Membrane Binding of Neuronal Calcium Sensor-1: Highly Specific Interaction with Phosphatidylinositol-3-Phosphate
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cloning of NCS-1 N-Terminal Mutants
2.3. Purification and Characterization of Myristoylated and Unmyristoylated Recombinant NCS-1 and Its Mutant Forms
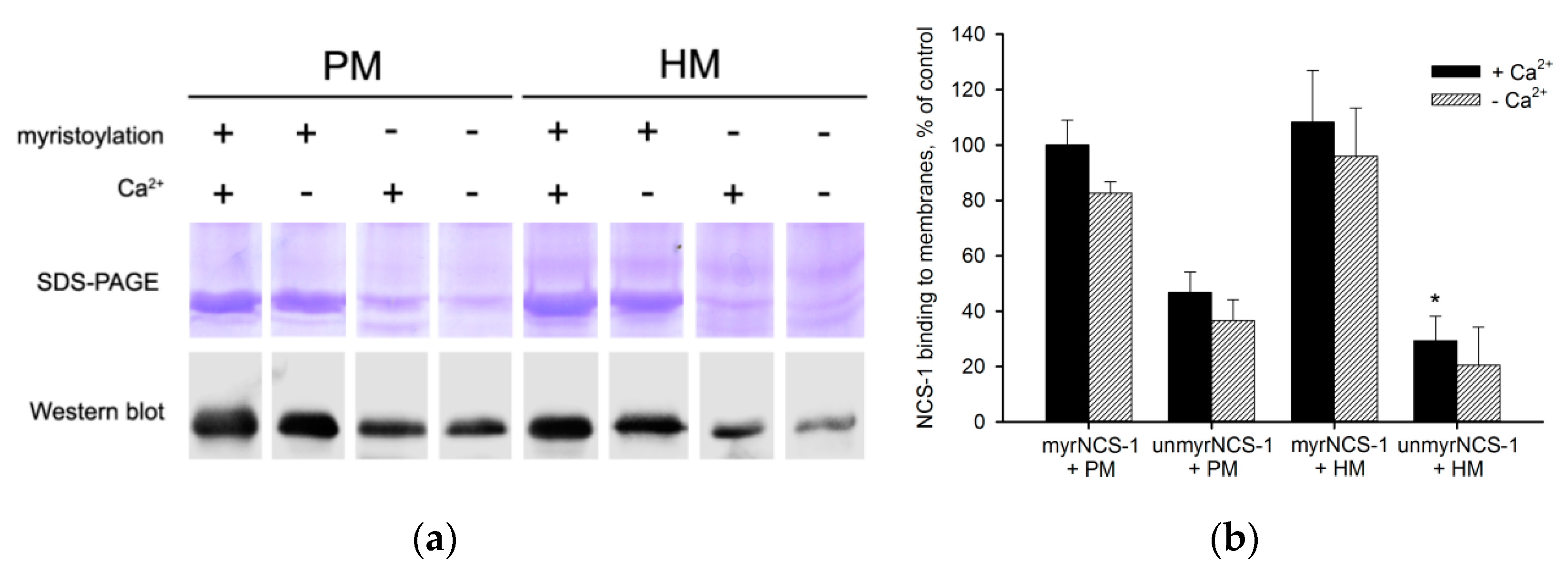
2.4. Preparation of Urea-Washed Photoreceptor and Hippocampal Membranes
2.5. Membrane Binding Assay
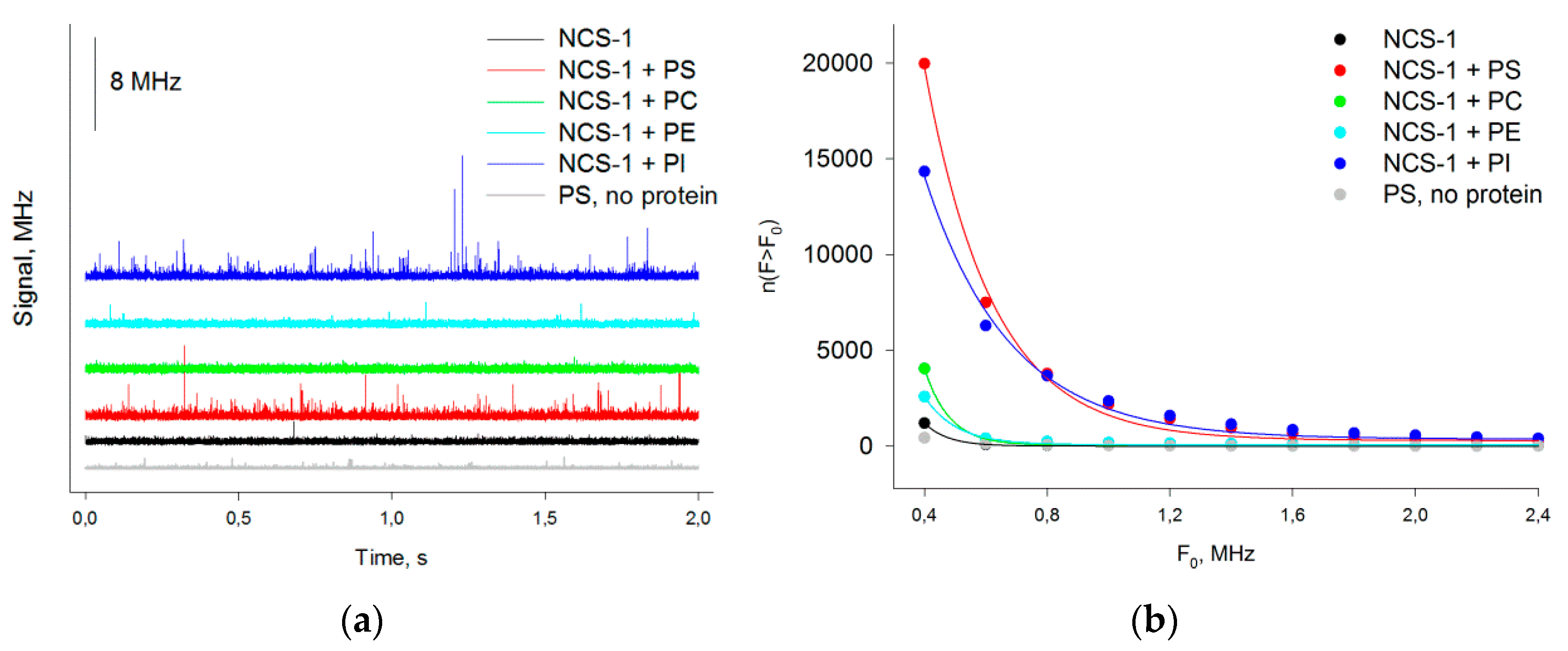
2.6. Analysis of NCS-1 Binding to Liposomes by Fluorescence Correlation Spectroscopy
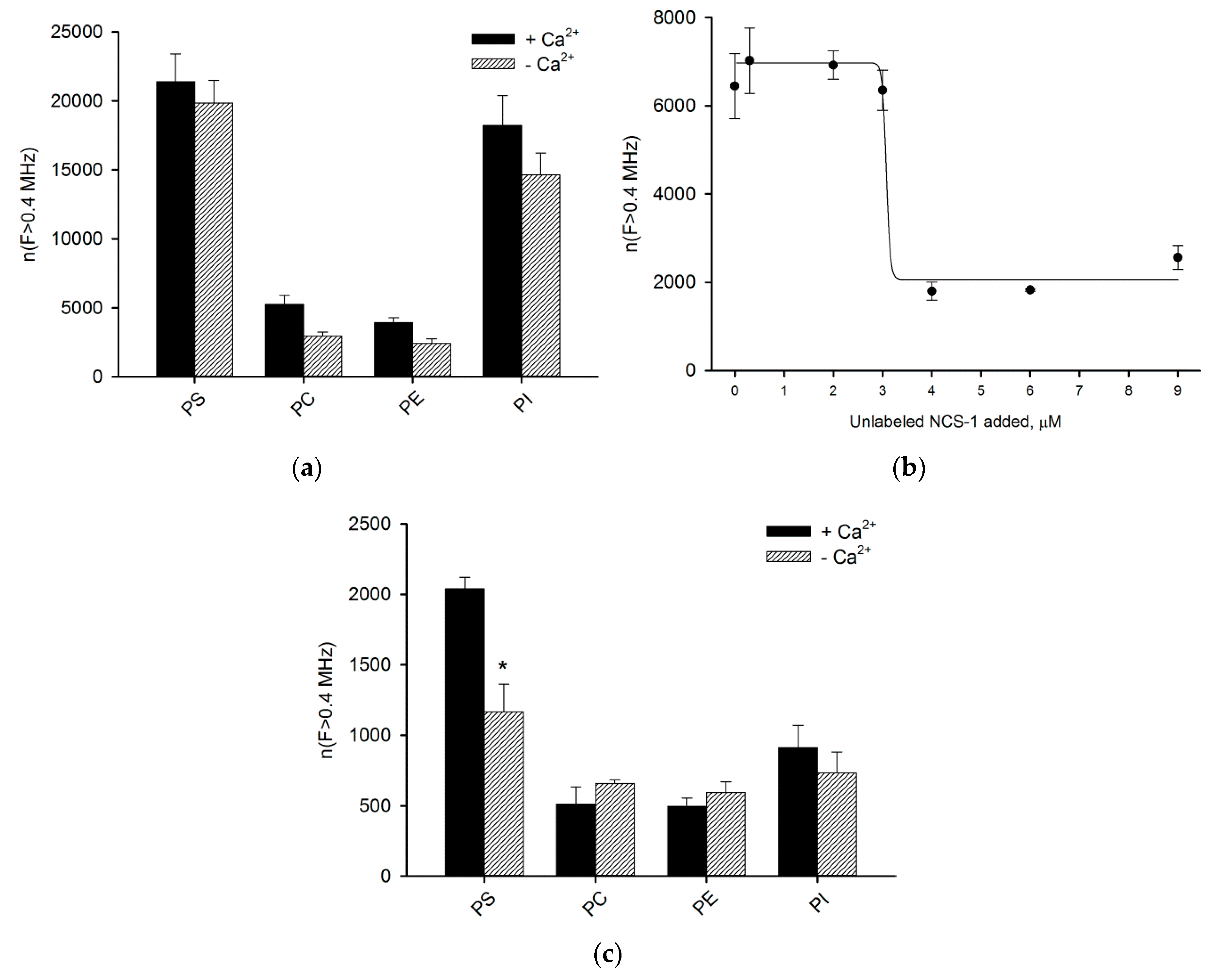
2.7. Phospholipid Binding Assay
2.8. Western Blotting
2.9. Molecular Docking
2.10. Statistical Analysis
3. Results
3.1. Binding of NCS-1 to Cellular Membranes
3.2. Binding of NCS-1 to Liposomes
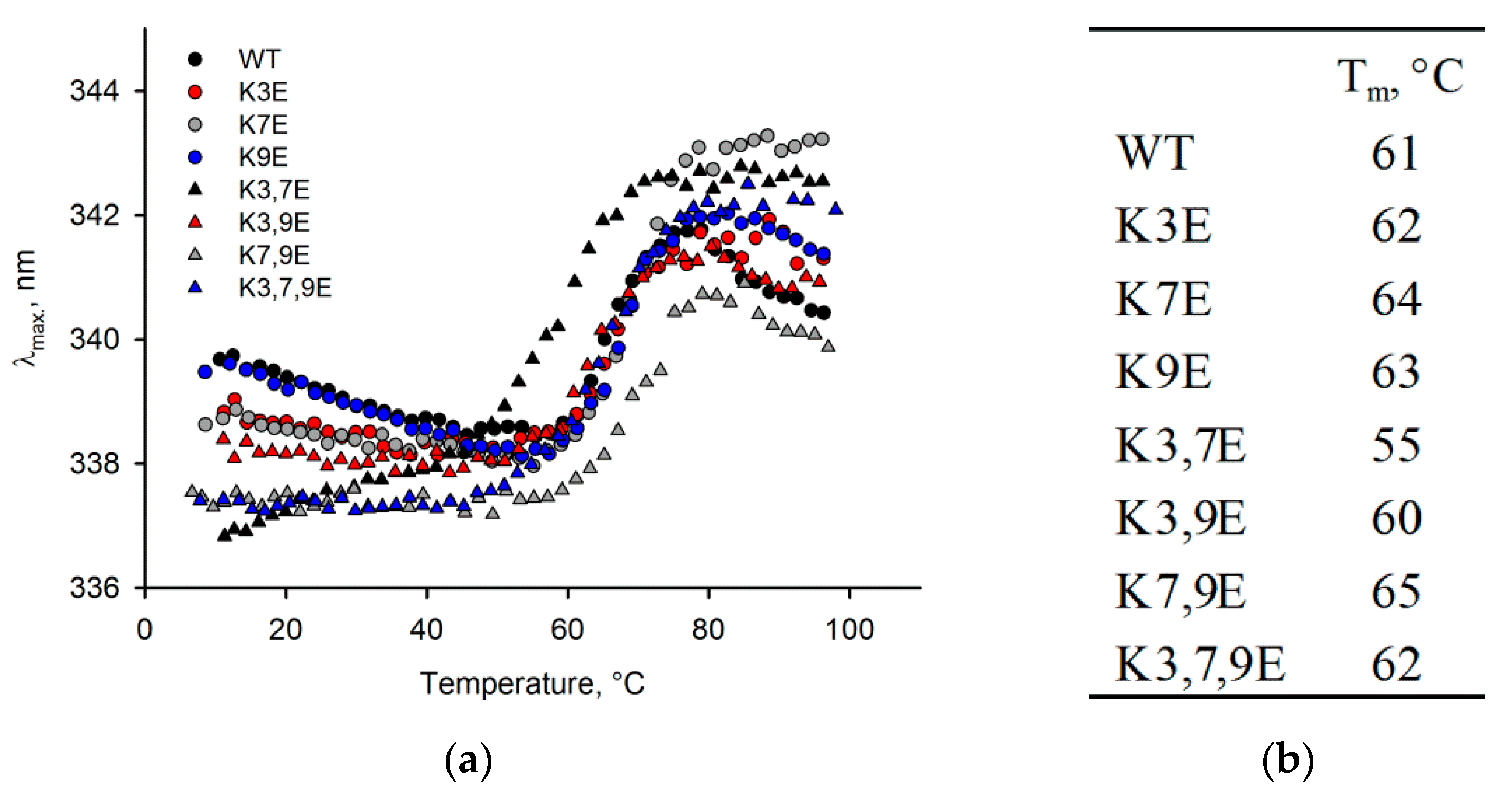
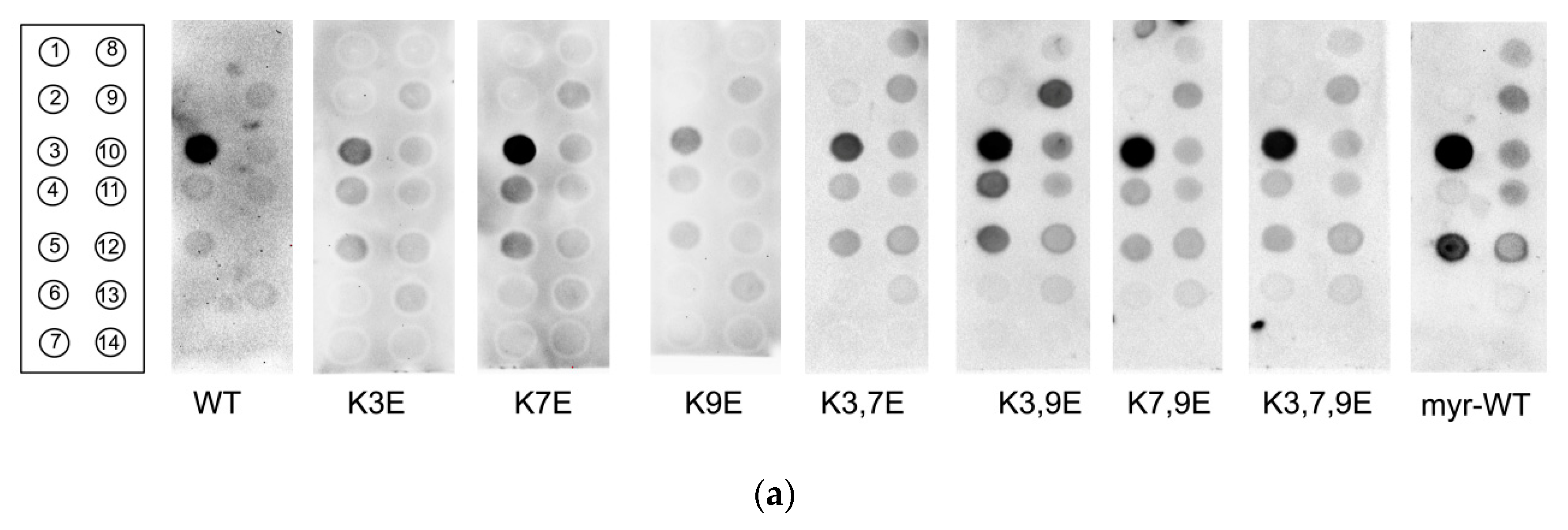
3.3. Obtaining and Characterization of NCS-1 N-Terminal Mutants
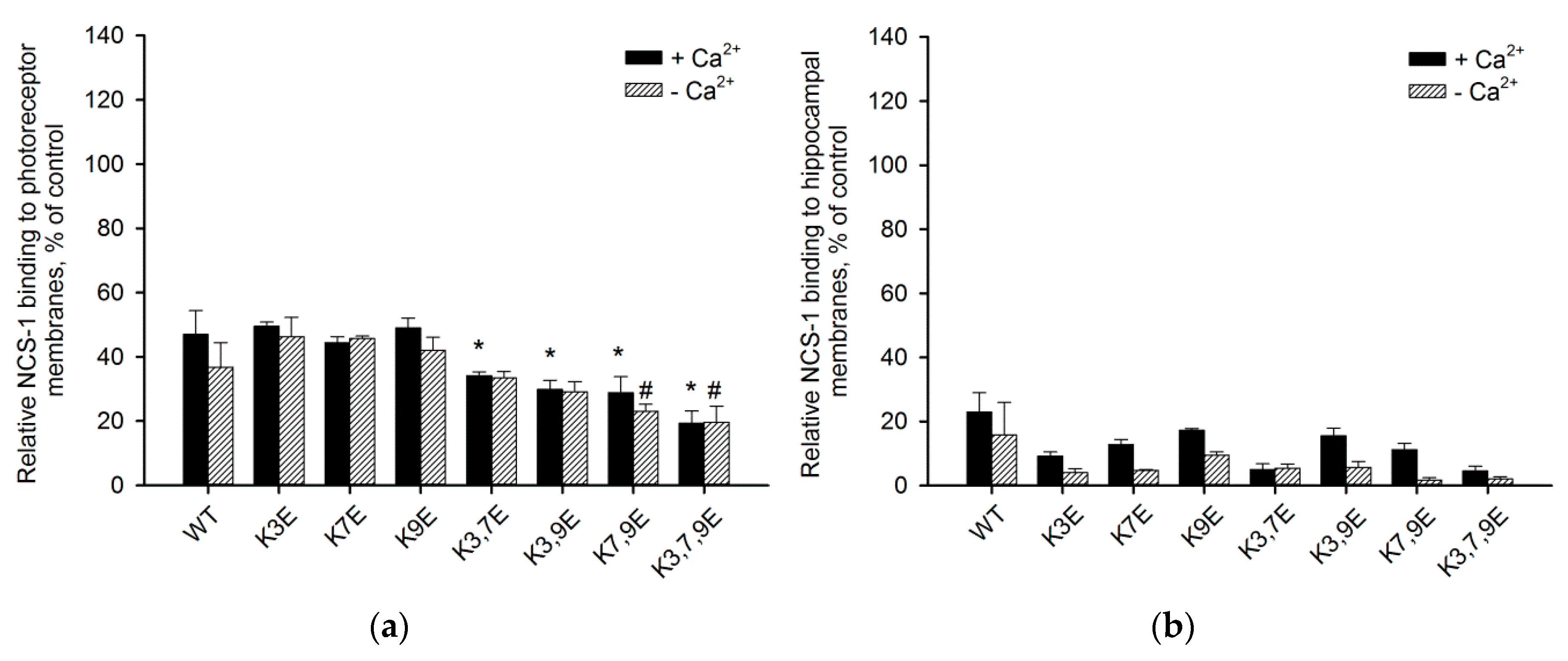
3.4. Binding of NCS-1 N-Terminal Mutants to Cellular Membranes
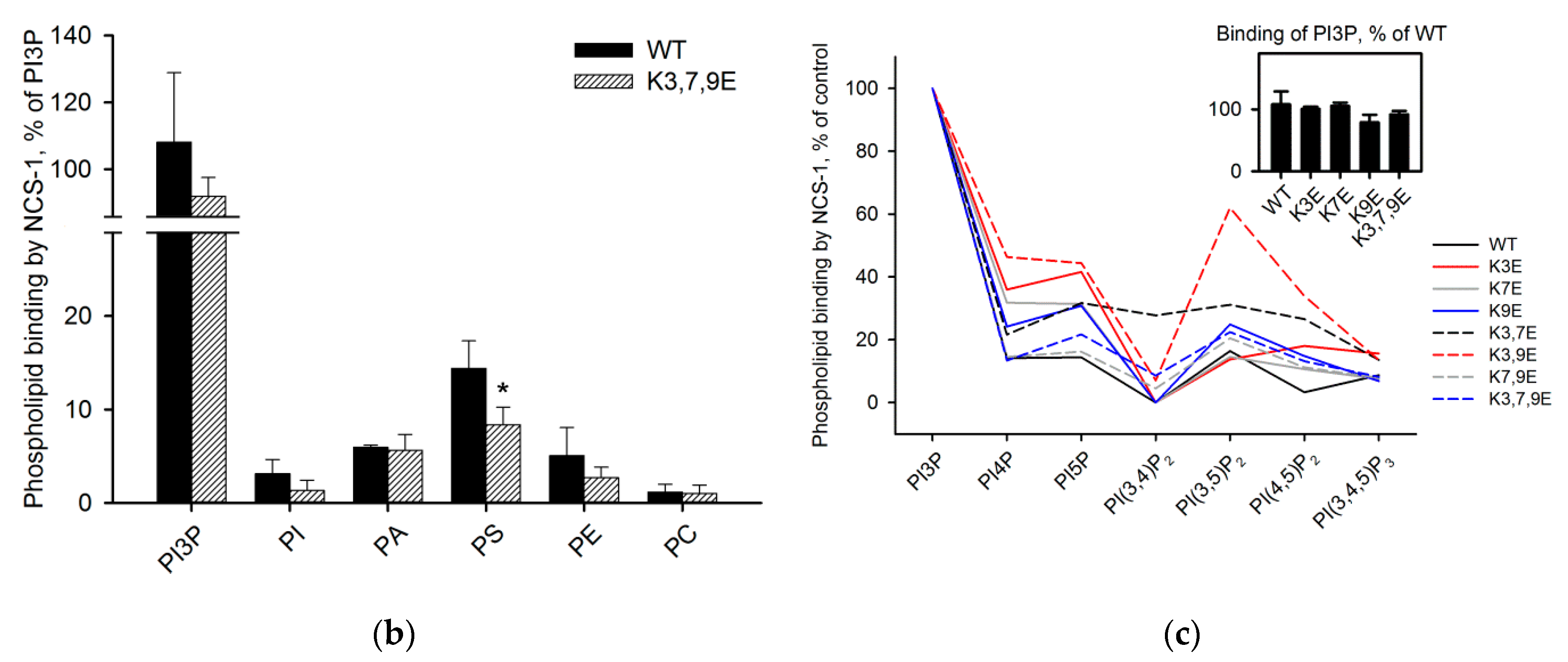
3.5. Binding of NCS-1 and Its N-Terminal Mutants to Individual Phospholipids
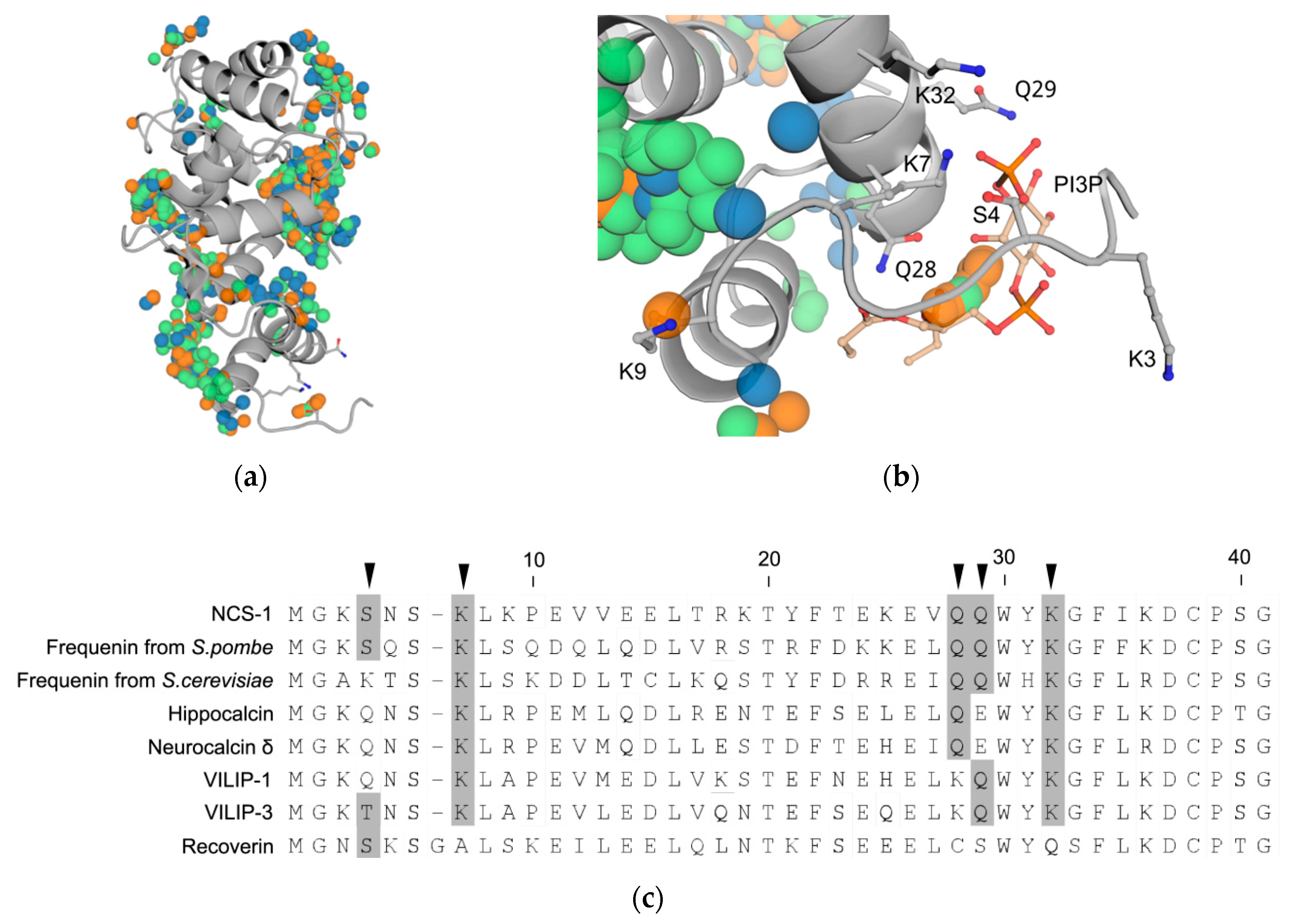
3.6. Prediction of PI3P-Binding Sites in Ca2+-Loaded NCS-1
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NCS | Neuronal calcium sensor |
| NCS-1 | Neuronal calcium sensor-1 |
| PS | Phosphatidylserine |
| PI | Phosphatidylinositol |
| PI(4,5)P2 | Phosphatidylinositol-4,5-bisphosphate |
| PI3P | Phosphatidylinositol-3-phosphate |
| PI4P | Phosphatidylinositol-4-phosphate |
| PI5P | Phosphatidylinositol-5-phosphate |
| NMT | Myristoyl-CoA:protein N-myristoyltransferase |
| FCS | Fluorescence correlation spectroscopy |
Appendix A
References
- Hurley, J.H.; Meyer, T. Subcellular targeting by membrane lipids. Curr. Opin. Cell Biol. 2001, 13, 146–152. [Google Scholar] [CrossRef]
- McCabe, J.B.; Berthiaume, L.G. Functional Roles for Fatty Acylated Amino-terminal Domains in Subcellular Localization. Mol. Biol. Cell 1999, 10, 3771–3786. [Google Scholar] [CrossRef] [PubMed]
- Burgoyne, R.D.; Haynes, L.P. Understanding the physiological roles of the neuronal calcium sensor proteins. Mol. Brain 2012, 5, 2. [Google Scholar] [CrossRef] [PubMed]
- Ames, J.B.; Ishima, R.; Tanaka, T.; Gordon, J.I.; Stryer, L.; Ikura, M. Molecular mechanics of calcium-myristoyl switches. Nature 1997, 389, 198. [Google Scholar] [CrossRef] [PubMed]
- Spilker, C.; Dresbach, T.; Braunewell, K.-H. Reversible translocation and activity-dependent localization of the calcium–myristoyl switch protein VILIP-1 to different membrane compartments in living hippocampal neurons. J. Neurosci. 2002, 22, 7331–7339. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, D.W.; Tepikin, A.V.; Burgoyne, R.D. Dynamics and calcium sensitivity of the Ca2+/myristoyl switch protein hippocalcin in living cells. J. Cell Biol. 2003, 163, 715–721. [Google Scholar] [CrossRef]
- Ivings, L.; Pennington, S.R.; Jenkins, R.; Weiss, J.L.; Burgoyne, R.D. Identification of Ca2+-dependent binding partners for the neuronal calcium sensor protein neurocalcin δ: Interaction with actin, clathrin and tubulin. Biochem. J. 2002, 363, 599–608. [Google Scholar] [CrossRef]
- O’Callaghan, D.W.; Haynes, L.P.; Burgoyne, R.D. High-affinity interaction of the N-terminal myristoylation motif of the neuronal calcium sensor protein hippocalcin with phosphatidylinositol 4, 5-bisphosphate. Biochem. J. 2005, 391, 231–238. [Google Scholar] [CrossRef]
- De Raad, S.; Comte, M.; Nef, P.; Lenz, S.E.; Gundelfinger, E.D.; Cox, J.A. Distribution pattern of three neural calcium-binding proteins (NCS-1, VILIP and recoverin) in chicken, bovine and rat retina. Histochem. J. 1995, 27, 524–535. [Google Scholar] [CrossRef]
- O’Callaghan, D.W.; Ivings, L.; Weiss, J.L.; Ashby, M.C.; Tepikin, A.V.; Burgoyne, R.D. Differential use of myristoyl groups on neuronal calcium sensor proteins as a determinant of spatio-temporal aspects of Ca2+ signal transduction. J. Biol. Chem. 2002, 277, 14227–14237. [Google Scholar] [CrossRef]
- Kachi, S.; Nishizawa, Y.; Olshevskaya, E.; Yamazaki, A.; Miyake, Y.; Wakabayashi, T.; Dizhoor, A.; Usukura, J. Detailed localization of photoreceptor guanylate cyclase activating protein-1 and-2 in mammalian retinas using light and electron microscopy. Exp. Eye Res. 1999, 68, 465–473. [Google Scholar] [CrossRef]
- Murray, D.; Hermida-Matsumoto, L.; Buser, C.A.; Tsang, J.; Sigal, C.T.; Ben-Tal, N.; Honig, B.; Resh, M.D.; McLaughlin, S. Electrostatics and the membrane association of Src: Theory and experiment. Biochemistry 1998, 37, 2145–2159. [Google Scholar] [CrossRef] [PubMed]
- Valentine, K.G.; Mesleh, M.F.; Opella, S.J.; Ikura, M.; Ames, J.B. Structure, topology, and dynamics of myristoylated recoverin bound to phospholipid bilayers. Biochemistry 2003, 42, 6333–6340. [Google Scholar] [CrossRef] [PubMed]
- Bentham, M.; Mazaleyrat, S.; Harris, M. Role of myristoylation and N-terminal basic residues in membrane association of the human immunodeficiency virus type 1 Nef protein. J. Gen. Virol. 2006, 87, 563–571. [Google Scholar] [CrossRef]
- Boesze-Battaglia, K.; Albert, A.D. Phospholipid distribution among bovine rod outer segment plasma membrane and disk membranes. Exp. Eye Res. 1992, 54, 821. [Google Scholar] [CrossRef]
- Boesze-Battaglia, K.; Organisciak, D.T.; Albert, A.D. RCS rat retinal rod outer segment membranes exhibit different cholesterol distributions than those of normal rats. Exp. Eye Res. 1994, 58, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, H.; Grant, M.A. Relationship of membrane phospholipid composition, lactosylceramide molecular species, and the specificity of CMP-N-acetylneuraminate: Lactosylceramide alpha 2, 3-sialyltransferase to the molecular species composition of GM3 ganglioside. J. Lipid Res. 1995, 36, 1274–1282. [Google Scholar]
- Wells, K.; Farooqui, A.A.; Liss, L.; Horrocks, L.A. Neural membrane phospholipids in Alzheimer disease. Neurochem. Res. 1995, 20, 1329–1333. [Google Scholar] [CrossRef]
- O’Callaghan, D.W.; Hasdemir, B.; Leighton, M.; Burgoyne, R.D. Residues within the myristoylation motif determine intracellular targeting of the neuronal Ca2+ sensor protein KChIP1 to post-ER transport vesicles and traffic of Kv4 K+ channels. J. Cell Sci. 2003, 116, 4833–4845. [Google Scholar] [CrossRef]
- Weiss, J.L.; Hui, H.; Burgoyne, R.D. Neuronal calcium sensor-1 regulation of calcium channels, secretion, and neuronal outgrowth. Cell. Mol. Neurobiol. 2010, 30, 1283–1292. [Google Scholar] [CrossRef]
- Negyessy, L.; Goldman-Rakic, P.S. Subcellular localization of the dopamine D2 receptor and coexistence with the calcium-binding protein neuronal calcium sensor-1 in the primate prefrontal cortex. J. Comp. Neurol. 2005, 488, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Martone, M.E.; Edelmann, V.M.; Ellisman, M.H.; Nef, P. Cellular and subcellular distribution of the calcium-binding protein NCS-1 in the central nervous system of the rat. Cell Tissue Res. 1999, 295, 395–407. [Google Scholar] [CrossRef]
- Vladimirov, V.I.; Zernii, E.Y.; Baksheeva, V.E.; Wimberg, H.; Kazakov, A.S.; Tikhomirova, N.K.; Nemashkalova, E.L.; Mitkevich, V.A.; Zamyatnin, A.A., Jr.; Lipkin, V.M. Photoreceptor calcium sensor proteins in detergent-resistant membrane rafts are regulated via binding to caveolin-1. Cell Calcium 2018, 73, 55–69. [Google Scholar] [CrossRef] [PubMed]
- Baksheeva, V.E.; Nazipova, A.A.; Zinchenko, D.V.; Serebryakova, M.V.; Senin, I.; Permyakov, S.E.; Philippov, P.P.; Li, Y.; Zamyatnin, A.A.; Zernii, E.Y.; et al. Ca2+-myristoyl switch in neuronal calcium sensor-1: A role of C-terminal segment. CNS Neurol. Disord. Drug Targets 2015, 14, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Rajebhosale, M.; Greenwood, S.; Vidugiriene, J.; Jeromin, A.; Hilfiker, S. Phosphatidylinositol 4-OH kinase is a downstream target of neuronal calcium sensor-1 in enhancing exocytosis in neuroendocrine cells. J. Biol. Chem. 2003, 278, 6075–6084. [Google Scholar] [CrossRef] [PubMed]
- Torres, K.; Souza, B.; Miranda, D.; Sampaio, A.; Nicolato, R.; Neves, F.; Barros, A.; Dutra, W.; Gollob, K.; Correa, H. Expression of neuronal calcium sensor-1 (NCS-1) is decreased in leukocytes of schizophrenia and bipolar disorder patients. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2009, 33, 229–234. [Google Scholar] [CrossRef] [PubMed]
- McFerran, B.W.; Weiss, J.L.; Burgoyne, R.D. Neuronal Ca2+ Sensor 1: Characterization of the myristoylated protein, its cellular effects in permeabilized adrenal chromaffin cells, Ca2+-independent membrane association, and interaction with binding proteins, suggesting a role in rapid Ca2+ signal transduction. J. Biol. Chem. 1999, 274, 30258–30265. [Google Scholar] [PubMed]
- O’Callaghan, D.W.; Burgoyne, R.D. Identification of residues that determine the absence of a Ca2+/myristoyl switch in Neuronal Calcium Sensor-1. J. Biol. Chem. 2004, 279, 14347–14354. [Google Scholar] [CrossRef]
- O’Callaghan, D.W.; Burgoyne, R.D. Role of myristoylation in the intracellular targeting of neuronal calcium sensor (NCS) proteins. Biochem. Soc. Trans. 2003, 31, 963–965. [Google Scholar] [CrossRef]
- Zernii, E.Y.; Tikhomirova, N.; Philippov, P.; Senin, I. Detection of annexin IV in bovine retinal rods. Biochemistry 2003, 68, 129–134. [Google Scholar]
- Tsvetkov, P.O.; Roman, A.Y.; Baksheeva, V.E.; Nazipova, A.A.; Shevelyova, M.P.; Vladimirov, V.I.; Buyanova, M.F.; Zinchenko, D.V.; Zamyatnin, A.A., Jr.; Devred, F.; et al. Functional Status of Neuronal Calcium Sensor-1 Is Modulated by Zinc Binding. Front. Mol. Neurosci. 2018, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Zernii, E.Y.; Komolov, K.E.; Permyakov, S.E.; Kolpakova, T.; Dell’orco, D.; Poetzsch, A.; Knyazeva, E.L.; Grigoriev, I.I.; Permyakov, E.A.; Senin, I.I.; et al. Involvement of the recoverin C-terminal segment in recognition of the target enzyme rhodopsin kinase. Biochem. J. 2011, 435, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Grigoriev, I.I.; Senin, I.I.; Tikhomirova, N.K.; Komolov, K.E.; Permyakov, S.E.; Zernii, E.Y.; Koch, K.-W.; Philippov, P.P. Synergetic effect of recoverin and calmodulin on regulation of rhodopsin kinase. Front. Mol. Neurosci. 2012, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Permyakov, S.E.; Cherskaya, A.M.; Wasserman, L.A.; Khokhlova, T.I.; Senin, I.I.; Zargarov, A.A.; Zinchenko, D.V.; Zernii, E.Y.; Lipkin, V.M.; Philippov, P.P.; et al. Recoverin is a zinc-binding protein. J. Proteome Res. 2003, 2, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Machaidze, G.G.; Mikeladze, D. Different effects of lectins on the ligand binding of the NMDA receptors and sigma sites in rat brain hippocampus synaptic membranes. Neurochem. Res. 2001, 26, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Weiergraber, O.H.; Senin, I.I.; Zernii, E.Y.; Churumova, V.A.; Kovaleva, N.A.; Nazipova, A.A.; Permyakov, S.E.; Permyakov, E.A.; Philippov, P.P.; Granzin, J.; et al. Tuning of a neuronal calcium sensor. J. Biol. Chem. 2006, 281, 37594–37602. [Google Scholar] [CrossRef] [PubMed]
- GelAnalyzer. Available online: http://www.gelanalyzer.com/ (accessed on 20 January 2020).
- Perevoshchikova, I.V.; Zorov, D.B.; Antonenko, Y.N. Peak intensity analysis as a method for estimation of fluorescent probe binding to artificial and natural nanoparticles: Tetramethylrhodamine uptake by isolated mitochondria. Biochim. Biophys. Acta 2008, 1778, 2182–2190. [Google Scholar] [CrossRef]
- Hess, S.T.; Huang, S.; Heikal, A.A.; Webb, W.W. Biological and chemical applications of fluorescence correlation spectroscopy: A review. Biochemistry 2002, 41, 697–705. [Google Scholar] [CrossRef]
- Petrasek, Z.; Schwille, P. Precise measurement of diffusion coefficients using scanning fluorescence correlation spectroscopy. Biophys. J. 2008, 94, 1437–1448. [Google Scholar] [CrossRef]
- Antonenko, Y.N.; Lapashina, A.S.; Kotova, E.A.; Ramonova, A.A.; Moisenovich, M.M.; Agapov, I.I. Application of Peak Intensity Analysis to Measurements of Protein Binding to Lipid Vesicles and Erythrocytes Using Fluorescence Correlation Spectroscopy: Dependence on Particle Size. J. Membr. Biol. 2017, 250, 77–87. [Google Scholar] [CrossRef]
- Zernii, E.Y.; Nazipova, A.A.; Gancharova, O.S.; Kazakov, A.S.; Serebryakova, M.V.; Zinchenko, D.V.; Tikhomirova, N.K.; Senin, I.I.; Philippov, P.P.; Permyakov, E.A.; et al. Light-induced disulfide dimerization of recoverin under ex vivo and in vivo conditions. Free Radic. Biol. Med. 2015, 83, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Hanwell, M.D.; Curtis, D.E.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminf. 2012, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Wójcikowski, M.; Zielenkiewicz, P.; Siedlecki, P. Open Drug Discovery Toolkit (ODDT): A new open-source player in the drug discovery field. J. Cheminf. 2015, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Söderberg, M.; Edlund, C.; Kristensson, K.; Dallner, G. Lipid compositions of different regions of the human brain during aging. J. Neurochem. 1990, 54, 415–423. [Google Scholar] [CrossRef]
- Maurer-Stroh, S.; Eisenhaber, B.; Eisenhaber, F. N-terminal N-myristoylation of proteins: Refinement of the sequence motif and its taxon-specific differences. J. Mol. Biol. 2002, 317, 523–540. [Google Scholar] [CrossRef]
- Lemmon, M.A. Phosphoinositide recognition domains. Traffic 2003, 4, 201–213. [Google Scholar] [CrossRef]
- Jinno, S.; Jeromin, A.; Roder, J.; Kosaka, T. Immunocytochemical localization of neuronal calcium sensor-1 in the hippocampus and cerebellum of the mouse, with special reference to presynaptic terminals. Neuroscience 2002, 113, 449–461. [Google Scholar] [CrossRef]
- Schaad, N.C.; De Castro, E.; Nef, S.; Hegi, S.; Hinrichsen, R.; Martone, M.E.; Ellisman, M.H.; Sikkink, R.; Rusnak, F.; Sygush, J. Direct modulation of calmodulin targets by the neuronal calcium sensor NCS-1. Proc. Natl. Acad. Sci. USA 1996, 93, 9253–9258. [Google Scholar] [CrossRef]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Stace, C.L.; Ktistakis, N.T. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta 2006, 1761, 913–926. [Google Scholar] [CrossRef]
- Krieger, M.; Krieger, J. Structures and functions of multiligand lipoprotein receptors: Macrophage scavenger receptors and LDL receptor-related protein (LRP). Annu. Rev. Biochem. 1994, 63, 601–637. [Google Scholar] [CrossRef] [PubMed]
- Bouma, B.; de Groot, P.G.; van den Elsen, J.M.; Ravelli, R.B.; Schouten, A.; Simmelink, M.J.; Derksen, R.H.; Kroon, J.; Gros, P. Adhesion mechanism of human β2-glycoprotein I to phospholipids based on its crystal structure. EMBO J. 1999, 18, 5166–5174. [Google Scholar] [CrossRef] [PubMed]
- Nakaoka, T.; Kojima, N.; Ogita, T.; Tsuji, S. Characterization of the Phosphatidylserine-binding Region of Rat MARCKS (Myristoylated, Alanine-rich Protein Kinase C Substrate) its regulation through phosphorylation of serine 152. J. Biol. Chem. 1995, 270, 12147–12151. [Google Scholar] [CrossRef] [PubMed]
- Yagisawa, H.; Sakuma, K.; Paterson, H.F.; Cheung, R.; Allen, V.; Hirata, H.; Watanabe, Y.; Hirata, M.; Williams, R.L.; Katan, M. Replacements of single basic amino acids in the pleckstrin homology domain of phospholipase C-δ1 alter the ligand binding, phospholipase activity, and interaction with the plasma membrane. J. Biol. Chem. 1998, 273, 417–424. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki-Katagiri, N.; Ames, J.B. Neuronal calcium sensor-1 (Ncs1p) is up-regulated by calcineurin to promote Ca2+ tolerance in fission yeast. J. Biol. Chem. 2010, 285, 4405–4414. [Google Scholar] [CrossRef]
- Huttner, I.G.; Strahl, T.; Osawa, M.; King, D.S.; Ames, J.B.; Thorner, J. Molecular interactions of yeast frequenin (Frq1) with the phosphatidylinositol 4-kinase isoform, Pik1. J. Biol. Chem. 2003, 278, 4862–4874. [Google Scholar] [CrossRef]
- Raghu, P.; Joseph, A.; Krishnan, H.; Singh, P.; Saha, S. Phosphoinositides: Regulators of nervous system function in health and disease. Front. Mol. Neurosci. 2019, 12, 208. [Google Scholar] [CrossRef]
- Zheng, Q.; Bobich, J.A.; Vidugiriene, J.; McFadden, S.C.; Thomas, F.; Roder, J.; Jeromin, A. Neuronal calcium sensor-1 facilitates neuronal exocytosis through phosphatidylinositol 4-kinase. J. Neurochem. 2005, 92, 442–451. [Google Scholar] [CrossRef]
- Stephens, L.; Jackson, T.; Hawkins, P. Agonist-stimulated synthesis of phosphatidylinositol (3, 4, 5)-trisphosphate: A new intracellular signalling system? Biochim. Biophys. Acta Mol. Cell Res. 1993, 1179, 27–75. [Google Scholar] [CrossRef]
- Kolay, S.; Basu, U.; Raghu, P. Control of diverse subcellular processes by a single multi-functional lipid phosphatidylinositol 4, 5-bisphosphate [PI (4, 5) P2]. Biochem. J. 2016, 473, 1681–1692. [Google Scholar] [CrossRef]
- Gadi, D.; Wagenknecht-Wiesner, A.; Holowka, D.; Baird, B. Sequestration of phosphoinositides by mutated MARCKS effector domain inhibits stimulated Ca2+ mobilization and degranulation in mast cells. Mol. Biol. Cell 2011, 22, 4908–4917. [Google Scholar] [CrossRef] [PubMed]
- Caroni, P. Actin cytoskeleton regulation through modulation of PI (4, 5) P2 rafts. EMBO J. 2001, 20, 4332–4336. [Google Scholar] [CrossRef] [PubMed]
- Hilfiker, S. Neuronal calcium sensor-1: A multifunctional regulator of secretion. Biochem. Soc. Trans. 2003, 31, 828–832. [Google Scholar] [CrossRef] [PubMed]
- Iwata, K.; Ito, K.; Fukuzaki, A.; Inaki, K.; Haga, T. Dynamin and rab5 regulate GRK2-dependent internalization of dopamine D2 receptors. Eur. J. Biochem. 1999, 263, 596–602. [Google Scholar] [CrossRef] [PubMed]
- Gaidarov, I.; Smith, M.E.; Domin, J.; Keen, J.H. The class II phosphoinositide 3-kinase C2α is activated by clathrin and regulates clathrin-mediated membrane trafficking. Mol. Cell 2001, 7, 443–449. [Google Scholar] [CrossRef]
- Odorizzi, G.; Babst, M.; Emr, S.D. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem. Sci. 2000, 25, 229–235. [Google Scholar] [CrossRef]
- Papadopoulos, T.; Rhee, H.J.; Subramanian, D.; Paraskevopoulou, F.; Mueller, R.; Schultz, C.; Brose, N.; Rhee, J.-S.; Betz, H. Endosomal phosphatidylinositol 3-phosphate promotes gephyrin clustering and GABAergic neurotransmission at inhibitory postsynapses. J. Biol. Chem. 2017, 292, 1160–1177. [Google Scholar] [CrossRef]
- Karim, S.; Mirza, Z.; Ansari, S.A.; Rasool, M.; Iqbal, Z.; Sohrab, S.S.; Kamal, M.A.; Abuzenadah, A.M.; Al-Qahtani, M.H. Transcriptomics study of neurodegenerative disease: Emphasis on synaptic dysfunction mechanism in Alzheimer’s disease. CNS Neurol. Disord. Drug Targets 2014, 13, 1202–1212. [Google Scholar] [CrossRef]
- Morel, E.; Chamoun, Z.; Lasiecka, Z.M.; Chan, R.B.; Williamson, R.L.; Vetanovetz, C.; Dall’Armi, C.; Simoes, S.; Du Jour, K.S.P.; McCabe, B.D. Phosphatidylinositol-3-phosphate regulates sorting and processing of amyloid precursor protein through the endosomal system. Nat. Commun. 2013, 4, 2250. [Google Scholar] [CrossRef]
- Noda, T.; Matsunaga, K.; Taguchi-Atarashi, N.; Yoshimori, T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. In Seminars in Cell & Developmental Biology; Elsevier: Amsterdam, The Netherland, 2010; pp. 671–676. [Google Scholar]
- Chuang, J.-Z.; Zhao, Y.; Sung, C.-H. SARA-regulated vesicular targeting underlies formation of the light-sensing organelle in mammalian rods. Cell 2007, 130, 535–547. [Google Scholar] [CrossRef]
- Guo, X.; Ghalayini, A.J.; Chen, H.; Anderson, R.E. Phosphatidylinositol 3-kinase in bovine photoreceptor rod outer segments. Investig. Ophthalmol. Vis. Sci. 1997, 38, 1873–1882. [Google Scholar]
- Ivanovic, I.; Allen, D.T.; Dighe, R.; Le, Y.Z.; Anderson, R.E.; Rajala, R.V. Phosphoinositide 3-kinase signaling in retinal rod photoreceptors. Investig. Ophthalmol. Vis. Sci. 2011, 52, 6355–6362. [Google Scholar] [CrossRef] [PubMed]
- Sarkes, D.; Rameh, L.E. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem. J. 2010, 428, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Wills, R.C.; Goulden, B.D.; Hammond, G.R. Genetically encoded lipid biosensors. Mol. Biol. Cell 2018, 29, 1526–1532. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baksheeva, V.E.; Nemashkalova, E.L.; Firsov, A.M.; Zalevsky, A.O.; Vladimirov, V.I.; Tikhomirova, N.K.; Philippov, P.P.; Zamyatnin, A.A., Jr.; Zinchenko, D.V.; Antonenko, Y.N.; et al. Membrane Binding of Neuronal Calcium Sensor-1: Highly Specific Interaction with Phosphatidylinositol-3-Phosphate. Biomolecules 2020, 10, 164. https://doi.org/10.3390/biom10020164
Baksheeva VE, Nemashkalova EL, Firsov AM, Zalevsky AO, Vladimirov VI, Tikhomirova NK, Philippov PP, Zamyatnin AA Jr., Zinchenko DV, Antonenko YN, et al. Membrane Binding of Neuronal Calcium Sensor-1: Highly Specific Interaction with Phosphatidylinositol-3-Phosphate. Biomolecules. 2020; 10(2):164. https://doi.org/10.3390/biom10020164
Chicago/Turabian StyleBaksheeva, Viktoriia E., Ekaterina L. Nemashkalova, Alexander M. Firsov, Arthur O. Zalevsky, Vasily I. Vladimirov, Natalia K. Tikhomirova, Pavel P. Philippov, Andrey A. Zamyatnin, Jr., Dmitry V. Zinchenko, Yuri N. Antonenko, and et al. 2020. "Membrane Binding of Neuronal Calcium Sensor-1: Highly Specific Interaction with Phosphatidylinositol-3-Phosphate" Biomolecules 10, no. 2: 164. https://doi.org/10.3390/biom10020164
APA StyleBaksheeva, V. E., Nemashkalova, E. L., Firsov, A. M., Zalevsky, A. O., Vladimirov, V. I., Tikhomirova, N. K., Philippov, P. P., Zamyatnin, A. A., Jr., Zinchenko, D. V., Antonenko, Y. N., Permyakov, S. E., & Zernii, E. Y. (2020). Membrane Binding of Neuronal Calcium Sensor-1: Highly Specific Interaction with Phosphatidylinositol-3-Phosphate. Biomolecules, 10(2), 164. https://doi.org/10.3390/biom10020164







