [Google Scholar] [CrossRef] [PubMed]
- Devi, N. Indian tribe’s and villager’s health and habits: Popularity of apocynaceae plants as medicine. Int. J. Green Pharm. 2017, 11, 256–259. [Google Scholar]
- Bouslimani, A.; Sanchez, L.M.; Garg, N.; Dorrestein, P.C. Mass spectrometry of natural products: Current, emerging and future technologies. Nat. Prod. Rep. 2014, 31, 718–729. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, S.; Zhang, H.; Wang, Y.; Zhao, Z.; Chen, S.; Xu, H. Holistic quality evaluation of commercial white and red ginseng using a UPLC-QTOF-MS/MS-based metabolomics approach. J. Pharm. Biomed. Anal. 2012, 62, 258–273. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Chu, C.; Li, X.; Yao, S.; Yan, B.; Ren, H.; Xu, N.; Liang, Z.; Zhao, Z. Isolation and identification of antioxidant compounds in Vaccinium Bracteatum thunb. by UHPLC-Q-TOF LC/MS and their kidney damage protection. J. Funct. Foods 2014, 11, 62–70. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T. Sharing and community curation of mass spectrometry data with global natural products social molecular networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [PubMed]
- Galili, T. Dendextend: An R package for visualizing, adjusting and comparing trees of hierarchical clustering. Bioinformatics 2015, 31, 3718–3720. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.H.; Park, J.H.; Kwon, S.W. An anti-estrogenic lignan glycoside, tracheloside, from seeds of Carthamus tinctorius. Biosci. Biotechnol. Biochem. 2006, 70, 2783–2785. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, Y.; Yamagishi, M.; Okazaki, K.; Son, H.; Imazawa, T.; Nishikawa, A.; Iwata, T.; Yamauchi, Y.; Kasai, M.; Tsutsumi, K.; et al. Lack of significant inhibitory effects of a plant lignan tracheloside on 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP)-induced mammary carcinogenesis in female sprague-dawley rats. Cancer Lett. 2003, 200, 133–139. [Google Scholar] [CrossRef]
- Rajalekshmi, D.S.; Kabeer, F.A.; Madhusoodhanan, A.R.; Bahulayan, A.K.; Prathapan, R.; Prakasan, N.; Varughese, S.; Nair, M.S. Anticancer activity studies of cubebin isolated from Piper Cubeba and its synthetic derivatives. bioorg. Med. Chem. Lett. 2016, 26, 1767–1771. [Google Scholar] [CrossRef] [PubMed]
- John, L.M.; Tinto, W.F. Revised 13C-NMR assignments for the biologically active butyrolactone (-)-trachelogenin. J. Nat. Prod. 1992, 55, 1313–1314. [Google Scholar] [CrossRef]
- Liu, X.; Wang, Z.; Yang, Y.; Wang, L.; Sun, R.; Zhao, Y.; Yu, N. Active components with inhibitory activities on IFN-Γ/STAT1 and IL-6/STAT3 signaling pathways from caulis trachelospermi. Molecules 2014, 19, 11560–11571. [Google Scholar] [CrossRef] [PubMed]
- Abe, F.; Yamauchi, T. Lignans from Trachelospermum Asiaticum (Tracheolospermum. II). Chem. Pharm. Bull. 1986, 34, 4340–4345. [Google Scholar] [CrossRef]
- Kato, A.; Hashimoto, Y.; Kidokor, M. (+)-Nortrachelogenin, a new pharmacologically active lignan from Wikstroemia indica. J. Nat. Prod. 1979, 42, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Nishibe, S.; Hisada, S.; Inagaki, I. Structures of tracheloside and nortracheloside from Trachelospermum asiaticum NAKAI var. intermedium NAKAI. Chem. Pharm. Bull. 1971, 19, 866–868. [Google Scholar] [CrossRef][Green Version]
- Nichibe, S.; Hisada, S.; Inagaki, I. Lignan diglucosides from Trachelospermum asiticum. Phytochemistry 1972, 11, 3084–3085. [Google Scholar]
- Nishibe, S.; Hisada, S.; Inagaki, I. Lignans of Trachelospermum asiaticum. var. intermedium. V. Isolation of nortrache1ogenin-4, 4′-Di-O-β-D-glucopyranoside. Chem. Pharm. Bull. 1973, 21, 1114–1117. [Google Scholar] [CrossRef]
- Inagaki, I.; Hisada, S.; Nishibe, S. Triterpenoids of Trachelospermum asiaticum var. intermedium. Phytochemistry 1970, 9, 2241. [Google Scholar] [CrossRef]
| Compound | IC50 (μM) 1 | |||
|---|---|---|---|---|
| A549 | SKOV3 | PC3 | HEP2 | |
| 1 | 19.5 | 63.3 | 27.8 | 18.3 |
| 2 | 67.9 | 23.8 | 19.3 | >100 |
| 3 | 33.7 | 72.7 | 48.7 | 46.7 |
| 4 | 20.6 | 24.7 | 55.6 | 43.1 |
| 5 | 46.9 | 23.2 | 40.8 | 47.2 |
| Etoposide 2 | 6.56 | 6.57 | 7.78 | 4.14 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.; Yang, H.S.; Jeong, H.; Kim, J.-H.; Yang, H. Targeted Isolation of Lignans from Trachelospermum asiaticum Using Molecular Networking and Hierarchical Clustering Analysis. Biomolecules 2020, 10, 378. https://doi.org/10.3390/biom10030378
Lee J, Yang HS, Jeong H, Kim J-H, Yang H. Targeted Isolation of Lignans from Trachelospermum asiaticum Using Molecular Networking and Hierarchical Clustering Analysis. Biomolecules. 2020; 10(3):378. https://doi.org/10.3390/biom10030378
Chicago/Turabian StyleLee, Jiho, Hong Seok Yang, Hyogeun Jeong, Jung-Hwan Kim, and Heejung Yang. 2020. "Targeted Isolation of Lignans from Trachelospermum asiaticum Using Molecular Networking and Hierarchical Clustering Analysis" Biomolecules 10, no. 3: 378. https://doi.org/10.3390/biom10030378
APA StyleLee, J., Yang, H. S., Jeong, H., Kim, J.-H., & Yang, H. (2020). Targeted Isolation of Lignans from Trachelospermum asiaticum Using Molecular Networking and Hierarchical Clustering Analysis. Biomolecules, 10(3), 378. https://doi.org/10.3390/biom10030378





