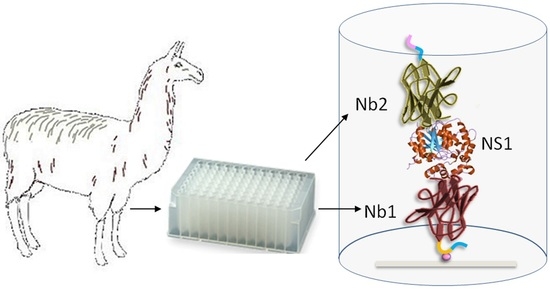
Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Llama Immunization and Library Construction
2.3. Purification of Immune Llama Immunoglobulins
2.4. Panning Strategies for the Selection of ZVNS1-Specific Antibodies
2.5. Nanobody Expression
2.6. ELISA Method for Selection of Capturing Nanobodies
2.7. Large-Scale Production of Biotinylated and HA-Tagged Nbs
2.8. Pairwise Selection of Nanobodies
2.9. Nanobody Sandwich ELISA for the Detection of ZVNS1 in Serum Samples
3. Results and Discussion
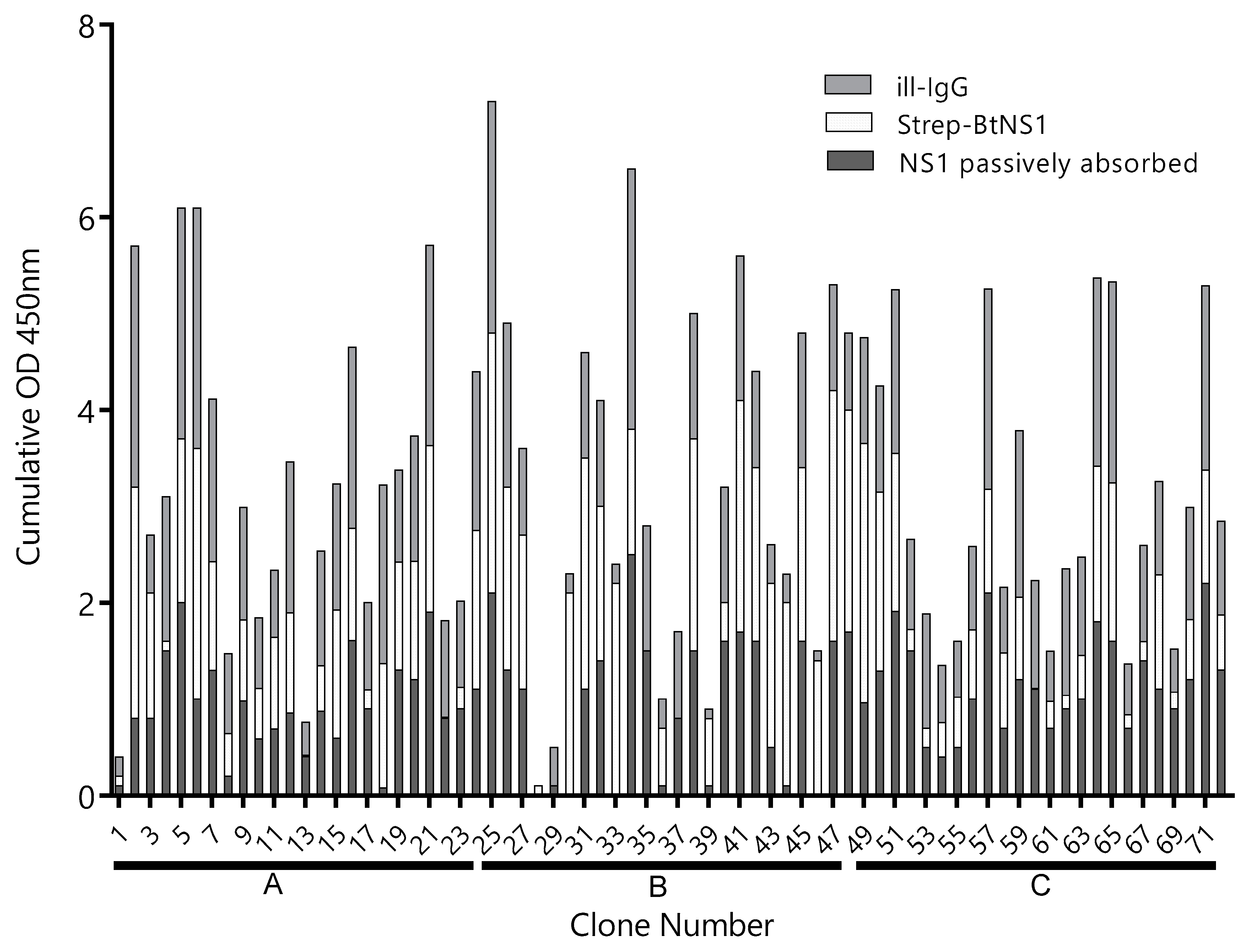
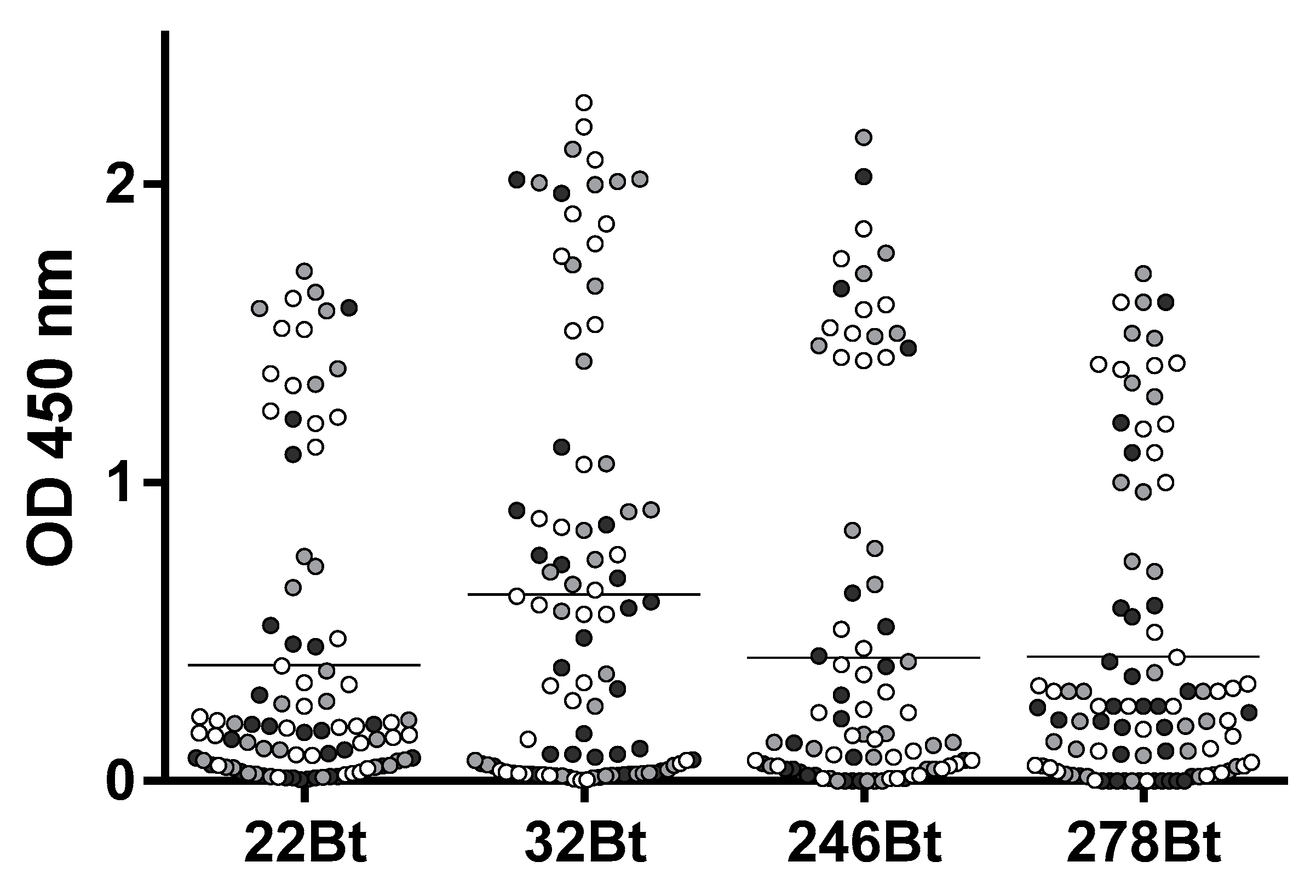
3.1. To Promote a Broad Representation of the NS1 Epitopes, Different Antigen Immobilization Strategies Were Used to Pan the Nb Library
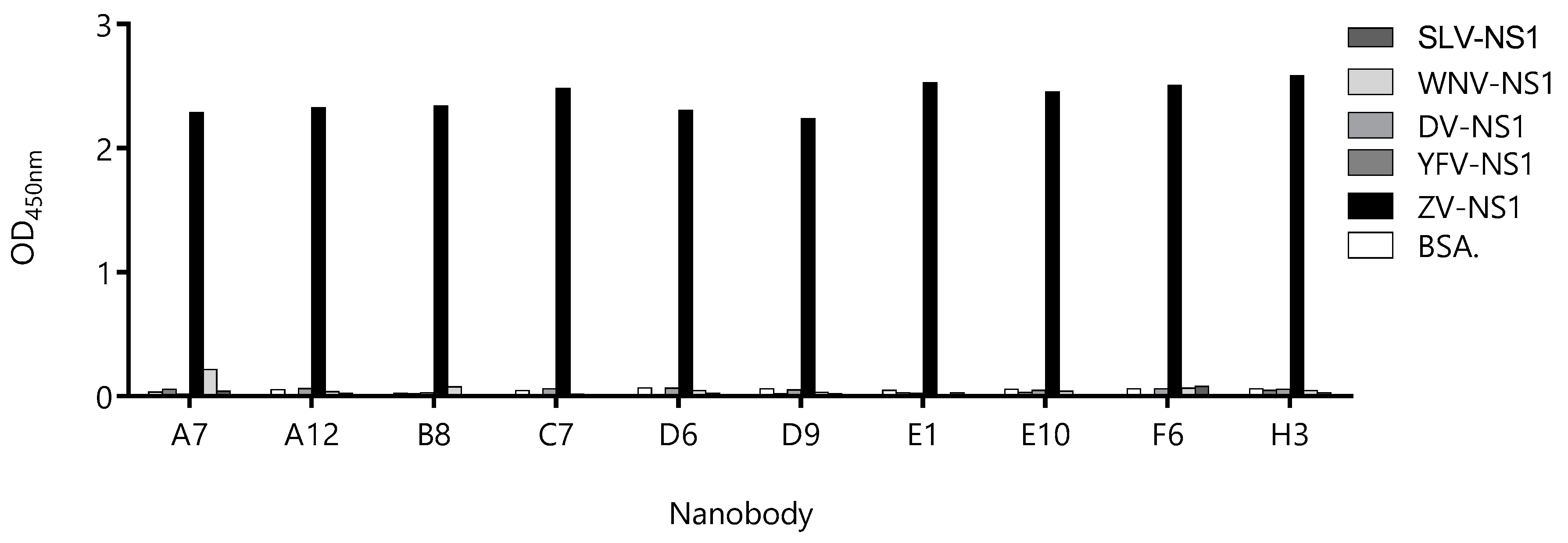
3.2. Most Capture Nbs Had Negligible Cross-Reactivity with Other Flavivirus NS1 Proteins
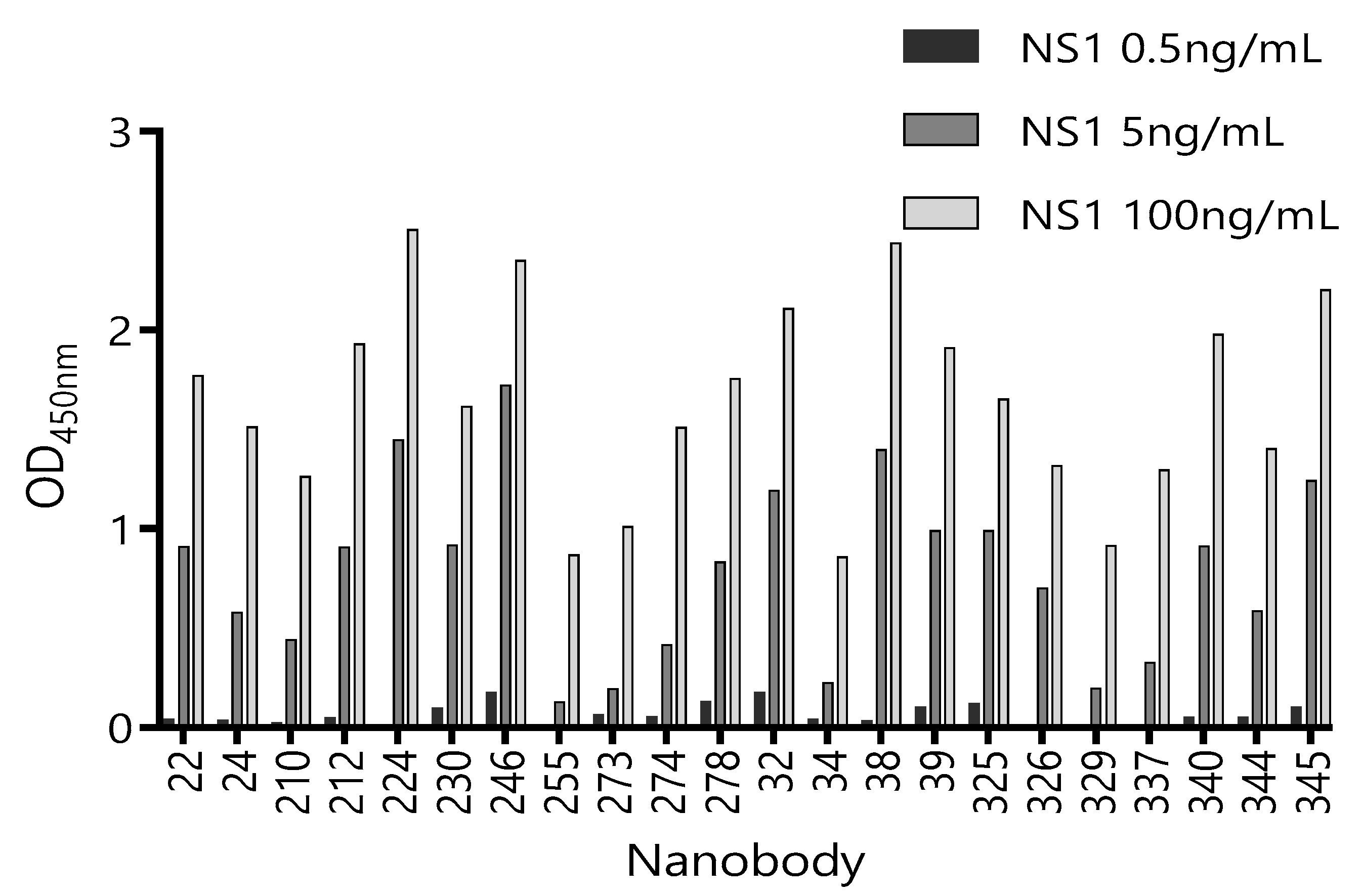
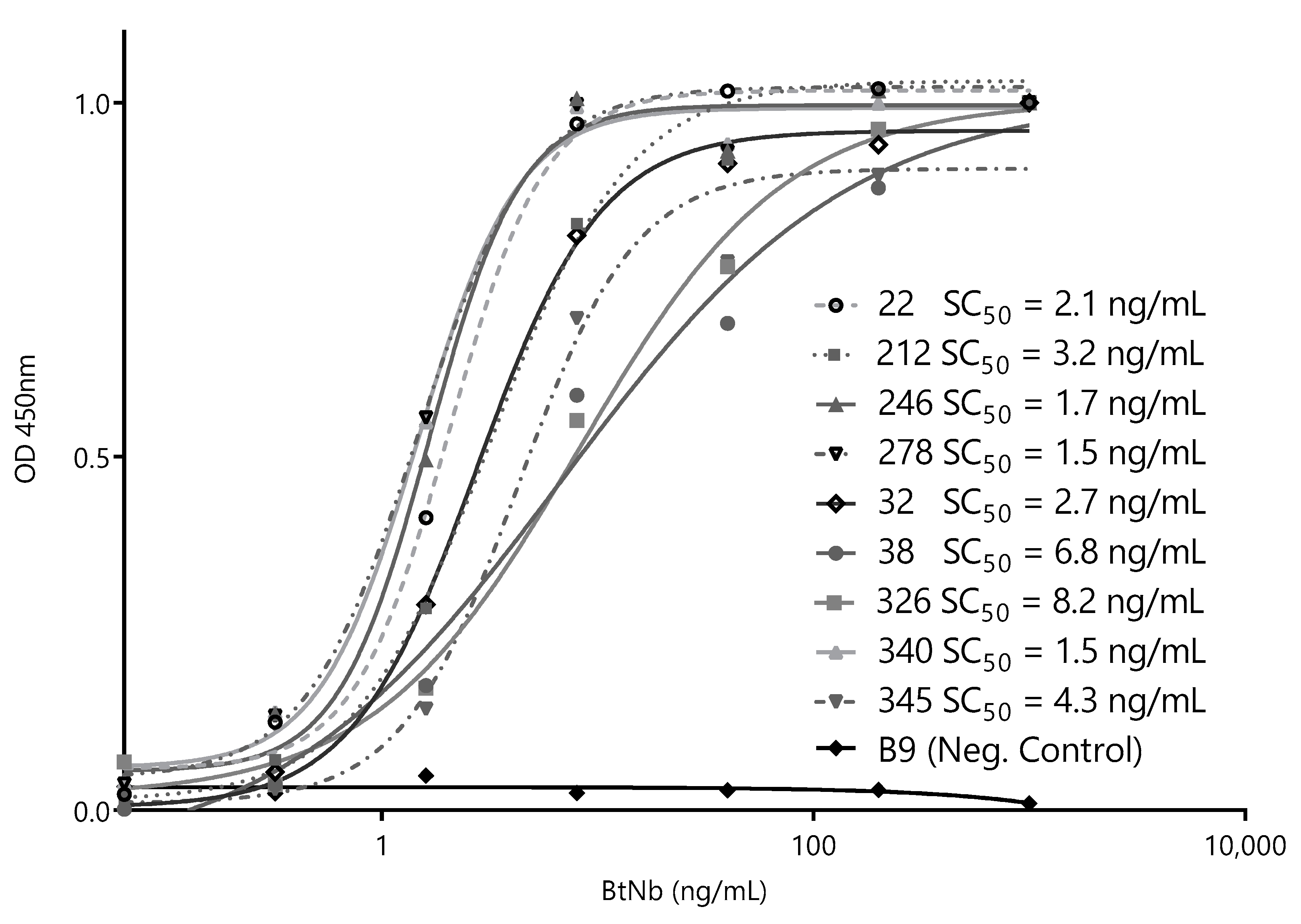
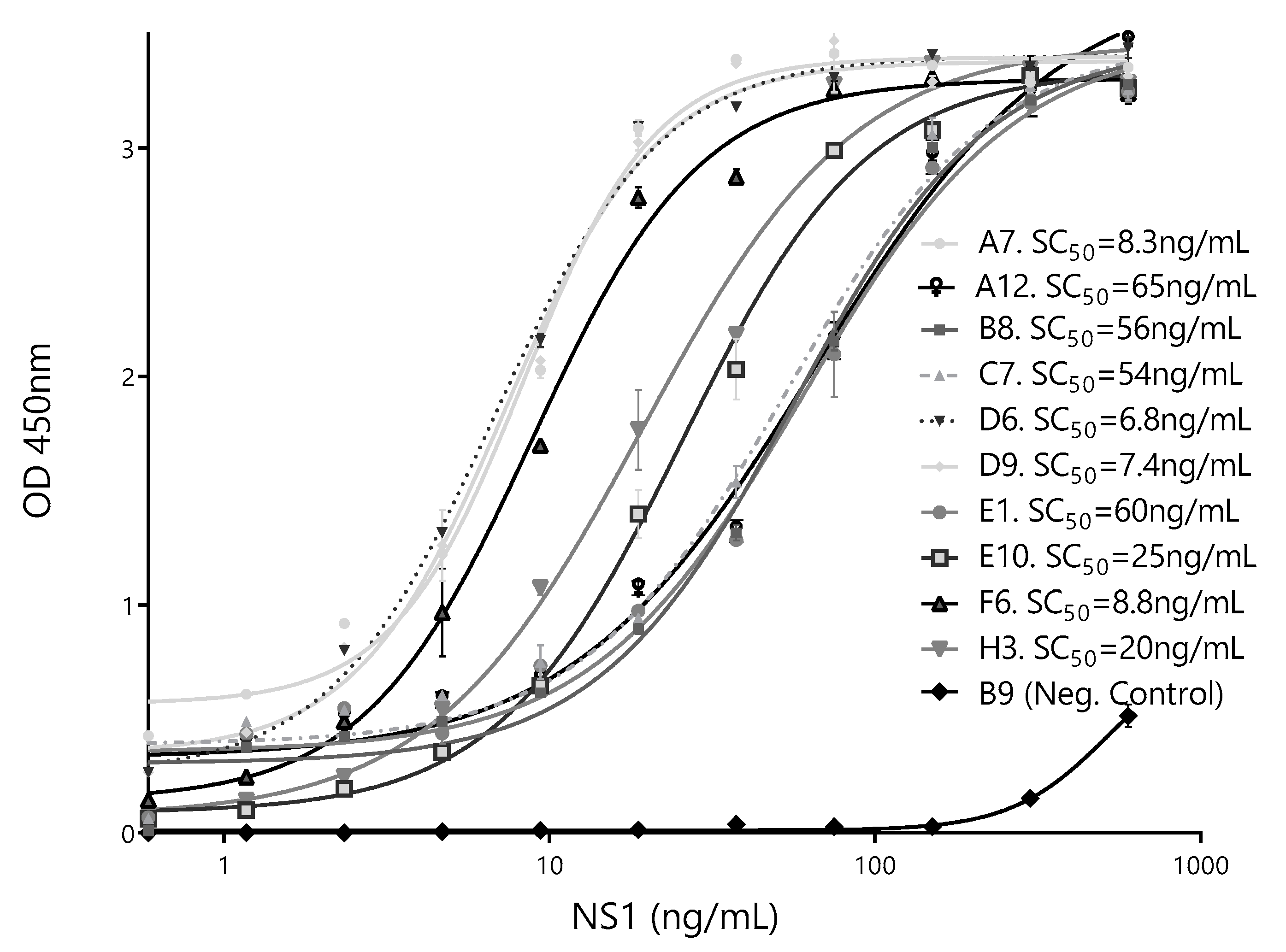
3.3. Selection of Best Nanobody Pair for the Detection of ZVNS1
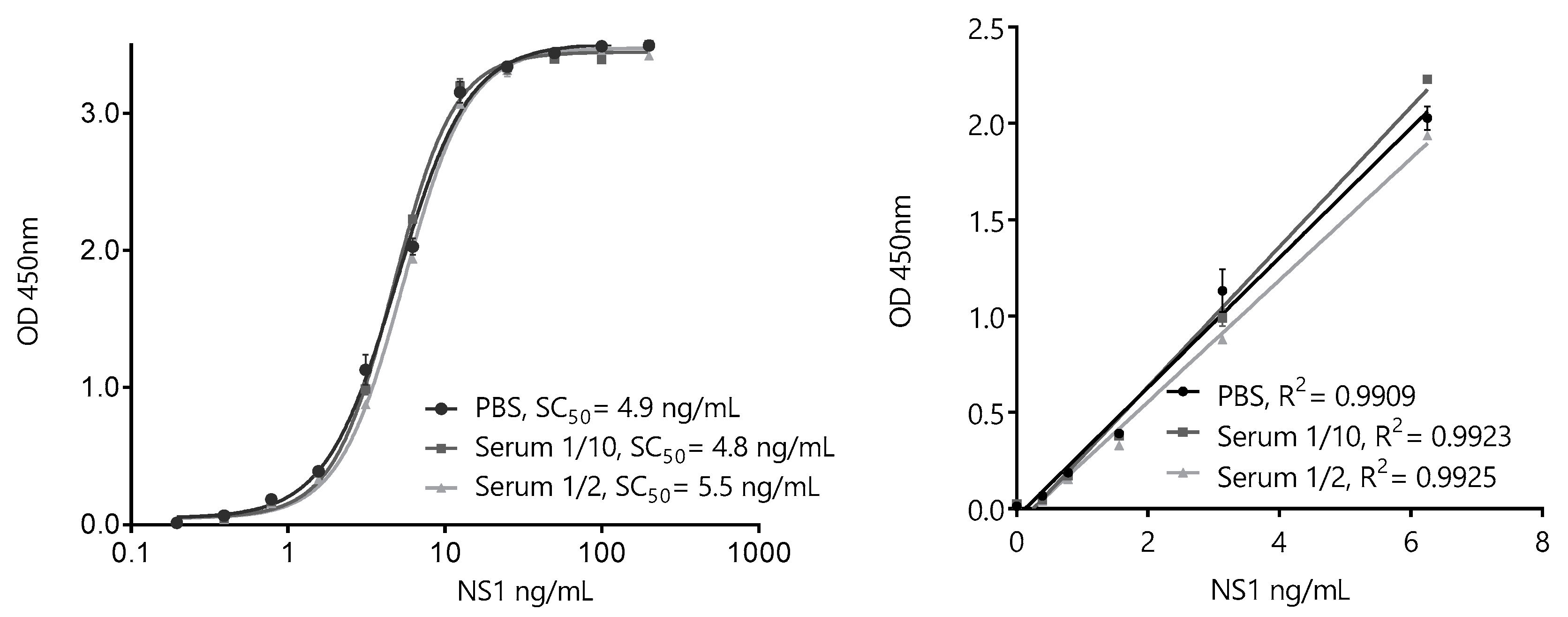
3.4. Development of a Nano-Sandwich ELISA for Quantification of ZVNS1
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Heukelbach, J.; Alencar, C.H.; Kelvin, A.A.; de Oliveira, W.K.; de Goes Cavalcanti, L.P. Zika virus outbreak in Brazil. J. Infect. Dev. Ctries. 2016, 10, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Azevedo, R.; Kraemer, M.U.G.; Souza, R.; Cunha, M.S.; Hill, S.C.; Theze, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; et al. Zika virus in the Americas: Early epidemiological and genetic findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [PubMed]
- Eppes, C.; Rac, M.; Dunn, J.; Versalovic, J.; Murray, K.O.; Suter, M.A.; Sanz Cortes, M.; Espinoza, J.; Seferovic, M.D.; Lee, W.; et al. Testing for Zika virus infection in pregnancy: Key concepts to deal with an emerging epidemic. Am. J. Obstet. Gynecol. 2017, 216, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef]
- Landry, M.L.; St George, K. Laboratory diagnosis of Zika virus infection. Arch. Pathol. Lab. Med. 2017, 141, 60–67. [Google Scholar] [CrossRef]
- Rossini, G.; Gaibani, P.; Vocale, C.; Cagarelli, R.; Landini, M.P. Comparison of Zika virus (ZIKV) RNA detection in plasma, whole blood and urine—Case series of travel-associated ZIKV infection imported to Italy, 2016. J. Infect. 2017, 75, 242–245. [Google Scholar] [CrossRef]
- Priyamvada, L.; Quicke, K.M.; Hudson, W.H.; Onlamoon, N.; Sewatanon, J.; Edupuganti, S.; Pattanapanyasat, K.; Chokephaibulkit, K.; Mulligan, M.J.; Wilson, P.C.; et al. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc. Natl. Acad. Sci. USA 2016, 113, 7852–7857. [Google Scholar] [CrossRef]
- Zaidi, M.B.; Cedillo-Barron, L.; Gonzalez, Y.A.M.E.; Garcia-Cordero, J.; Campos, F.D.; Namorado-Tonix, K.; Perez, F. Serological tests reveal significant cross-reactive human antibody responses to Zika and Dengue viruses in the Mexican population. Acta Trop. 2020, 201, 105201. [Google Scholar] [CrossRef]
- Felix, A.C.; Souza, N.C.S.; Figueiredo, W.M.; Costa, A.A.; Inenami, M.; da Silva, R.M.G.; Levi, J.E.; Pannuti, C.S.; Romano, C.M. Cross reactivity of commercial anti-dengue immunoassays in patients with acute Zika virus infection. J. Med. Virol. 2017, 89, 1477–1479. [Google Scholar] [CrossRef]
- Muller, D.A.; Young, P.R. The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antivir. Res. 2013, 98, 192–208. [Google Scholar] [CrossRef] [PubMed]
- Gutsche, I.; Coulibaly, F.; Voss, J.E.; Salmon, J.; d’Alayer, J.; Ermonval, M.; Larquet, E.; Charneau, P.; Krey, T.; Megret, F.; et al. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. USA 2011, 108, 8003–8008. [Google Scholar] [CrossRef] [PubMed]
- Alcon, S.; Talarmin, A.; Debruyne, M.; Falconar, A.; Deubel, V.; Flamand, M. Enzyme-linked immunosorbent assay specific to Dengue virus type 1 nonstructural protein NS1 reveals circulation of the antigen in the blood during the acute phase of disease in patients experiencing primary or secondary infections. J. Clin. Microbiol. 2002, 40, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Allonso, D.; Meneses, M.D.; Fernandes, C.A.; Ferreira, D.F.; Mohana-Borges, R. Assessing positivity and circulating levels of NS1 in samples from a 2012 dengue outbreak in Rio de Janeiro, Brazil. PLoS ONE 2014, 9, e113634. [Google Scholar] [CrossRef] [PubMed]
- Russo, F.B.; Jungmann, P.; Beltrao-Braga, P.C.B. Zika infection and the development of neurological defects. Cell Microbiol. 2017, 19. [Google Scholar] [CrossRef] [PubMed]
- Andreata-Santos, R.; Pereira, S.S.; Pereira, L.R.; Felix, A.C.; Romano, C.M.; Ferreira, L.C.S. Specificity of NS1-based immunochromatographic tests for dengue virus with regard to the Zika virus protein. Int. J. Infect. Dis. 2020, 95, 276–278. [Google Scholar] [CrossRef]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef]
- Gonzalez-Sapienza, G.; Rossotti, M.A.; Tabares-da Rosa, S. Single-domain antibodies as versatile affinity reagents for analytical and diagnostic applications. Front. Immunol. 2017, 8, 977. [Google Scholar] [CrossRef]
- Muyldermans, S. Nanobodies: Natural single-domain antibodies. Annu. Rev. Biochem. 2013, 82, 775–797. [Google Scholar] [CrossRef]
- van der Linden, R.H.; Frenken, L.G.; de Geus, B.; Harmsen, M.M.; Ruuls, R.C.; Stok, W.; de Ron, L.; Wilson, S.; Davis, P.; Verrips, C.T. Comparison of physical chemical properties of llama VHH antibody fragments and mouse monoclonal antibodies. Biochim. Biophys. Acta 1999, 1431, 37–46. [Google Scholar] [CrossRef]
- Rossotti, M.A.; Pirez, M.; Gonzalez-Techera, A.; Cui, Y.; Bever, C.S.; Lee, K.S.; Morisseau, C.; Leizagoyen, C.; Gee, S.; Hammock, B.D.; et al. Method for sorting and pairwise selection of nanobodies for the development of highly sensitive sandwich immunoassays. Anal. Chem. 2015, 87, 11907–11914. [Google Scholar] [CrossRef]
- Delfin-Riela, T.; Rossotti, M.A.; Echaides, C.; Gonzalez-Sapienza, G. A nanobody-based test for highly sensitive detection of hemoglobin in fecal samples. Anal. Bioanal. Chem. 2020, 412, 389–396. [Google Scholar] [CrossRef]
- Tabares-da Rosa, S.; Rossotti, M.; Carleiza, C.; Carrion, F.; Pritsch, O.; Ahn, K.C.; Last, J.A.; Hammock, B.D.; Gonzalez-Sapienza, G. Competitive selection from single domain antibody libraries allows isolation of high-affinity antihapten antibodies that are not favored in the llama immune response. Anal. Chem. 2011, 83, 7213–7220. [Google Scholar] [CrossRef] [PubMed]
- Rossotti, M.; Tabares, S.; Alfaya, L.; Leizagoyen, C.; Moron, G.; Gonzalez-Sapienza, G. Streamlined method for parallel identification of single domain antibodies to membrane receptors on whole cells. Biochim. Biophys. Acta 2015, 1850, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Chapman-Smith, A.; Turner, D.L.; Cronan, J.E., Jr.; Morris, T.W.; Wallace, J.C. Expression, biotinylation and purification of a biotin-domain peptide from the biotin carboxy carrier protein of Escherichia coli acetyl-CoA carboxylase. Biochem. J. 1994, 302, 881–887. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, M.M.; Ruuls, R.C.; Nijman, I.J.; Niewold, T.A.; Frenken, L.G.; de Geus, B. Llama heavy-chain V regions consist of at least four distinct subfamilies revealing novel sequence features. Mol. Immunol. 2000, 37, 579–590. [Google Scholar] [CrossRef]
- Song, H.; Qi, J.; Haywood, J.; Shi, Y.; Gao, G.F. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat. Struct. Mol. Biol. 2016, 23, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Bosch, I.; de Puig, H.; Hiley, M.; Carre-Camps, M.; Perdomo-Celis, F.; Narvaez, C.F.; Salgado, D.M.; Senthoor, D.; O’Grady, M.; Phillips, E.; et al. Rapid antigen tests for dengue virus serotypes and Zika virus in patient serum. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz-Hernandez, S.I.; Flores-Aguilar, H.; Gonzalez-Mateos, S.; Lopez-Martinez, I.; Alpuche-Aranda, C.; Ludert, J.E.; del Angel, R.M. Determination of viremia and concentration of circulating nonstructural protein 1 in patients infected with dengue virus in Mexico. Am. J. Trop. Med. Hyg. 2013, 88, 446–454. [Google Scholar] [CrossRef]
- SANTE/11813/2017. Available online: https://ec.europa.eu/food/sites/food/files/plant/docs/pesticides_mrl_guidelines_wrkdoc_2017-11813.pdf (accessed on 24 November 2020).

| Unspiked | Spiked ZVNS1 1.50 ng/mL | Spiked ZVNS1 4.50 ng/mL | |||
|---|---|---|---|---|---|
| Sample | ZVNS1 ng/mL | ZVNS1 ng/mL | % Recovery | ZVNS1 ng/mL | % Recovery |
| 1 | <LOQ | 1.18 ± 0.10 | 78 | 4.62 ± 0.13 | 103 |
| 2 | <LOQ | 1.35 ± 0.03 | 90 | 3.54 ± 0.04 | 79 |
| 3 | <LOQ | 1.55 ± 0.01 | 103 | 4.35 ± 0.09 | 97 |
| 4 | <LOQ | 1.43 ± 0.19 | 95 | 3.97 ± 0.12 | 88 |
| 5 | <LOQ | 1.36 ± 0.01 | 90 | 4.20 ± 0.01 | 93 |
| 6 | <LOQ | 1.32 ± 0.20 | 88 | 3.68 ± 0.23 | 82 |
| 7 | <LOQ | 1.33 ± 0.16 | 89 | 4.14 ± 0.05 | 92 |
| 8 | <LOQ | 1.66 ± 0.09 | 110 | 5.26 ± 0.07 | 117 |
| 9 | <LOQ | 1.57 ± 0.12 | 105 | 4.96 ± 0.13 | 110 |
| 10 | <LOQ | 1.08 ± 0.05 | 72 | 4.02 ± 0.15 | 89 |
| 11 | <LOQ | 1.27 ± 0.10 | 84 | 3.91 ± 0.01 | 87 |
| 12 | <LOQ | 1.77 ± 0.09 | 118 | 4.10 ± 0.10 | 91 |
| 13 | <LOQ | 1.17 ± 0.21 | 78 | 3.96 ± 0.02 | 88 |
| 14 | <LOQ | 1.53 ± 0.14 | 102 | 3.46 ± 0.15 | 77 |
| 15 | <LOQ | 1.29 ± 0.18 | 86 | 4.58 ± 0.10 | 102 |
| 16 | <LOQ | 1.30 ± 0.06 | 87 | 4.20 ± 0.03 | 93 |
| 17 | <LOQ | 1.30 ± 0.30 | 86 | 4.17± 0.21 | 93 |
| 18 | <LOQ | 1.54 ± 0.11 | 102 | 4.77 ± 0.04 | 106 |
| 19 | <LOQ | 1.31 ± 0.02 | 87 | 4.49 ± 0.13 | 100 |
| 20 | <LOQ | 1.82 ± 0.09 | 121 | 4.22 ± 0.20 | 94 |
| 21 | <LOQ | 1.60 ± 0.14 | 107 | 4.19 ± 0.05 | 93 |
| 22 | <LOQ | 1.38 ± 0.25 | 92 | 4.47 ± 0.19 | 99 |
| 23 | <LOQ | 1.48 ± 0.06 | 99 | 4.62 ± 0.01 | 103 |
| 24 | <LOQ | 1.14 ± 0.10 | 76 | 4.19 ± 0.10 | 93 |
| 25 | <LOQ | 1.34 ± 0.19 | 89 | 3.88 ± 0.31 | 86 |
| 26 | <LOQ | 1.30 ± 0.01 | 86 | 3.14 ± 0.04 | 70 |
| 27 | <LOQ | 1.54 ± 0.27 | 103 | 3.89 ± 0.11 | 86 |
| Assay | Unspiked Serum | Spiking 0.80 ng/mL | Spiking 1.60 ng/mL | Spiking 3.10 ng/mL | |||
|---|---|---|---|---|---|---|---|
| Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | ||
| 1 | <LOQ | 0.81 | 102 | 1.51 | 94 | 3.16 | 102 |
| 2 | <LOQ | 0.81 | 102 | 1.52 | 95 | 3.18 | 102 |
| 3 | <LOQ | 0.69 | 86 | 1.47 | 92 | 3.21 | 104 |
| 4 | <LOQ | 0.77 | 96 | 1.54 | 96 | 3.21 | 104 |
| 5 | <LOQ | 0.76 | 95 | 1.51 | 94 | 3.19 | 103 |
| Average | 0.77 | 96 | 1.51 | 94 | 3.2 | 102.8 | |
| CV% | 5.1 | 2.3 | 2.3 | ||||
| Day | Unspiked Serum | Spiking 0.80 ng/mL | Spiking 1.60 ng/mL | Spiking 3.10 ng/mL | |||
|---|---|---|---|---|---|---|---|
| Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | Measured (ng/mL) | Recovery % | ||
| 1 | <LOQ | 0.77 | 97 | 1.56 | 104 | 3.02 | 97 |
| 2 | <LOQ | 0.67 | 84 | 1.38 | 86 | 2.84 | 92 |
| 3 | <LOQ | 0.65 | 82 | 1.64 | 106 | 3.22 | 104 |
| 4 | <LOQ | 0.58 | 73 | 1.48 | 92 | 2.97 | 96 |
| 5 | <LOQ | 0.77 | 98 | 1.51 | 94 | 3.19 | 103 |
| Average | 0.69 | 86.8 | 1.51 | 96.4 | 3.05 | 98.4 | |
| CV% | 8.1 | 9.8 | 16.0 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Delfin-Riela, T.; Rossotti, M.; Alvez-Rosado, R.; Leizagoyen, C.; González-Sapienza, G. Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA. Biomolecules 2020, 10, 1652. https://doi.org/10.3390/biom10121652
Delfin-Riela T, Rossotti M, Alvez-Rosado R, Leizagoyen C, González-Sapienza G. Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA. Biomolecules. 2020; 10(12):1652. https://doi.org/10.3390/biom10121652
Chicago/Turabian StyleDelfin-Riela, Triana, Martín Rossotti, Romina Alvez-Rosado, Carmen Leizagoyen, and Gualberto González-Sapienza. 2020. "Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA" Biomolecules 10, no. 12: 1652. https://doi.org/10.3390/biom10121652
APA StyleDelfin-Riela, T., Rossotti, M., Alvez-Rosado, R., Leizagoyen, C., & González-Sapienza, G. (2020). Highly Sensitive Detection of Zika Virus Nonstructural Protein 1 in Serum Samples by a Two-Site Nanobody ELISA. Biomolecules, 10(12), 1652. https://doi.org/10.3390/biom10121652