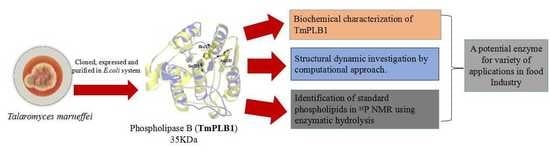
A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study
Abstract
1. Introduction
2. Material and methods
2.1. Reagents and Chemicals
2.2. Cloning, Expression and Purification of TmPLB1
2.3. Biochemical Characterization of TmPLB1
2.3.1. Hydrolytic Assay
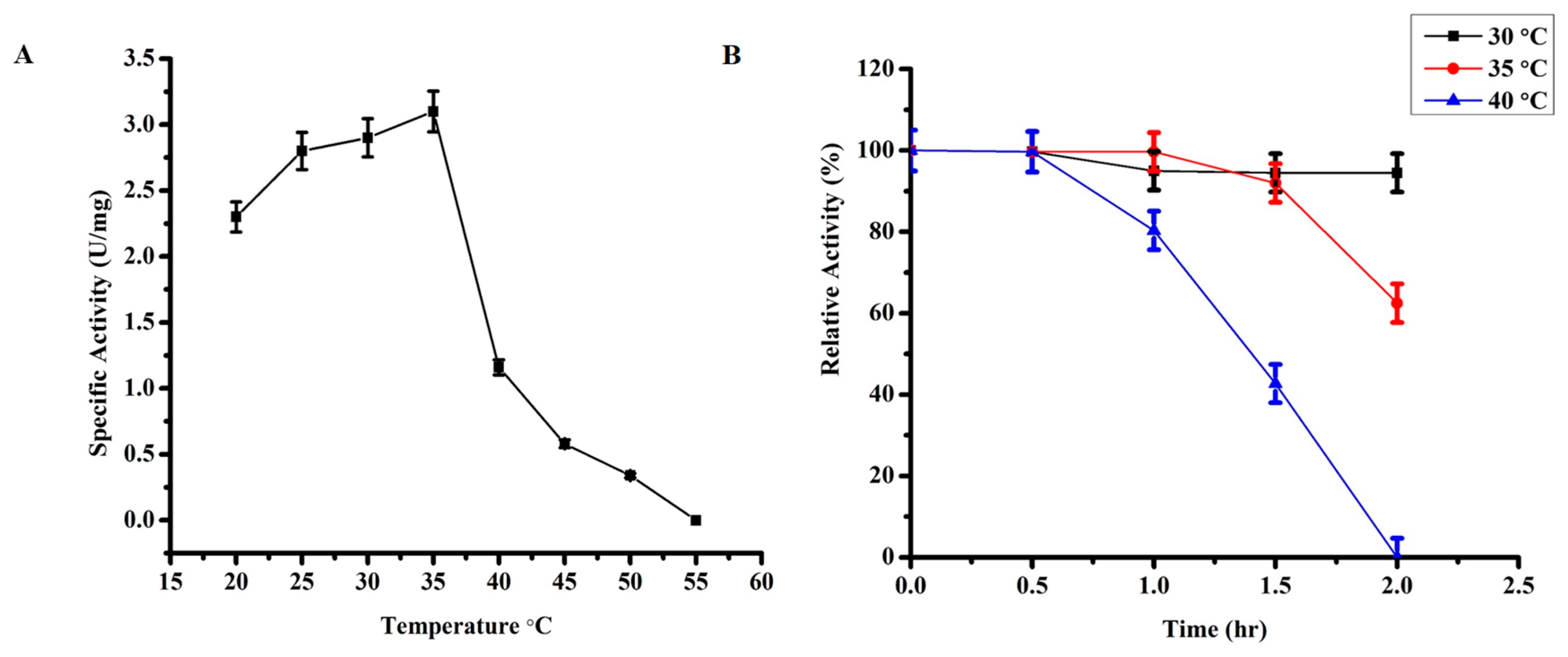
2.3.2. Effect of Temperature on TmPLB1 Activity
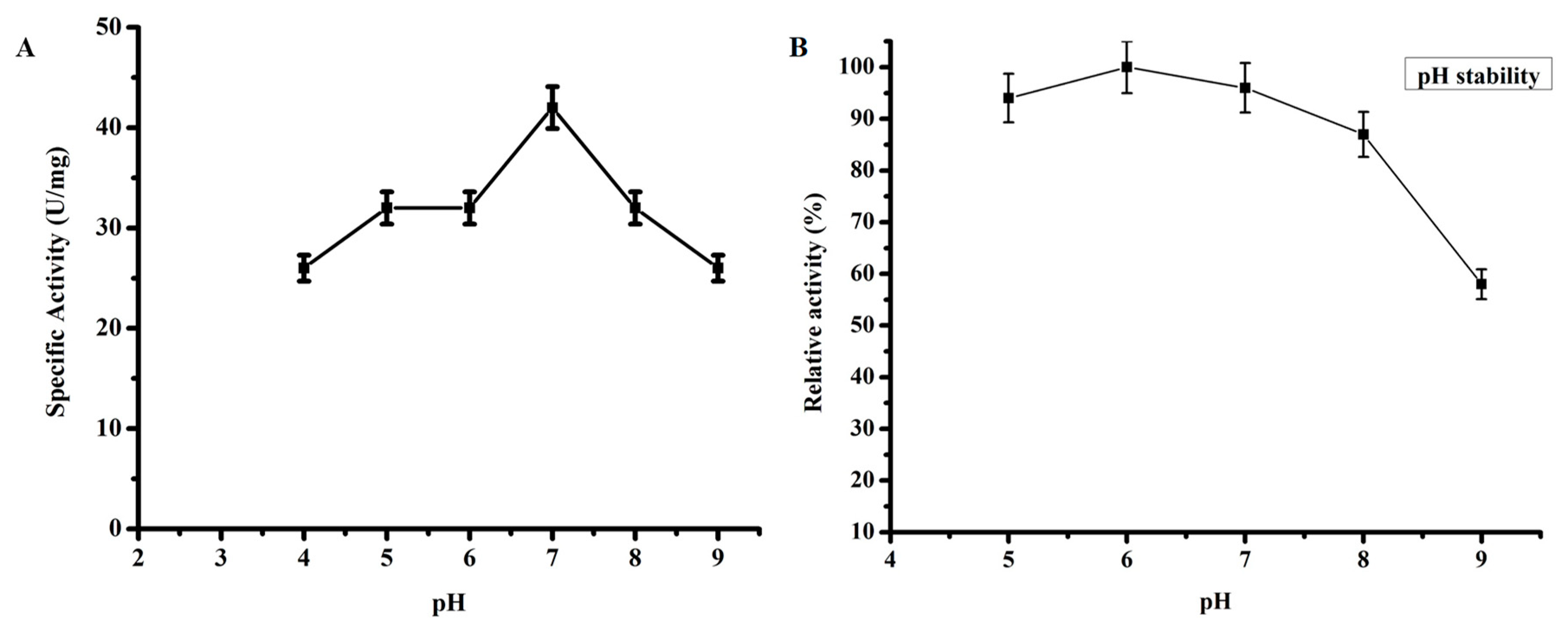
2.3.3. Effect of pH on TmPLB1 Activity
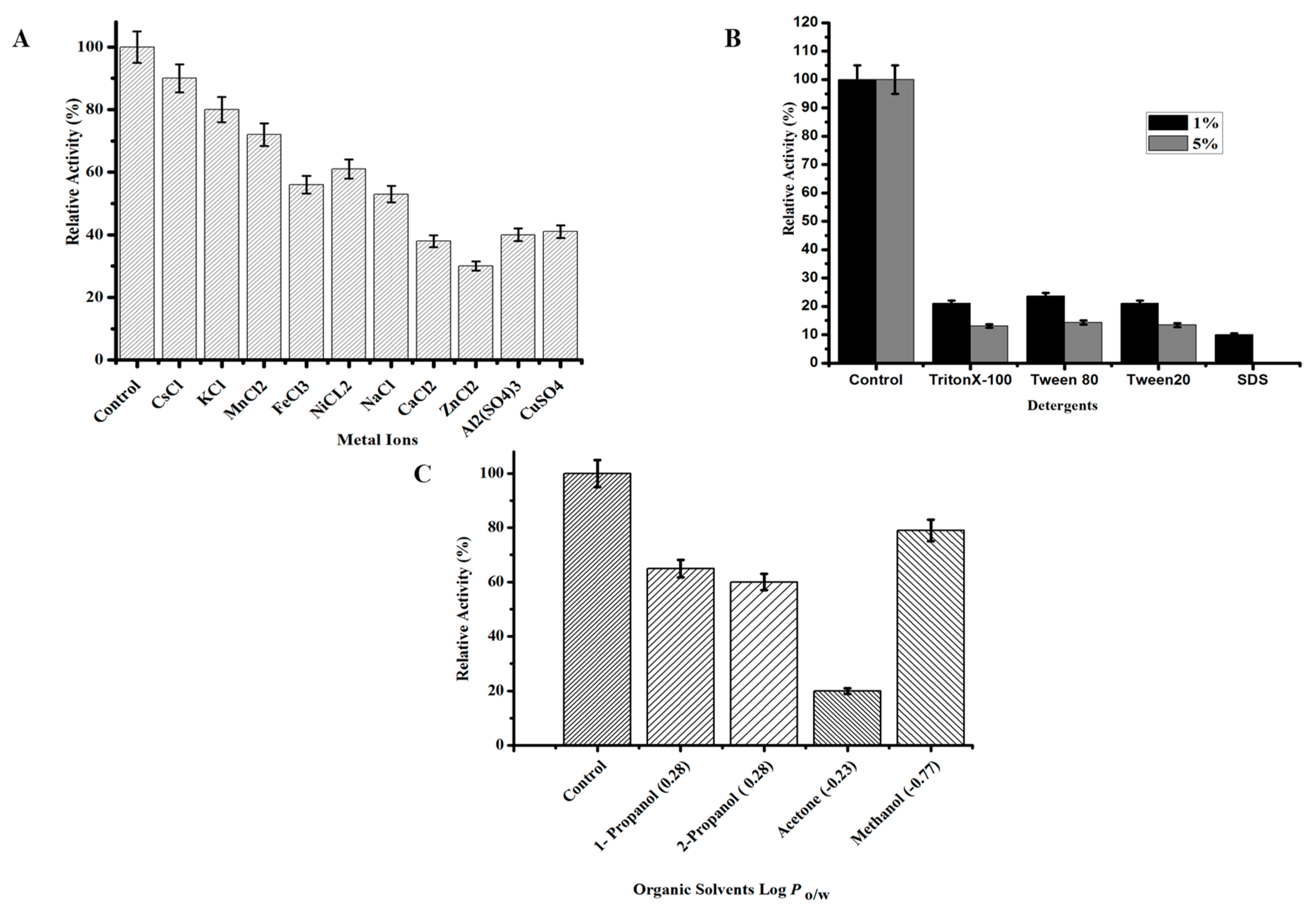
2.3.4. Effect of Chemicals on TmPLB1 Activity and Stability
2.3.5. Circular Dichroism Spectroscopy Analysis
2.4. Analysis of Phospholipids in Lecithin Using 31P NMR
2.5. Protein Sequence and Structure Analysis
2.6. Molecular Dynamics Simulation
3. Results and Discussion
3.1. Recombinant TmPLB1 Expression and p Simple Point Charge (SPCPurification)
3.2. Biochemical Characterization
3.2.1. Optimum Temperature
3.2.2. Optimum pH
3.2.3. Effect of Chemicals on the Activity and Stability of TmPLB1
3.2.4. Circular Dichroism Spectral Analysis
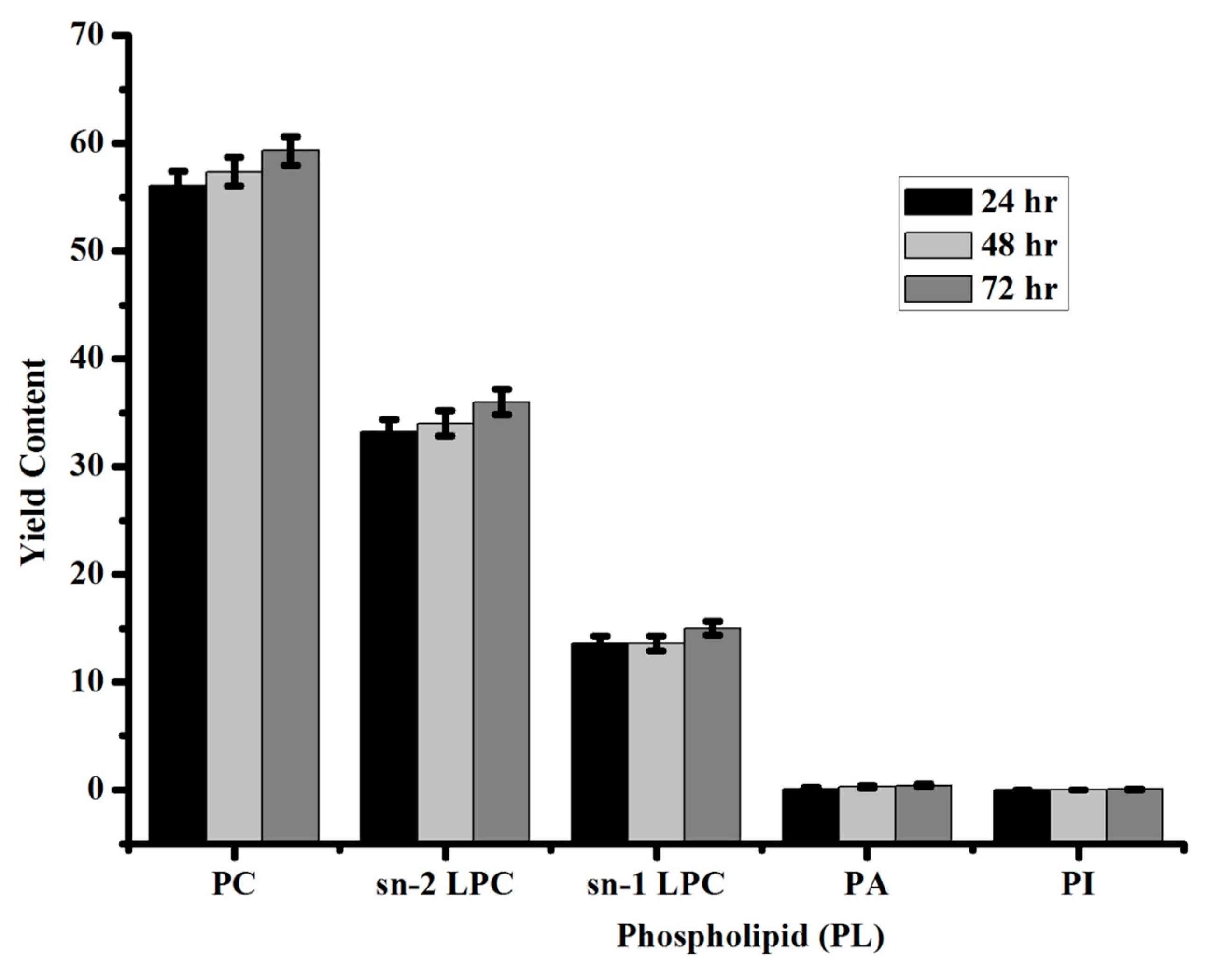
3.3. Identification of Standard Phospholipids by 31P NMR
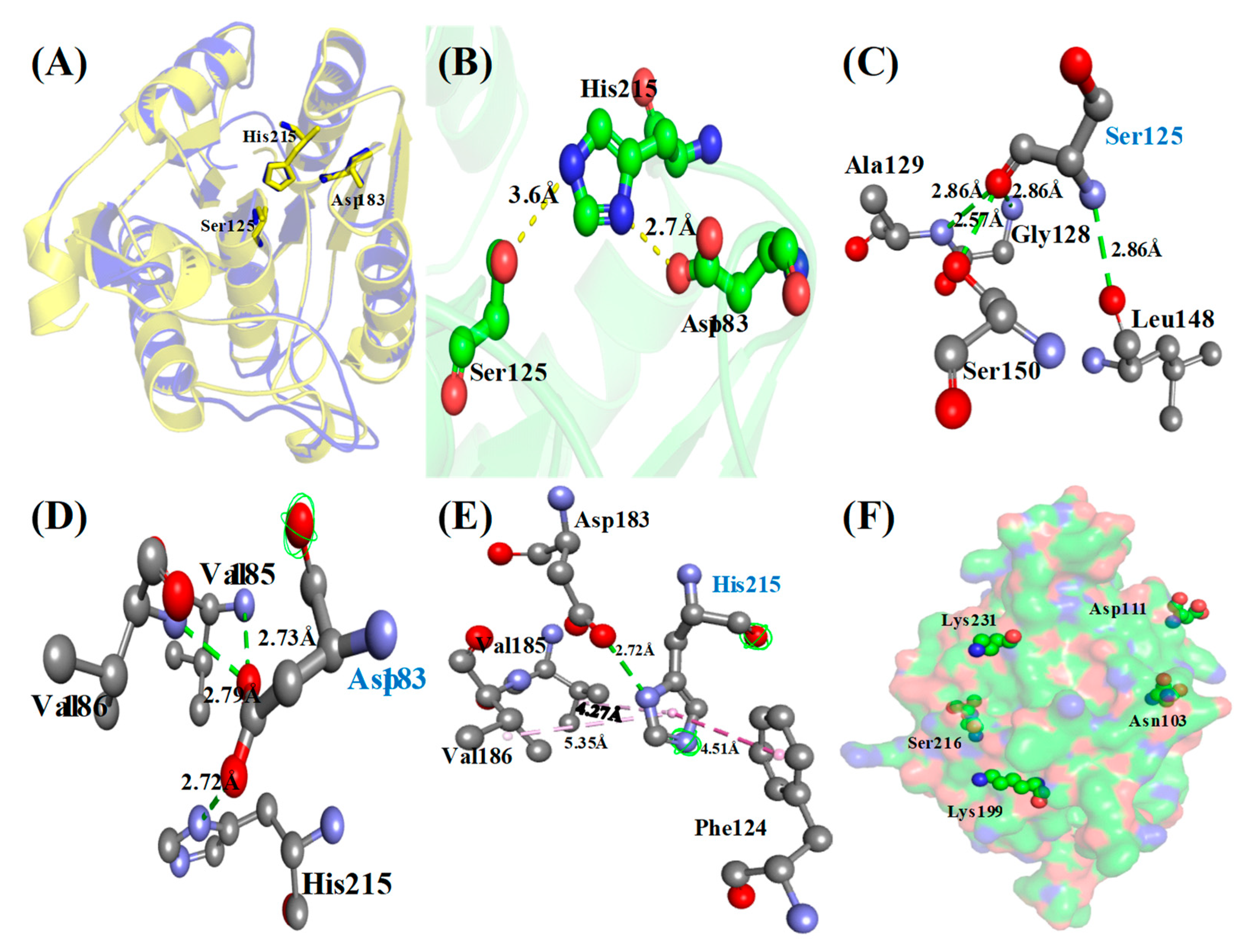
3.4. Protein Sequence, Structure Analysis and Identification of Catalytic Triad
Identification of Phosphosites
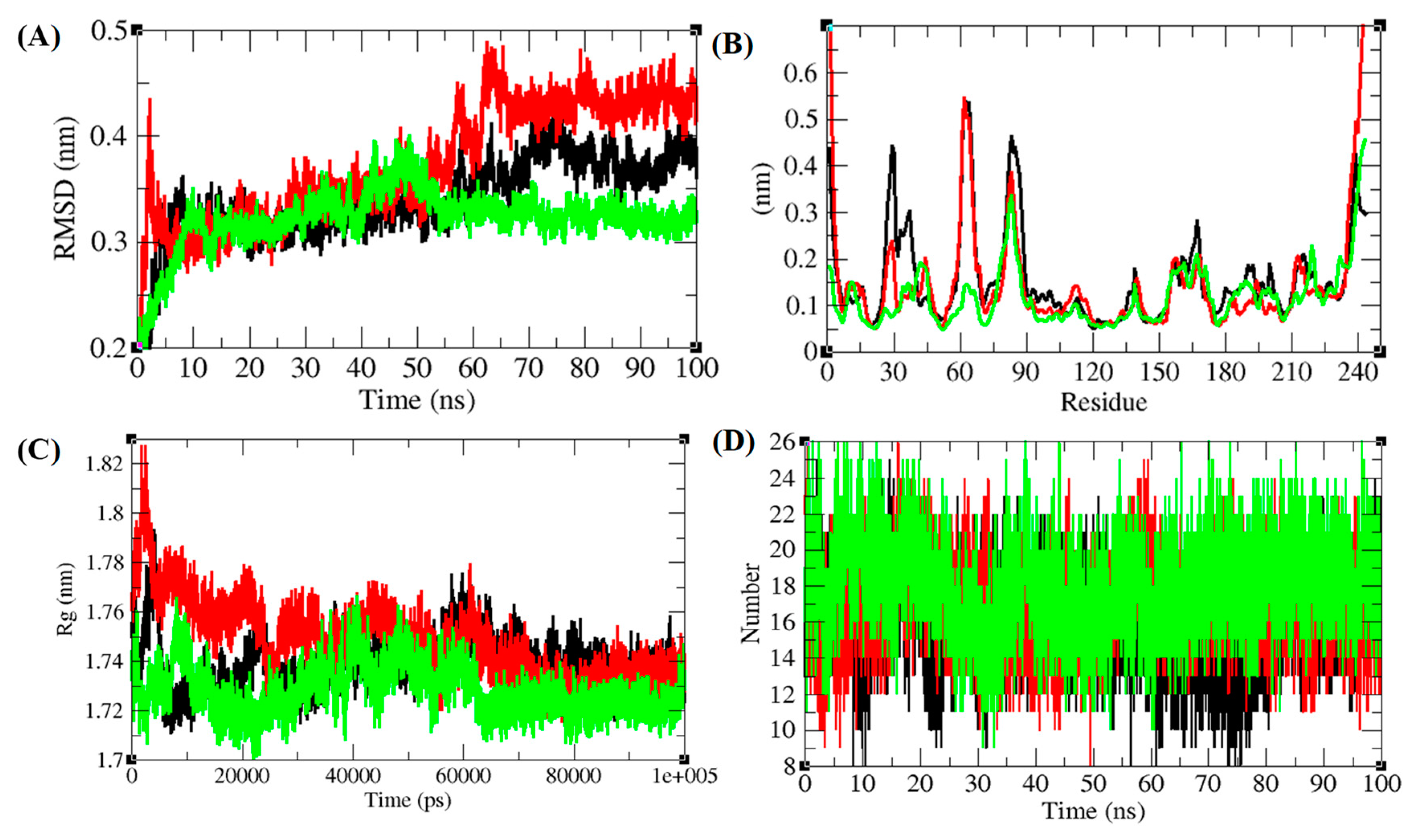
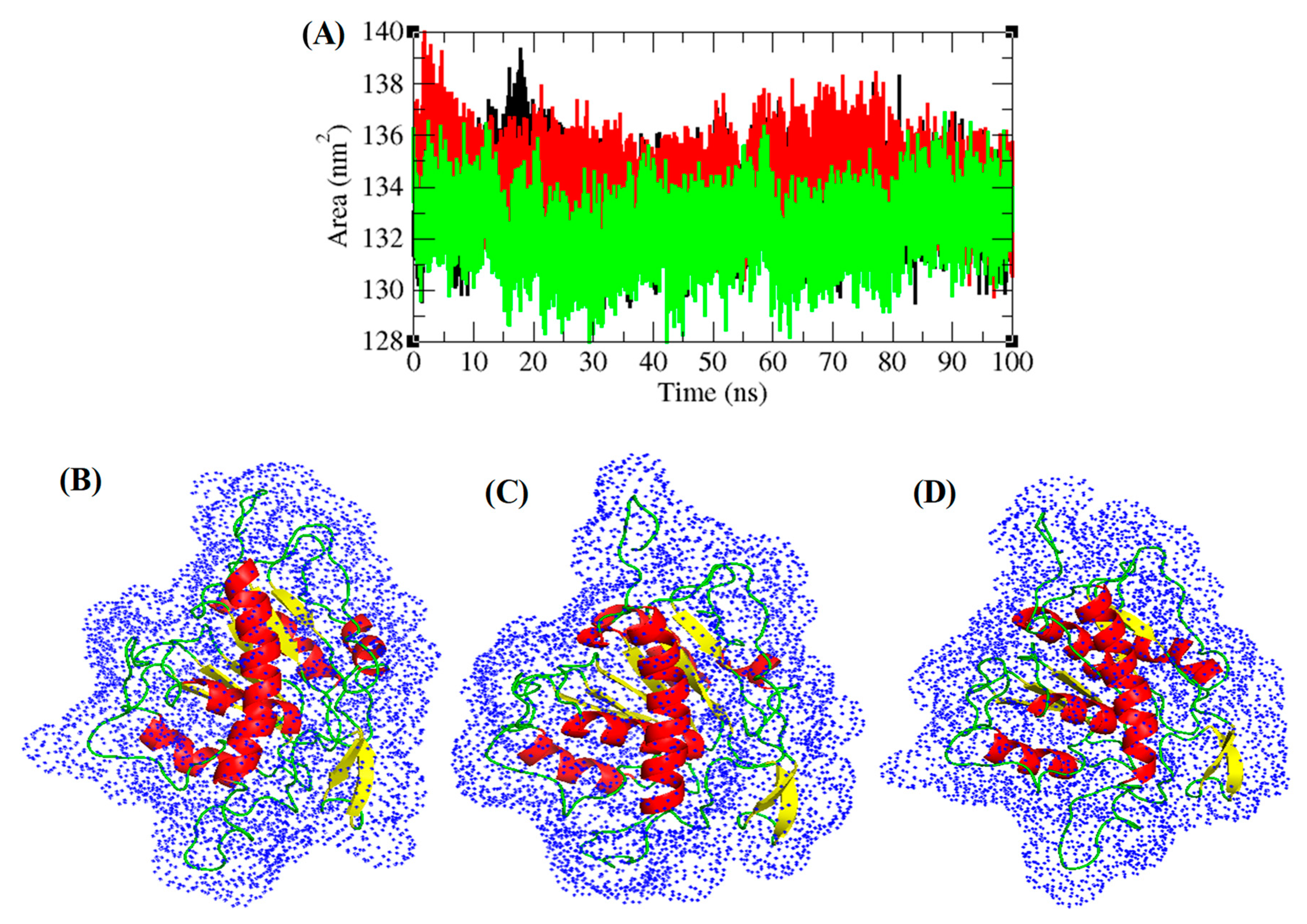
3.5. Structural Deviations, Hydrogen Bonding, and SASA Analysis
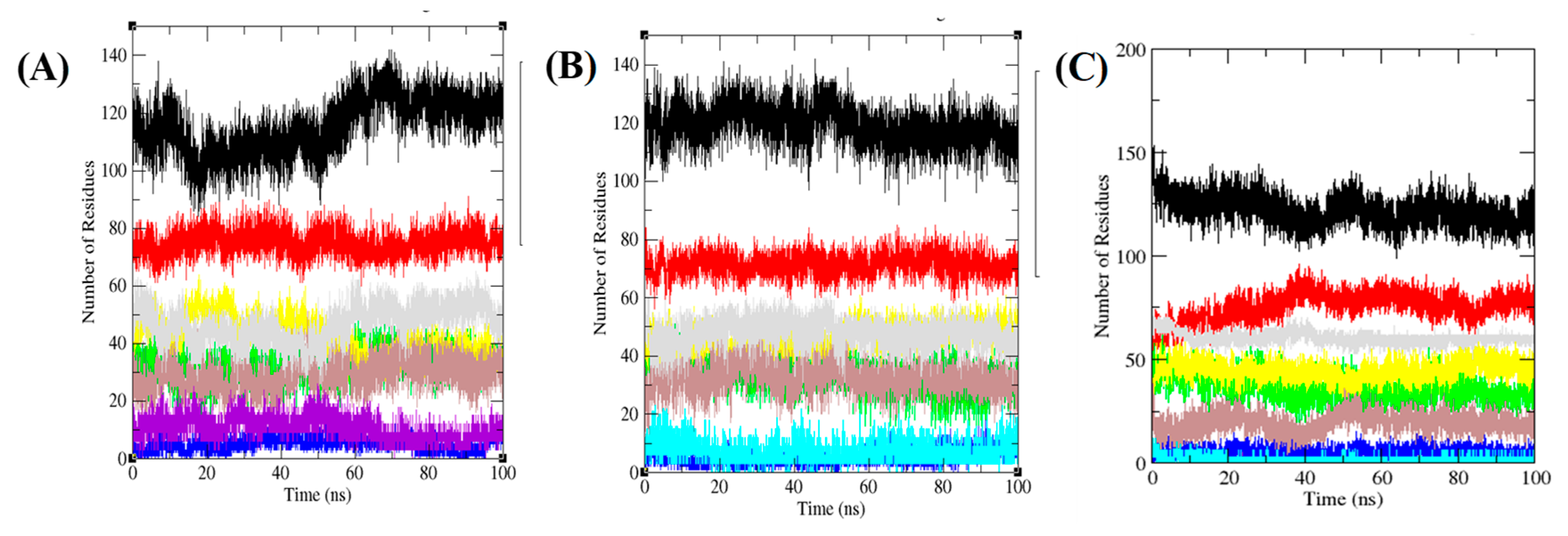
3.6. Secondary Structure Analysis
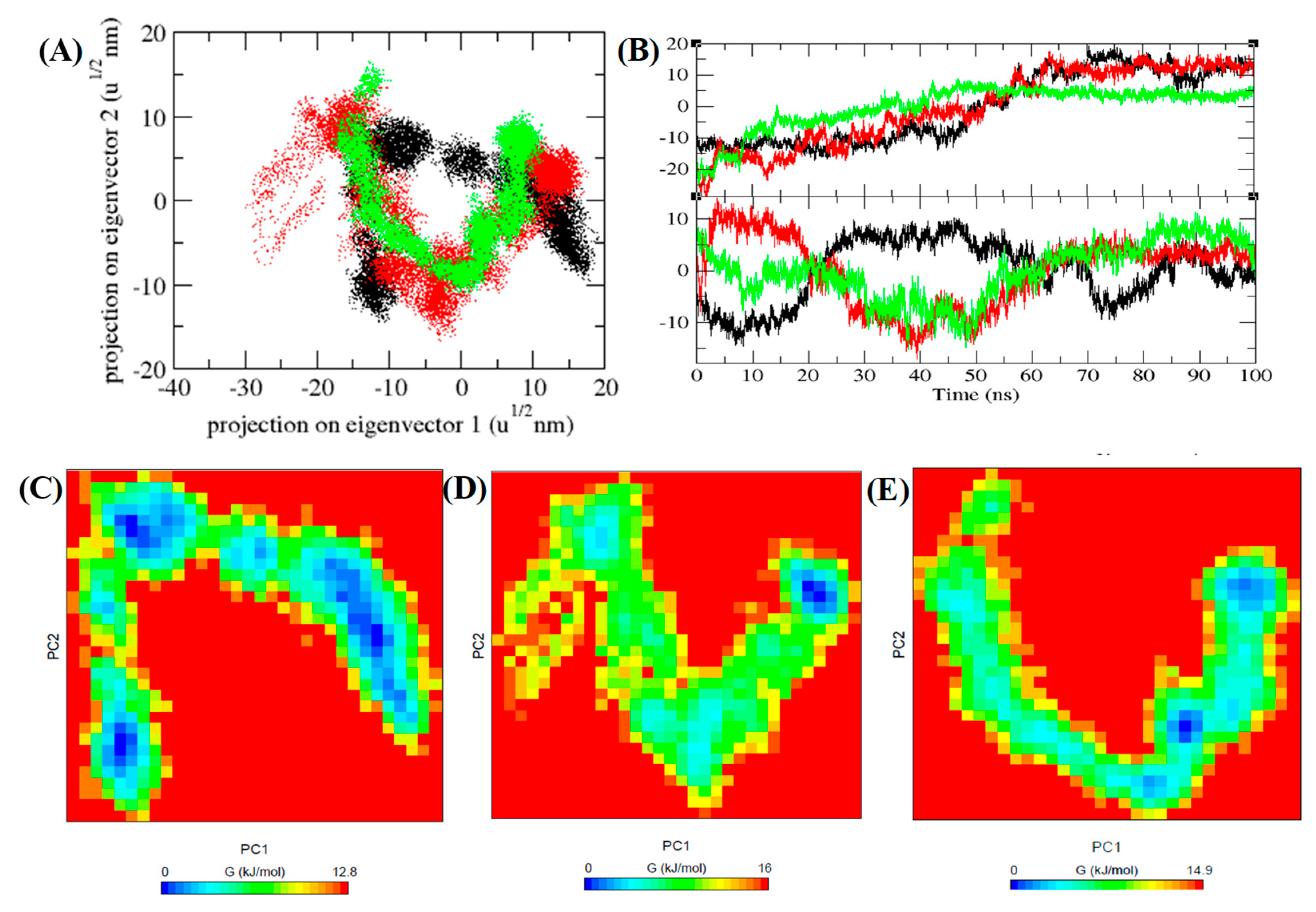
3.7. PCA and Gibbs Free Energy Landscape
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Borrelli, G.M.; Trono, D. Recombinant Lipases and Phospholipases and Their Use as Biocatalysts for Industrial Applications. Int. J. Mol. Sci. 2015, 16, 20774–20840. [Google Scholar] [CrossRef] [PubMed]
- Ulbrich-Hofmann, R. Methods in Molecular Biology, Volume 861: Lipases and Phospholipases: Methods and Protocols. Edited by Georgina Sandoval. ChemBioChem 2012, 13, 2148–2149. [Google Scholar] [CrossRef]
- Pryor, W. Biochemistry of Lipids, Lipoproteins and Membranes. New Comprehensive Biochemistry. Free Radic. Biol. Med. 1995, 18, 629. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Mineta, S.; Murayama, K.; Sugimori, D. A novel phospholipase B from Streptomyces sp. NA684—Purification, characterization, gene cloning, extracellular production and prediction of the catalytic residues. FEBS J. 2013, 280, 3780–3796. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.P.; Insall, R.; Haynes, L.; Cockcroft, S. Identification of phospholipase B from Dictyostelium discoideum reveals a new lipase family present in mammals, flies and nematodes, but not yeast. Biochem. J. 2004, 382, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Michel, N.; Lauriane, G.; Brigitte, C.; Christine, P.; Françoise, H.-M.; Bertrand, P.; Hugues, C.; Ama, G.-D. Guinea Pig Phospholipase B, Identification of the Catalytic Serine and the Proregion Involved in Its Processing and Enzymatic Activity. J. Biol. Chem. 2002, 277, 44093–44099. [Google Scholar]
- Shuji, F.; Daigo, A.; Satoko, A.; Tomonari, F.; Yasuo, W.; Youichi, T. Purification and Characterization of Phospholipase B from Candida utilis. Biosci. Biotechnol. Biochem. 2006, 70, 377–386. [Google Scholar]
- Ramrakhiani, L.; Chand, S. Recent Progress on Phospholipases: Different Sources, Assay Methods, Industrial Potential and Pathogenicity. Appl. Biochem. Biotechnol. 2011, 164, 991–1022. [Google Scholar] [CrossRef]
- Sandoval, G. Lipases and Phospholipases: Methods and Protocols; Humana Press: New York, NY, USA, 2012. [Google Scholar]
- Liu, Y.; Li, M.; Huang, L.; Gui, S.; Jia, L.; Zheng, D.; Fu, Y.; Zhang, Y.; Rui, J.; Lu, F. Cloning, expression and characterisation of phospholipase B from Saccharomyces cerevisiae and its application in the synthesis of l-alpha-glycerylphosphorylcholine and peanut oil degumming. Biotechnol. Biotechnol. Equip. 2018, 32, 968–973. [Google Scholar] [CrossRef]
- Su, L.; Ji, D.; Tao, X.; Yu, L.; Wu, J.; Xia, Y. Recombinant expression, characterization, and application of a phospholipase B from Fusarium oxysporum. J. Biotechnol. 2016, 242, 92–100. [Google Scholar] [CrossRef]
- Shen, D.-K.; Noodeh, A.D.; Kazemi, A.; Grillot, R.; Robson, G.; Brugère, J.-F. Characterisation and expression of phospholipase B from the opportunistic fungus Aspergillus fumigatus. FEMS Microbiol. Lett. 2004, 239, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zhang, H.; Zhao, Z.; Wei, R.; Yang, B.; Wang, Y. Recombinant Lipase from Gibberella zeae Exhibits Broad Substrate Specificity: A Comparative Study on Emulsified and Monomolecular Substrate. Int. J. Mol. Sci. 2017, 18, 1535. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wang, J.-H.; Yang, B.; Mainda, G.; Guo, Y. Degumming of Vegetable Oil by a New Microbial Lipase. Food Technol. Biotechnol. 2006, 44, 101–104. [Google Scholar]
- Tang, W.; Lan, D.; Zhao, Z.; Li, S.; Li, X.; Wang, Y. A Thermostable Monoacylglycerol Lipase from Marine Geobacillus sp. 12AMOR1: Biochemical Characterization and Mutagenesis Study. Int. J. Mol. Sci. 2019, 20, 780. [Google Scholar] [CrossRef] [PubMed]
- Tyukhtenko, S.; Rajarshi, G.; Karageorgos, I. Effects of Distal Mutations on the Structure, Dynamics and Catalysis of Human Monoacylglycerol Lipase. Sci. Rep. 2018, 8, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Anwer, K.; Rahman, S.; Sonani, R.R.; Khan, F.I.; Islam, A.; Madamwar, D.; Ahmad, F.; Hassan, M.I. Probing pH sensitivity of C-phycoerythrin and its natural truncant: A comparative study. Int. J. Biol. Macromol. 2016, 86, 18–27. [Google Scholar] [CrossRef]
- Das, S.; Islam, M.M.; Jana, G.C.; Patra, A.; Jha, P.K.; Hossain, M. Molecular binding of toxic phenothiazinium derivatives, azures to bovine serum albumin: A comparative spectroscopic, calorimetric, and in silico study. J. Mol. Recognit. 2017, 30, e2609. [Google Scholar] [CrossRef]
- Yang, Y.; Hiserodt, R.; Li, J. Rapid Identification and Relative Quantification of the Phospholipid Composition in Commercial Lecithins by 31P NMR. J. Am. Oil Chem. Soc. 2017, 94, 1–8. [Google Scholar] [CrossRef]
- Brinkmann-Trettenes, U.; Stein, P.C.; Klösgen, B.; Bauer-Brandl, A. A method for simultaneous quantification of phospholipid species by routine P-31 NMR. J. Pharm. Biomed. Anal. 2012, 70, 708–712. [Google Scholar] [CrossRef]
- Monakhova, Y.; Diehl, B. Automated multicomponent phospholipid analysis using 31P NMR spectroscopy: Example of vegetable lecithin and krill oil. Anal. Bioanal. Chem. 2018, 410, 7891–7900. [Google Scholar] [CrossRef]
- Zailer, E.; Monakhova, Y.; Diehl, B. 31 P NMR Method for Phospholipid Analysis in Krill Oil: Proficiency Testing-A Step toward Becoming an Official Method. J. Am. Oil Chem. Soc. 2018, 95, 1467–1474. [Google Scholar] [CrossRef]
- Balsgart, N.M.; Mulbjerg, M.; Guo, Z.; Bertelsen, K.; Vosegaard, T. High Throughput Identification and Quantification of Phospholipids in Complex Mixtures. Anal. Chem. 2016, 88, 2170–2176. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Jung, S. 31P NMR Phospholipid Profiling of Soybean Emulsion Recovered from Aqueous Extraction. J. Agric. Food Chem. 2010, 58, 4866–4872. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Finn, R.D.; Clements, J.; Arndt, W.; Miller, B.L.; Wheeler, T.J.; Schreiber, F.; Bateman, A.; Eddy, S.R. HMMER web server: 2015 update. Nucleic Acids Res. 2015, 43, W30–W38. [Google Scholar] [CrossRef]
- Rodriguez, R.; Chinea, G.; Lopez, N.; Pons, T.; Vriend, G. Homology modeling, model and software evaluation: Three related resources. Bioinformatics 1998, 14, 523–528. [Google Scholar] [CrossRef]
- Khan, F.I.; Nizami, B.; Anwer, R.; Gu, K.-R.; Bisetty, K.; Imtaiyaz Hassan, M.; Wei, D.-Q. Structure prediction and functional analyses of a thermostable lipase obtained from Shewanella putrefaciens. J. Biomol. Struct. Dyn. 2017, 35, 2123–2135. [Google Scholar] [CrossRef]
- Khan, F.I.; Wei, D.-Q.; Gu, K.-R.; Imtaiyaz Hassan, M.; Tabrez, S. Current updates on computer aided protein modeling and designing. Int. J. Biol. Macromol. 2016, 85, 48–62. [Google Scholar] [CrossRef]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.Y.; Sali, A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2008, 426, 145–159. [Google Scholar]
- Wang, Q.; Canutescu, A.A.; Dunbrack, R.L., Jr. SCWRL and MolIDE: Computer programs for side-chain conformation prediction and homology modeling. Nat. Protoc. 2008, 3, 1832–1847. [Google Scholar] [CrossRef]
- Kaplan, W.; Littlejohn, T.G. Swiss-PDB Viewer (Deep View). Brief Bioinform. 2001, 2, 195–197. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Luthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzym. 1997, 277, 396–404. [Google Scholar]
- Wiederstein, M.; Sippl, M.J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007, 35, W407–W410. [Google Scholar] [CrossRef] [PubMed]
- Gopal, S.M.; Kuhn, A.B.; Schäfer, L.V. Systematic evaluation of bundled SPC water for biomolecular simulations. Phys. Chem. Chem. Phys. 2015, 17, 8393–8406. [Google Scholar] [CrossRef] [PubMed]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Norberto de Souza, O.; Ornstein, R.L. Molecular dynamics simulations of a protein-protein dimer: Particle-mesh Ewald electrostatic model yields far superior results to standard cutoff model. J. Biomol. Struct. Dyn. 1999, 16, 1205–1218. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Zykova-Timan, T.; Parrinello, M. Isothermal-isobaric molecular dynamics using stochastic velocity rescaling. J. Chem. Phys. 2009, 130, 074101. [Google Scholar] [CrossRef]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Hess, B.; Bekker, H.; Berendsen, H.J.C.; Fraaije, J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Miyamoto, S.; Kollman, P.A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. [Google Scholar] [CrossRef]
- The PyMOL Molecular Graphics System. 2002. Available online: https://pymol.org/2/ (accessed on 30 January 2020).
- He, Y.; Li, L.; Hu, F.; Chen, W.; Lei, H.; Chen, X.; Cai, W.; Tang, X. Expression and characterization of a Talaromyces marneffei active phospholipase B expressed in a Pichia pastoris expression system. Emerg. Microbes Infect. 2016, 5, 1–6. [Google Scholar]
- Doi, O.; Nojima, S. Lysophospholipase of Escherichia coli. J. Biol. Chem. 1975, 250, 5208–5214. [Google Scholar] [PubMed]
- Altenbach, C.; Seelig, J. Calcium binding to phosphatidylcholine bilayers as studied by deuterium magnetic resonance. Evidence for the formation of a calcium complex with two phospholipid molecules. Biochemistry 1984, 23, 3913–3920. [Google Scholar] [CrossRef] [PubMed]
- Binder, H.; Arnold, K.; Ulrich, A.S.; Zschornig, O. Interaction of Zn2+ with phospholipid membranes. Biophys. Chem. 2001, 90, 57–74. [Google Scholar] [CrossRef]
- Huster, D.; Arnold, K.; Gawrisch, K. Strength of Ca2+ Binding to Retinal Lipid Membranes: Consequences for Lipid Organization. Biophys. J. 2000, 78, 3011–3018. [Google Scholar] [CrossRef]
- Uttatree, S.; Winayanuwattikun, P.; Charoenpanich, J. Isolation and Characterization of a Novel Thermophilic-Organic Solvent Stable Lipase from Acinetobacter baylyi. Appl. Biochem. Biotechnol. 2010, 162, 1362–1376. [Google Scholar] [CrossRef]
- Wang, F.; Wu, Z.; Abousalham, A.; Yang, B.; Wang, Y. Deletion the C-terminal peptides of Vibrio harveyi phospholipase D significantly improved its enzymatic properties. Int. J. Biol. Macromol. 2018, 129, 1140–1147. [Google Scholar] [CrossRef]
- Beer, T.A.P.; Berka, K.; Thornton, J.M.; Laskowski, R.A. PDBsum additions. Nucleic Acids Res. 2013, 42, D292–D296. [Google Scholar] [CrossRef]
- Samuel, M.P.; Serber, Z.; Ferrell, J.E. A Mechanism for the Evolution of Phosphorylation Sites. Cell 2011, 147, 934–946. [Google Scholar]
- Leslie, C.C. Properties and Regulation of Cytosolic Phospholipase A2. J. Biol. Chem. 1997, 272, 16709–16712. [Google Scholar] [CrossRef]
- Laub, M.T.; Goulian, M. Specificity in Two-Component Signal Transduction Pathways. Annu. Rev. Genet. 2007, 41, 121–145. [Google Scholar] [CrossRef] [PubMed]
- Meisel, P.; Margaret, O. Dayhoff: Atlas of Protein Sequence and Structure 1969 (Volume 4) XXIV u. 361 S., 21 Ausklapptafeln, 68 Abb. und zahlreiche Tabellen. National Biomedical Research Foundation, Silver Spring/Maryland 1969. Preis $ 12,50. Mol. Nutr. Food Res. 2010, 15, 217–218. [Google Scholar] [CrossRef]
- Huang, H.D.; Lee, T.-Y.; Tzeng, S.W.; Horng, G.-T. KinasePhos: A web tool for identifying protein kinase-specific phosphorylation sites. Nucleic Acids Res. 2005, 33, W226–W229. [Google Scholar] [CrossRef] [PubMed]
- Gramany, V.; Khan, F.I.; Govender, A.; Bisetty, K.; Singh, S.; Permaul, K. Cloning, expression, and molecular dynamics simulations of a xylosidase obtained from Thermomyces lanuginosus. J. Biomol. Struct. Dyn. 2016, 34, 1681–1692. [Google Scholar] [CrossRef]
- Khan, F.I.; Shahbaaz, M.; Bisetty, K.; Waheed, A.; Sly, W.S.; Ahmad, F.; Imtaiyaz Hassan, M. Large scale analysis of the mutational landscape in beta-glucuronidase: A major player of mucopolysaccharidosis type VII. Gene 2016, 576, 36–44. [Google Scholar] [CrossRef]
- Kuzmanic, A.; Zagrovic, B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys. J. 2010, 98, 861–871. [Google Scholar] [CrossRef]
- Mazola, Y.; Guirola, O.; Palomares, S. A comparative molecular dynamics study of thermophilic and mesophilic beta-fructosidase enzymes. J. Mol. Model 2015, 21, 2772. [Google Scholar] [CrossRef]
- Maisuradze, G.G.; Liwo, A.; Scheraga, H.A. Principal component analysis for protein folding dynamics. J. Mol. Biol. 2009, 385, 312–329. [Google Scholar] [CrossRef]
- David, C.C.; Jacobs, D.J. Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol. Biol. 2014, 1084, 193–226. [Google Scholar]
| 185–260 nm | 190–260 nm | 195–260 nm | 200–260 nm | 205–260 nm | 210–260 nm | |
|---|---|---|---|---|---|---|
| Helix | 20 | 20 | 12 | 3 | 2 | 1 |
| Antiparallel | 2 | 1 | 8 | 15 | 16 | 22 |
| Parallel | 14 | 15 | 17 | 22 | 25 | 20 |
| β-turn | 10 | 10 | 13 | 13 | 15 | 18 |
| Random coil | 54 | 54 | 50 | 47 | 42 | 39 |
| Total Sum | 100% | 100% | 100% | 100% | 100% | 100% |
| Phospholipid (PL) | Yield Content |
|---|---|
| Phosphotidylcholine (PC) | 59 ± 1 |
| sn-2 lysophosphatidylcholine (2-LPC) | 36 ± 1 |
| sn-1 lysophosphatidylcholine (1-LPC) | 15 ± 0.6 |
| Phosphatidic acid (PA) | 0.4 ± 0.1 |
| Phosphatidylinositol (PI) | 0.05 ± 0.01 |
| Secondary Structure (SS%) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Temp | Structure * | coil | β-Sheet | β-Bridge | Bend | Turn | α-Helix | 3-Helix |
| 30 °C | 48% | 31% | 14% | 2% | 17% | 12% | 19% | 4% |
| 35 °C | 49% | 29% | 14% | 2% | 19% | 14% | 20% | 3% |
| 40 °C | 44% | 30% | 13% | 4% | 21% | 10% | 17% | 5% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Durrani, R.; Khan, F.I.; Ali, S.; Wang, Y.; Yang, B. A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study. Biomolecules 2020, 10, 231. https://doi.org/10.3390/biom10020231
Durrani R, Khan FI, Ali S, Wang Y, Yang B. A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study. Biomolecules. 2020; 10(2):231. https://doi.org/10.3390/biom10020231
Chicago/Turabian StyleDurrani, Rabia, Faez Iqbal Khan, Shahid Ali, Yonghua Wang, and Bo Yang. 2020. "A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study" Biomolecules 10, no. 2: 231. https://doi.org/10.3390/biom10020231
APA StyleDurrani, R., Khan, F. I., Ali, S., Wang, Y., & Yang, B. (2020). A Thermolabile Phospholipase B from Talaromyces marneffei GD-0079: Biochemical Characterization and Structure Dynamics Study. Biomolecules, 10(2), 231. https://doi.org/10.3390/biom10020231