Structural Analyses on the Deamidation of N-Terminal Asn in the Human N-Degron Pathway
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning, Protein Expression, and Purification
2.2. Mutagenesis
2.3. Crystallography
2.4. Deamidation Assay Quantified by LC-MS Analysis
2.5. Surface Plasmon Resonance (SPR)
2.6. Statistical Analysis
2.7. Data Availability
3. Results
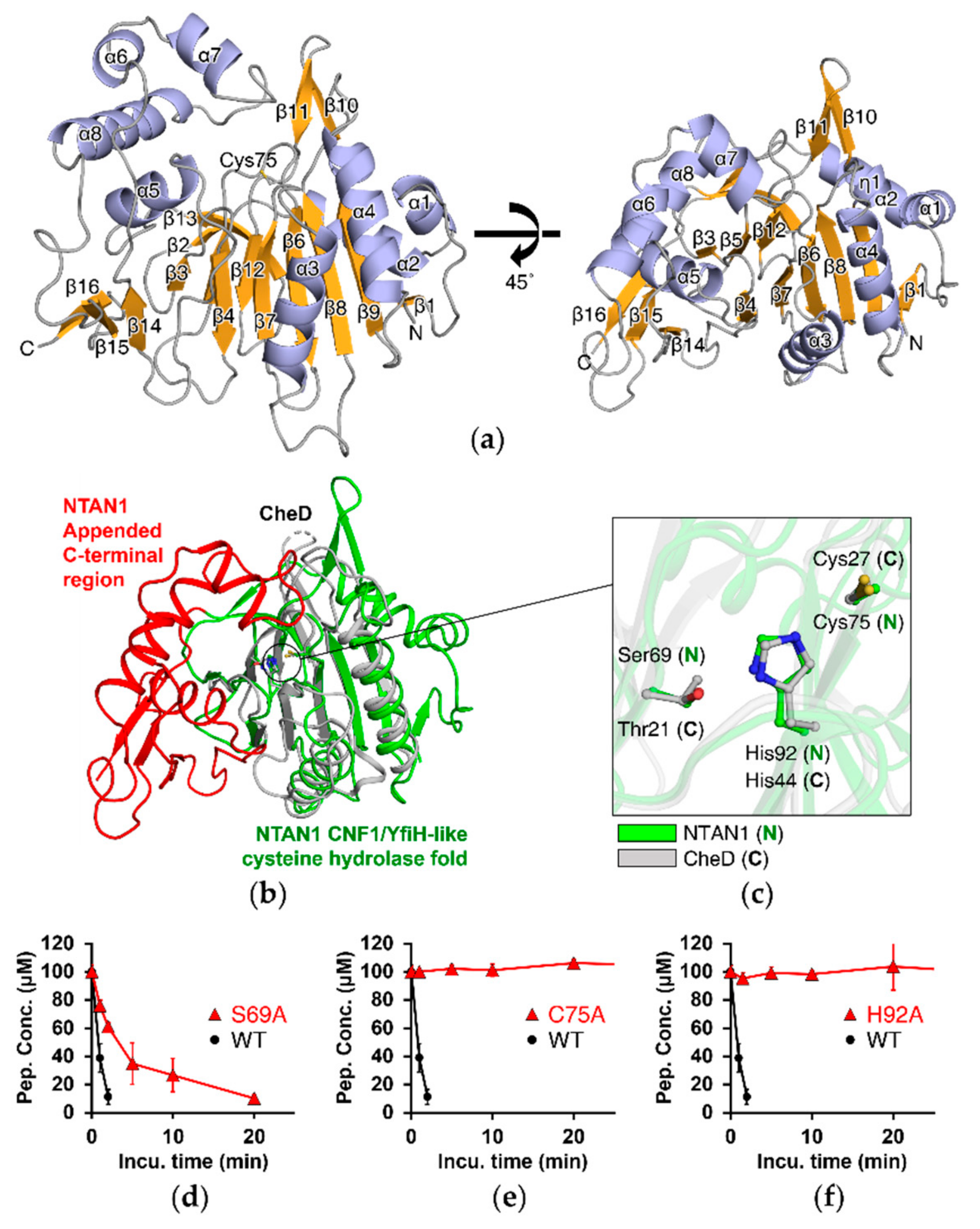
3.1. NTAN1 Structurally Belongs to the CNF1/YfiH-Like Cysteine Hydrolase Family
3.2. Deamidation Activity of NTAN1 Is Mediated by a Canonical Catalytic Triad
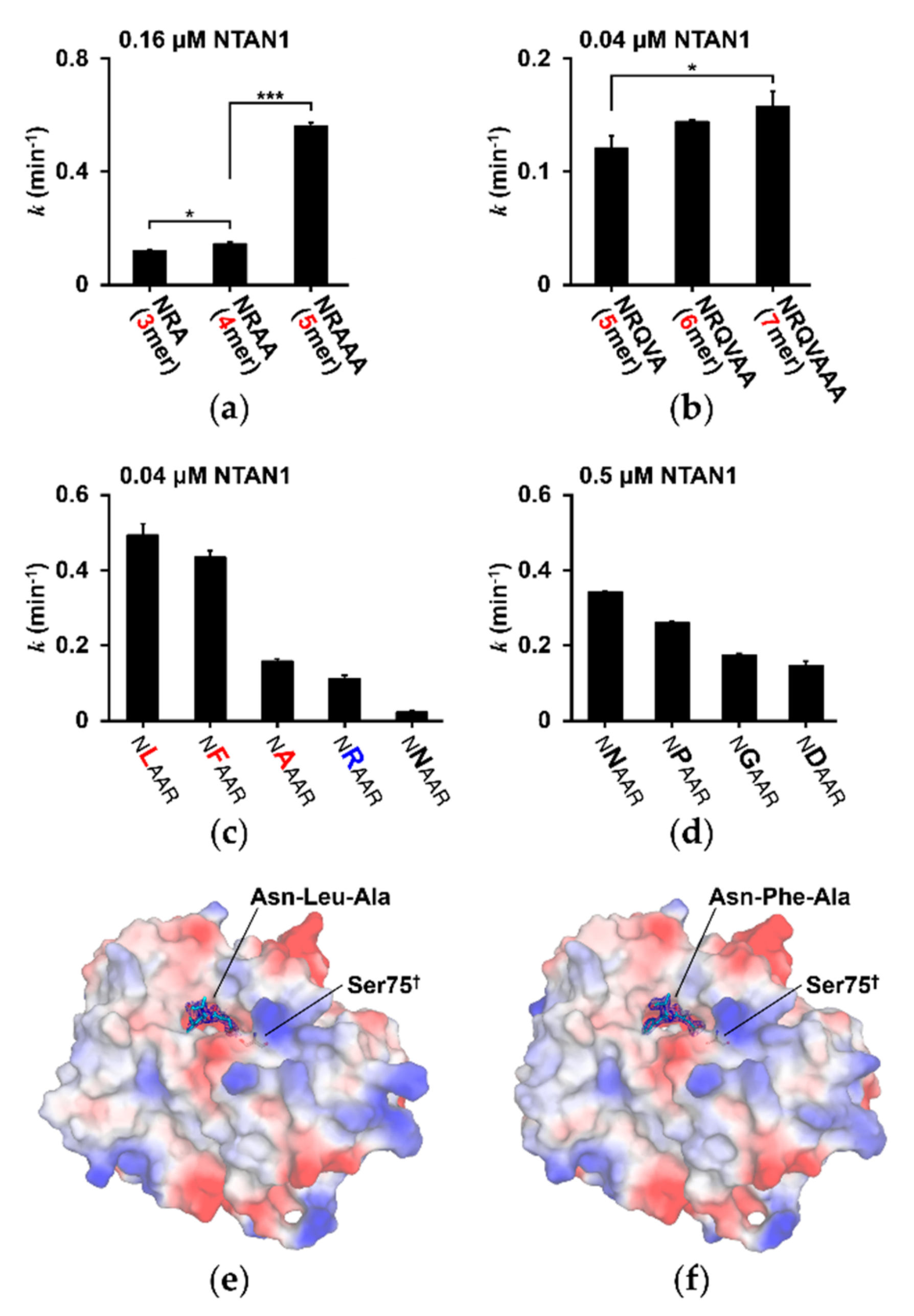
3.3. Substrate Specificity of NTAN1 Is Affected by the Second-Position Residue of Substrates
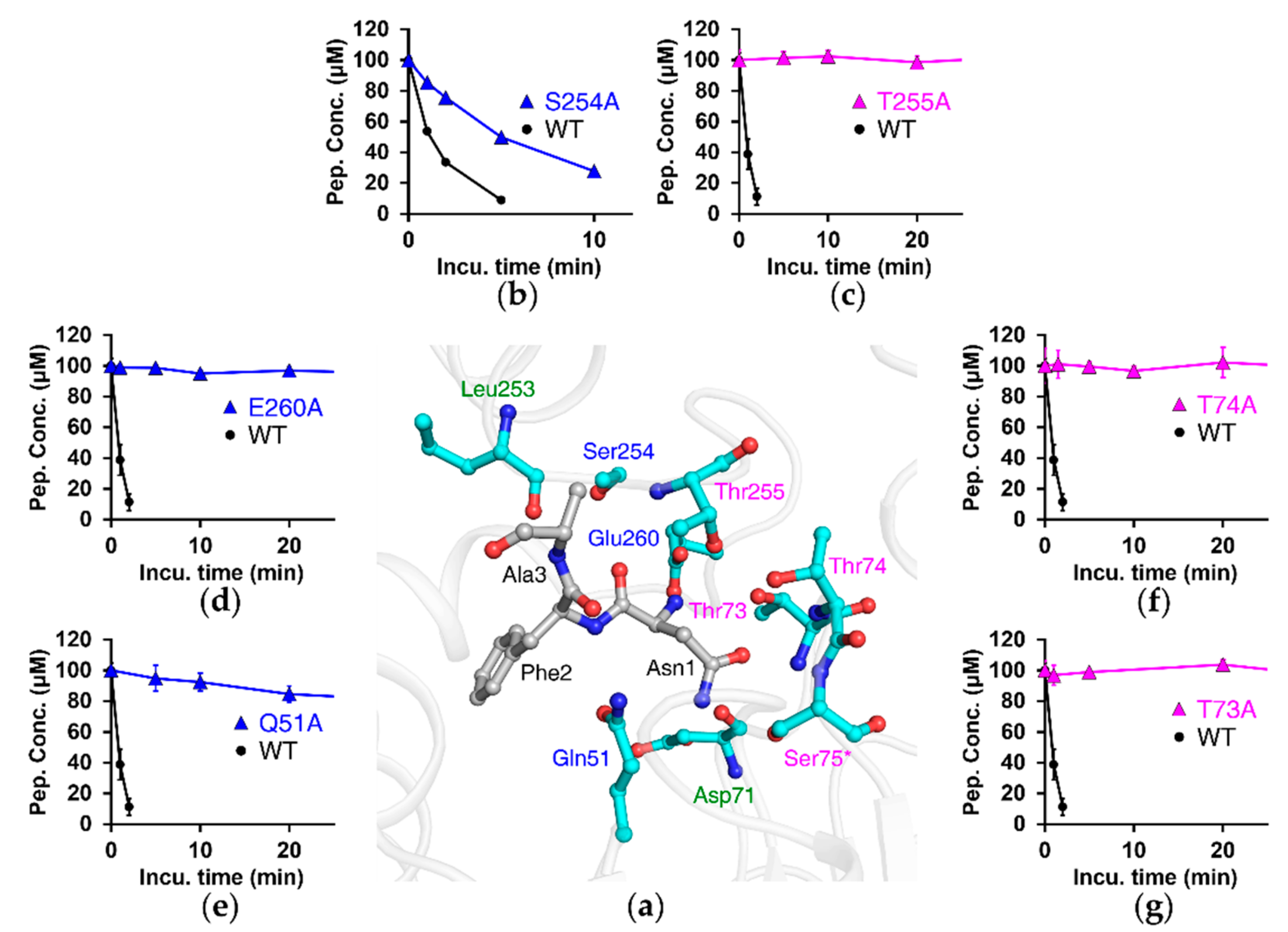
3.4. Substrate-Recognition Mode of NTAN1 to Nt-Asn
3.5. NTAN1-Peptide Complex Structures Also Reveal the Importance of Second-Position and, to a Lesser Degree, Third-Position Amino Acid of Substrates
3.6. The Appended C-Terminal Region of NTAN1 Is a Key Component in the Recognition of Both Nt-Asn and Subsequent Residues
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kayed, R.; Head, E.; Thompson, J.L.; McIntire, T.M.; Milton, S.C.; Cotman, C.W.; Glabe, C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science 2003, 300, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Ross, C.A.; Poirier, M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004, 10, S10–S17. [Google Scholar] [CrossRef] [PubMed]
- Chiti, F.; Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Ben-Zvi, A.; Miller, E.A.; Morimoto, R.I. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. USA 2009, 106, 14914–14919. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Klionsky, D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009, 43, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; McEwan, D.G.; Novak, I.; Dikic, I. A role for ubiquitin in selective autophagy. Mol. Cell 2009, 34, 259–269. [Google Scholar] [CrossRef]
- Nakatogawa, H.; Suzuki, K.; Kamada, Y.; Ohsumi, Y. Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat. Rev. Mol. Cell Biol. 2009, 10, 458–467. [Google Scholar] [CrossRef]
- Akerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef]
- Stolz, A.; Wolf, D.H. Endoplasmic reticulum associated protein degradation: A chaperone assisted journey to hell. Biochim. Biophys. Acta 2010, 1803, 694–705. [Google Scholar] [CrossRef]
- Tyedmers, J.; Mogk, A.; Bukau, B. Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 2010, 11, 777–788. [Google Scholar] [CrossRef]
- Voisine, C.; Pedersen, J.S.; Morimoto, R.I. Chaperone networks: Tipping the balance in protein folding diseases. Neurobiol. Dis. 2010, 40, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Fredrickson, E.K.; Gardner, R.G. Selective destruction of abnormal proteins by ubiquitin-mediated protein quality control degradation. Semin. Cell Dev. Biol. 2012, 23, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1843, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Bachmair, A.; Varshavsky, A. The degradation signal in a short-lived protein. Cell 1989, 56, 1019–1032. [Google Scholar] [CrossRef]
- Johnson, E.S.; Gonda, D.K.; Varshavsky, A. Cis-trans recognition and subunit-specific degradation of short-lived proteins. Nature 1990, 346, 287–291. [Google Scholar] [CrossRef]
- Varshavsky, A. N-degron and C-degron pathways of protein degradation. Proc. Natl. Acad. Sci. USA 2019, 116, 358–366. [Google Scholar] [CrossRef]
- Varshavsky, A. The N-end rule pathway and regulation by proteolysis. Protein Sci. 2011, 20, 1298–1345. [Google Scholar] [CrossRef]
- Lee, K.E.; Heo, J.E.; Kim, J.M.; Hwang, C.S. N-Terminal Acetylation-Targeted N-End Rule Proteolytic System: The Ac/N-End Rule Pathway. Mol. Cells 2016, 39, 169–178. [Google Scholar] [CrossRef]
- Chen, S.J.; Wu, X.; Wadas, B.; Oh, J.H.; Varshavsky, A. An N-end rule pathway that recognizes proline and destroys gluconeogenic enzymes. Science 2017, 355. [Google Scholar] [CrossRef]
- Gonda, D.K.; Bachmair, A.; Wunning, I.; Tobias, J.W.; Lane, W.S.; Varshavsky, A. Universality and structure of the N-end rule. J. Biol. Chem. 1989, 264, 16700–16712. [Google Scholar] [PubMed]
- Stewart, A.E.; Arfin, S.M.; Bradshaw, R.A. Protein NH2-terminal asparagine deamidase. Isolation and characterization of a new enzyme. J. Biol. Chem. 1994, 269, 23509–23517. [Google Scholar] [PubMed]
- Stewart, A.E.; Arfin, S.M.; Bradshaw, R.A. The sequence of porcine protein NH2-terminal asparagine amidohydrolase. A new component of the N-end Rule pathway. J. Biol. Chem. 1995, 270, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Grigoryev, S.; Stewart, A.E.; Kwon, Y.T.; Arfin, S.M.; Bradshaw, R.A.; Jenkins, N.A.; Copeland, N.G.; Varshavsky, A. A mouse amidase specific for N-terminal asparagine. The gene, the enzyme, and their function in the N-end rule pathway. J. Biol. Chem. 1996, 271, 28521–28532. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Balogh, S.A.; Davydov, I.V.; Kashina, A.S.; Yoon, J.K.; Xie, Y.; Gaur, A.; Hyde, L.; Denenberg, V.H.; Varshavsky, A. Altered activity, social behavior, and spatial memory in mice lacking the NTAN1p amidase and the asparagine branch of the N-end rule pathway. Mol. Cell. Biol. 2000, 20, 4135–4148. [Google Scholar] [CrossRef]
- Wang, H.; Piatkov, K.I.; Brower, C.S.; Varshavsky, A. Glutamine-specific N-terminal amidase, a component of the N-end rule pathway. Mol. Cell 2009, 34, 686–695. [Google Scholar] [CrossRef]
- Park, M.S.; Bitto, E.; Kim, K.R.; Bingman, C.A.; Miller, M.D.; Kim, H.J.; Han, B.W.; Phillips, G.N., Jr. Crystal structure of human protein N-terminal glutamine amidohydrolase, an initial component of the N-end rule pathway. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Vicente, J.; Mendiondo, G.M.; Pauwels, J.; Pastor, V.; Izquierdo, Y.; Naumann, C.; Movahedi, M.; Rooney, D.; Gibbs, D.J.; Smart, K.; et al. Distinct branches of the N-end rule pathway modulate the plant immune response. New Phytol. 2019, 221, 988–1000. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Kashina, A.S.; Davydov, I.V.; Hu, R.G.; An, J.Y.; Seo, J.W.; Du, F.; Varshavsky, A. An essential role of N-terminal arginylation in cardiovascular development. Science 2002, 297, 96–99. [Google Scholar] [CrossRef]
- Hu, R.G.; Sheng, J.; Qi, X.; Xu, Z.; Takahashi, T.T.; Varshavsky, A. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 2005, 437, 981–986. [Google Scholar] [CrossRef]
- Lee, M.J.; Tasaki, T.; Moroi, K.; An, J.Y.; Kimura, S.; Davydov, I.V.; Kwon, Y.T. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 15030–15035. [Google Scholar] [CrossRef] [PubMed]
- White, M.D.; Klecker, M.; Hopkinson, R.J.; Weits, D.A.; Mueller, C.; Naumann, C.; O’Neill, R.; Wickens, J.; Yang, J.; Brooks-Bartlett, J.C.; et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, T.; Sriram, S.M.; Park, K.S.; Kwon, Y.T. The N-end rule pathway. Annu. Rev. Biochem. 2012, 81, 261–289. [Google Scholar] [CrossRef] [PubMed]
- Gibbs, D.J.; Bacardit, J.; Bachmair, A.; Holdsworth, M.J. The eukaryotic N-end rule pathway: Conserved mechanisms and diverse functions. Trends Cell Biol. 2014, 24, 603–611. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Xia, Z.; Davydov, I.V.; Lecker, S.H.; Varshavsky, A. Construction and analysis of mouse strains lacking the ubiquitin ligase UBR1 (E3alpha) of the N-end rule pathway. Mol. Cell. Biol. 2001, 21, 8007–8021. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Xia, Z.; An, J.Y.; Tasaki, T.; Davydov, I.V.; Seo, J.W.; Sheng, J.; Xie, Y.; Varshavsky, A. Female lethality and apoptosis of spermatocytes in mice lacking the UBR2 ubiquitin ligase of the N-end rule pathway. Mol. Cell. Biol. 2003, 23, 8255–8271. [Google Scholar] [CrossRef]
- Tasaki, T.; Mulder, L.C.; Iwamatsu, A.; Lee, M.J.; Davydov, I.V.; Varshavsky, A.; Muesing, M.; Kwon, Y.T. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol. Cell. Biol. 2005, 25, 7120–7136. [Google Scholar] [CrossRef]
- Sriram, S.M.; Banerjee, R.; Kane, R.S.; Kwon, Y.T. Multivalency-assisted control of intracellular signaling pathways: Application for ubiquitin- dependent N-end rule pathway. Chem. Biol. 2009, 16, 121–131. [Google Scholar] [CrossRef]
- Sriram, S.M.; Kwon, Y.T. The molecular principles of N-end rule recognition. Nat. Struct. Mol. Biol. 2010, 17, 1164–1165. [Google Scholar] [CrossRef]
- Tasaki, T.; Kim, S.T.; Zakrzewska, A.; Lee, B.E.; Kang, M.J.; Yoo, Y.D.; Cha-Molstad, H.J.; Hwang, J.; Soung, N.K.; Sung, K.S.; et al. UBR box N-recognin-4 (UBR4), an N-recognin of the N-end rule pathway, and its role in yolk sac vascular development and autophagy. Proc. Natl. Acad. Sci. USA 2013, 110, 3800–3805. [Google Scholar] [CrossRef]
- Sriram, S.M.; Kim, B.Y.; Kwon, Y.T. The N-end rule pathway: Emerging functions and molecular principles of substrate recognition. Nat. Rev. Mol. Cell Biol. 2011, 12, 735–747. [Google Scholar] [CrossRef] [PubMed]
- Cha-Molstad, H.; Yu, J.E.; Feng, Z.; Lee, S.H.; Kim, J.G.; Yang, P.; Han, B.; Sung, K.W.; Yoo, Y.D.; Hwang, J.; et al. p62/SQSTM1/Sequestosome-1 is an N-recognin of the N-end rule pathway which modulates autophagosome biogenesis. Nat. Commun. 2017, 8, 102. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.D.; Mun, S.R.; Ji, C.H.; Sung, K.W.; Kang, K.Y.; Heo, A.J.; Lee, S.H.; An, J.Y.; Hwang, J.; Xie, X.Q.; et al. N-terminal arginylation generates a bimodal degron that modulates autophagic proteolysis. Proc. Natl. Acad. Sci. USA 2018, 115, E2716–E2724. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, D.; Jones, D.A. Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol. Plant Microbe Interact. 2005, 18, 1258–1268. [Google Scholar] [CrossRef]
- Piatkov, K.I.; Colnaghi, L.; Bekes, M.; Varshavsky, A.; Huang, T.T. The auto-generated fragment of the Usp1 deubiquitylase is a physiological substrate of the N-end rule pathway. Mol. Cell 2012, 48, 926–933. [Google Scholar] [CrossRef]
- Baker, R.T.; Varshavsky, A. Yeast N-terminal amidase. A new enzyme and component of the N-end rule pathway. J. Biol. Chem. 1995, 270, 12065–12074. [Google Scholar] [CrossRef]
- Kim, M.K.; Oh, S.J.; Lee, B.G.; Song, H.K. Structural basis for dual specificity of yeast N-terminal amidase in the N-end rule pathway. Proc. Natl. Acad. Sci. USA 2016, 113, 12438–12443. [Google Scholar] [CrossRef]
- Cantor, J.R.; Stone, E.M.; Georgiou, G. Expression and biochemical characterization of the human enzyme N-terminal asparagine amidohydrolase. Biochemistry 2011, 50, 3025–3033. [Google Scholar] [CrossRef]
- Balogh, S.A.; Kwon, Y.T.; Denenberg, V.H. Varying intertrial interval reveals temporally defined memory deficits and enhancements in NTAN1-deficient mice. Learn. Mem. 2000, 7, 279–286. [Google Scholar] [CrossRef][Green Version]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, T.C. SOLVE and RESOLVE: Automated structure solution and density modification. Methods Enzymol. 2003, 374, 22–37. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.J.; Grosse-Kunstleve, R.W.; Adams, P.D.; Winn, M.D.; Storoni, L.C.; Read, R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007, 40, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012, 68, 352–367. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Holm, L.; Laakso, L.M. Dali server update. Nucleic Acids Res. 2016, 44, W351–W355. [Google Scholar] [CrossRef]
- Buetow, L.; Flatau, G.; Chiu, K.; Boquet, P.; Ghosh, P. Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1. Nat. Struct. Biol. 2001, 8, 584–588. [Google Scholar] [CrossRef]
- Chao, X.; Muff, T.J.; Park, S.Y.; Zhang, S.; Pollard, A.M.; Ordal, G.W.; Bilwes, A.M.; Crane, B.R. A receptor-modifying deamidase in complex with a signaling phosphatase reveals reciprocal regulation. Cell 2006, 124, 561–571. [Google Scholar] [CrossRef][Green Version]
- Kim, Y.; Maltseva, N.; Dementieva, I.; Collart, F.; Holzle, D.; Joachimiak, A. Crystal structure of hypothetical protein YfiH from Shigella flexneri at 2 Å resolution. Proteins 2006, 63, 1097–1101. [Google Scholar] [CrossRef]
- Cruz-Migoni, A.; Hautbergue, G.M.; Artymiuk, P.J.; Baker, P.J.; Bokori-Brown, M.; Chang, C.T.; Dickman, M.J.; Essex-Lopresti, A.; Harding, S.V.; Mahadi, N.M.; et al. A Burkholderia pseudomallei toxin inhibits helicase activity of translation factor eIF4A. Science 2011, 334, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Pulido, L.; Ponting, C.P. Vasohibins: New transglutaminase-like cysteine proteases possessing a non-canonical Cys-His-Ser catalytic triad. Bioinformatics 2016, 32, 1441–1445. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, J.S.; Lee, J.-Y.; Nguyen, Y.T.K.; Kang, N.-W.; Oh, E.K.; Jang, D.M.; Kim, H.-J.; Kim, D.-D.; Han, B.W. Structural Analyses on the Deamidation of N-Terminal Asn in the Human N-Degron Pathway. Biomolecules 2020, 10, 163. https://doi.org/10.3390/biom10010163
Park JS, Lee J-Y, Nguyen YTK, Kang N-W, Oh EK, Jang DM, Kim H-J, Kim D-D, Han BW. Structural Analyses on the Deamidation of N-Terminal Asn in the Human N-Degron Pathway. Biomolecules. 2020; 10(1):163. https://doi.org/10.3390/biom10010163
Chicago/Turabian StylePark, Joon Sung, Jae-Young Lee, Yen Thi Kim Nguyen, Nae-Won Kang, Eun Kyung Oh, Dong Man Jang, Hyun-Jung Kim, Dae-Duk Kim, and Byung Woo Han. 2020. "Structural Analyses on the Deamidation of N-Terminal Asn in the Human N-Degron Pathway" Biomolecules 10, no. 1: 163. https://doi.org/10.3390/biom10010163
APA StylePark, J. S., Lee, J.-Y., Nguyen, Y. T. K., Kang, N.-W., Oh, E. K., Jang, D. M., Kim, H.-J., Kim, D.-D., & Han, B. W. (2020). Structural Analyses on the Deamidation of N-Terminal Asn in the Human N-Degron Pathway. Biomolecules, 10(1), 163. https://doi.org/10.3390/biom10010163





