The c.*52 A/G and c.*773 A/G Genetic Variants in the UTR′3 of the LDLR Gene Are Associated with the Risk of Acute Coronary Syndrome and Lower Plasma HDL-Cholesterol Concentration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of the Study Population
2.2. Laboratory Analyses
2.3. Genetic Analysis
2.4. Inheritance Models Analysis
2.5. Analysis of the Haplotypes
2.6. Functional Prediction Analysis
2.7. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
3.2. Allele and Genotype Frequencies
3.3. Linkage Disequilibrium Analysis
3.4. Functional Prediction
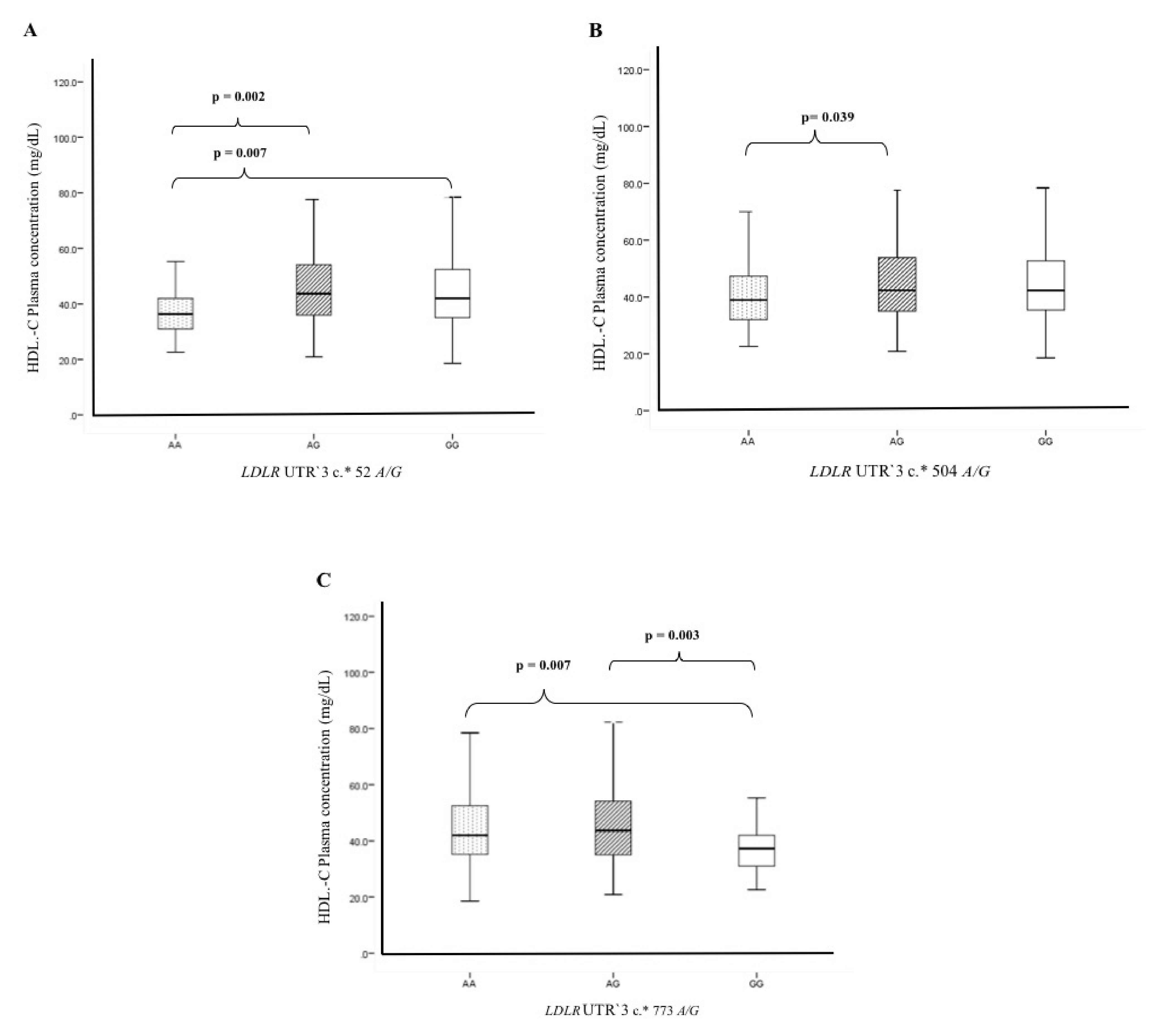
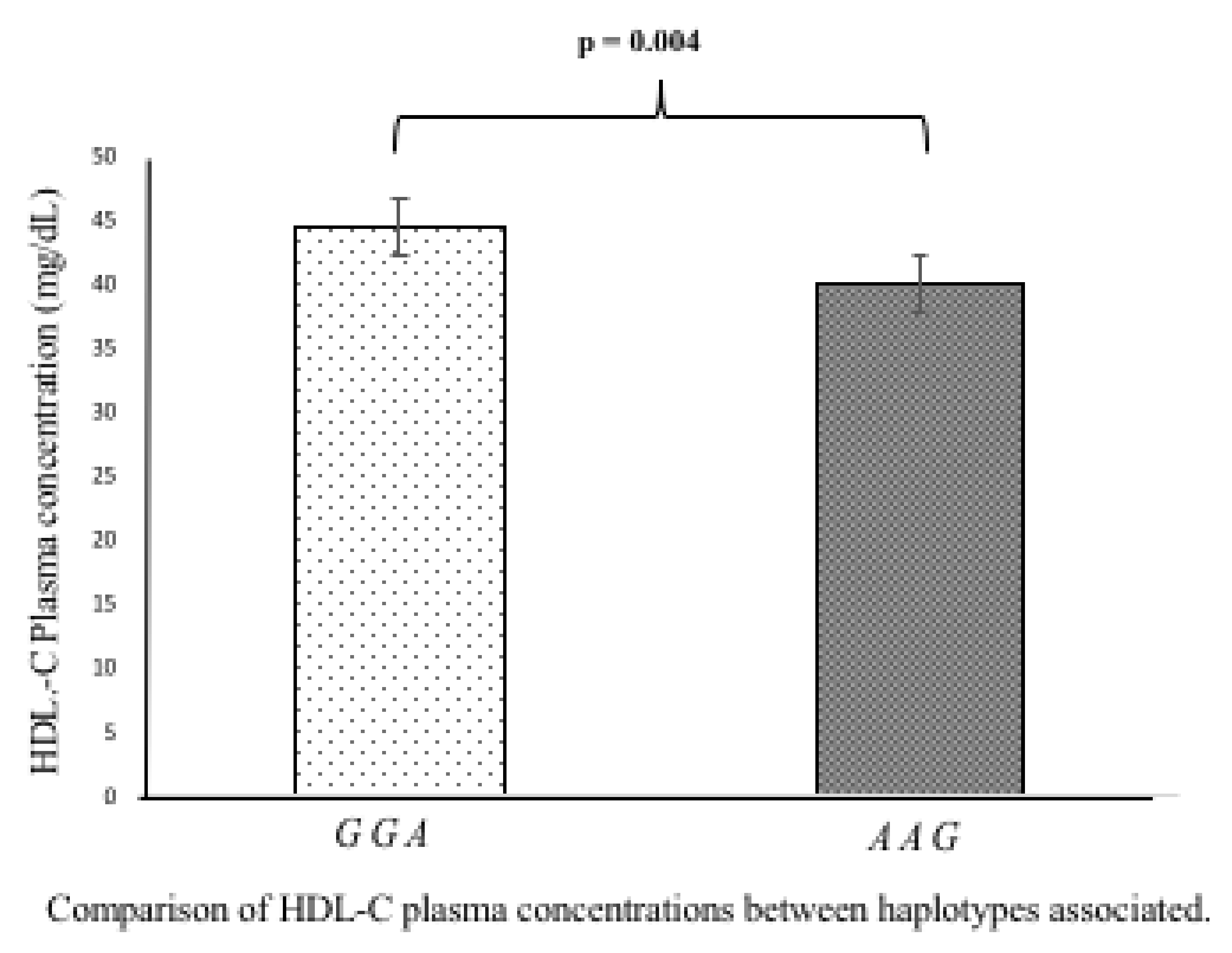
3.5. Association of Polymorphisms and Haplotypes with Plasma Lipids Levels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LDLR | Low-density lipoprotein receptor |
| UTR′3 | Untranslated region-′3 |
| HDL-C | High-density lipoprotein-cholesterol |
| LDL-C | Low-density lipoprotein-cholesterol |
| T2DM | Type 2 diabetes mellitus |
| SNP | Single nucleotide polymorphism |
| ACS | Acute coronary syndrome |
References
- Libby, P.; Ridker, P.M.; Maseri, A. Inflammation and Atherosclerosis. Circulation 2002, 105, 1135–1143. [Google Scholar] [CrossRef] [PubMed]
- Virmani, R.; Kolodgie, F.D.; Burke, A.; Finn, A.V.; Gold, H.K.; Tulenko, T.N.; Wrenn, S.P.; Narula, J. Atherosclerotic Plaque Progression and Vulnerability to Rupture. Arter. Thromb. Vasc. Biol. 2005, 25, 2054–2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Ma, K.; Ruan, X.Z.; Liu, B.C. Dysregulation of the Low-Density Lipoprotein Receptor Pathway Is Involved in Lipid Disorder-Mediated Organ Injury. Int. J. Biol. Sci. 2016, 12, 569–579. [Google Scholar] [CrossRef] [Green Version]
- Litvinov, D.Y.; Savushkin, E.V.; Dergunov, A.D. Intracellular and Plasma Membrane Events in Cholesterol Transport and Homeostasis. J. Lipids 2018, 2018, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Abisambra, J.F.; Fiorelli, T.; Padmanabhan, J.; Neame, P.; Wefes, I.; Potter, H. LDLR Expression and Localization Are Altered in Mouse and Human Cell Culture Models of Alzheimer’s Disease. PLoS ONE 2010, 5, e8556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolic, J.; Belot, L.; Raux, H.; Legrand, P.; Gaudin, Y.; Albertini, A.A. Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat. Commun. 2018, 9, 1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourbon, M.; Alves, A.C.; Sijbrands, E.J.G. Low-density lipoprotein receptor mutational analysis in diagnosis of familial hypercholesterolemia. Curr. Opin. Lipidol. 2017, 28, 120–129. [Google Scholar] [CrossRef] [Green Version]
- Defesche, J.C.; Gidding, S.S.; Harada-Shiba, M.; Hegele, R.A.; Santos, R.D.; Wierzbicki, A.S. Familial hypercholesterolaemia. Nat. Rev. Dis. Primers 2017, 3, 17093. [Google Scholar] [CrossRef]
- Zambrano, T.; Hirata, M.H.; Cerda, Á.; Dorea, E.L.; Pinto, G.A.; Gusukuma, M.C.; Bertolami, M.C.; Salazar, L.A.; Hirata, R.D.C. Impact of 3′UTR genetic variants in PCSK9 and LDLR genes on plasma lipid traits and response to atorvastatin in Brazilian subjects: A pilot study. Int. J. Clin. Exp. Med. 2015, 8, 5978–5988. [Google Scholar]
- De Castro-Orós, I.; Solà, R.; Valls, R.-M.; Brea, Á.; Mozas, P.; Puzo, J.; Pocovi, M. Genetic Variants of LDLR and PCSK9 Associated with Variations in Response to Antihypercholesterolemic Effects of Armolipid Plus with Berberine. PLoS ONE 2016, 11, e0150785. [Google Scholar] [CrossRef] [Green Version]
- Van Zyl, T.; Jerling, J.C.; Conradie, K.R.; Feskens, E.J. Common and rare single nucleotide polymorphisms in the LDLR gene are present in a black South African population and associate with low-density lipoprotein cholesterol levels. J. Hum. Genet. 2013, 59, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Openepi.com. Available online: http://www.openepi.com/SampleSize/SSCC.html (accessed on 1 April 2020).
- Cannon, C.P.; Battler, A.; Brindis, R.G.; Cox, J.L.; Ellis, S.G.; Every, N.R.; Flaherty, J.T.; Harrington, R.A.; Krumholz, H.M.; Simoons, M.L.; et al. American College of Cardiology key data elements and definitions for measuring the clinical management and outcomes of patients with acute coronary syndromes: A report of the American College of Cardiology Task Force on Clinical Data Standards (Acute Coronary Syndromes Writing Committee) Endorsed by the American Association of Cardiovascular and Pulmonary Rehabilitation, American College of Emergency Physicians, American Heart Association, Cardiac Society of Australia & New Zealand, National Heart Foundation of Australia, Society for Cardiac Angiography and Interventions, and the Taiwan Society of Cardiology. J. Am. Coll. Cardiol. 2001, 38, 2114–2130. [Google Scholar] [CrossRef] [Green Version]
- Hamm, C.W.; Bassand, J.-P.; Agewall, S.; Bax, J.; Boersma, E.; Bueno, H.; Caso, P.; Dudek, D.; Gielen, S.; Huber, K.; et al. ESC Guidelines for the management of acute coronary syndromes in patients presenting without persistent ST-segment elevation: The Task Force for the management of acute coronary syndromes (ACS) in patients presenting without persistent ST-segment elevation of the European Society of Cardiology (ESC). Eur. Heart J. 2011, 32, 2999–3054. [Google Scholar] [CrossRef] [Green Version]
- Posadas-Sánchez, R.; Pérez-Hernández, N.; Angeles-Martinez, J.; López-Bautista, F.; Villarreal-Molina, T.; Rodriguez-Perez, J.M.; Fragoso, J.M.; Posadas-Romero, C.; Vargas-Alarcón, G. Interleukin 35 Polymorphisms Are Associated with Decreased Risk of Premature Coronary Artery Disease, Metabolic Parameters, and IL-35 Levels: The Genetics of Atherosclerotic Disease (GEA) Study. Mediat. Inflamm. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Delong, D.M.; Delong, E.R.; Wood, P.D.; Lippel, K.; Rifkind, B.M. A Comparison of Methods for the Estimation of Plasma Low- and Very Low-Density Lipoprotein Cholesterol. JAMA 1986, 256, 2372–2377. [Google Scholar] [CrossRef] [PubMed]
- ATP III Guidelines At-A-Glance Quick Desk Reference. Available online: https://www.nhlbi.nih.gov/files/docs/guidelines/atglance.pdf (accessed on 29 April 2020).
- Lahiri, D.K.; Numberger, J.I. A rapid non-enzymatic method for the preparation of HMW DNA from blood for RFLP studies. Nucleic Acids Res. 1991, 19, 5444. [Google Scholar] [CrossRef]
- Smith, P.J.; Zhang, C.; Wang, J.; Chew, S.L.; Zhang, M.Q.; Krainer, A. An increased specificity score matrix for the prediction of SF2/ASF-specific exonic splicing enhancers. Hum. Mol. Genet. 2006, 15, 2490–2508. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Taylor, J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009, 37, W600–W605. [Google Scholar] [CrossRef] [Green Version]
- QUANTO. Available online: https://preventivemedicine.usc.edu/download-quanto/ (accessed on 3 April 2020).
- Chen, W.; Wang, S.; Ma, Y.; Zhou, Y.; Liu, H.; Strnad, P.; Kraemer, F.B.; Krauss, R.M.; Liu, J. Analysis of polymorphisms in the 3′ untranslated region of the LDL receptor gene and their effect on plasma cholesterol levels and drug response. Int. J. Mol. Med. 2008, 21, 345–353. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Zhao, T.-Y.; Tan, X.-H.; Lei, S.; Huang, L.; Yang, L. Polymorphisms in PCSK9, LDLR, BCMO1, SLC12A3, and KCNJ1 are Associated with Serum Lipid Profile in Chinese Han Population. Int. J. Environ. Res. Public Health 2019, 16, 3207. [Google Scholar] [CrossRef] [Green Version]
- Bashore, A.C.; Liu, M.; Key, C.-C.C.; Boudyguina, E.; Wang, X.; Carroll, C.M.; Sawyer, J.K.; Mullick, A.E.; Lee, R.G.; Macauley, S.L.; et al. Targeted Deletion of Hepatocyte Abca1 Increases Plasma HDL (High-Density Lipoprotein) Reverse Cholesterol Transport via the LDL (Low-Density Lipoprotein) Receptor. Arter. Thromb. Vasc. Biol. 2019, 39, 1747–1761. [Google Scholar] [CrossRef]
- Joyce, C.; Wagner, E.M.; Basso, F.; Amar, M.J.; Freeman, L.A.; Shamburek, R.D.; Knapper, C.L.; Syed, J.; Wu, J.; Vaisman, B.L.; et al. ABCA1 Overexpression in the Liver of LDLr-KO Mice Leads to Accumulation of Pro-atherogenic Lipoproteins and Enhanced Atherosclerosis. J. Biol. Chem. 2006, 281, 33053–33065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinninger, F.; Heine, M.; Singaraja, R.; Hayden, M.; Brundert, M.; Ramakrishnan, R.; Heeren, J. High density lipoprotein metabolism in low density lipoprotein receptor-deficient mice. J. Lipid Res. 2014, 55, 1914–1924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Zuo, H.; Liu, C.; Yang, Y. Overexpression of miR-200a protects cardiomyocytes against hypoxia-induced apoptosis by modulating the kelch-like ECH-associated protein 1-nuclear factor erythroid 2-related factor 2 signaling axis. Int. J. Mol. Med. 2016, 38, 1303–1311. [Google Scholar] [CrossRef]
- Li, P.; Liu, Y.; Yi, B.; Wang, G.; You, X.; Zhao, X.; Summer, R.; Qin, Y.; Sun, J. MicroRNA-638 is highly expressed in human vascular smooth muscle cells and inhibits PDGF-BB-induced cell proliferation and migration through targeting orphan nuclear receptor NOR1. Cardiovasc. Res. 2013, 99, 185–193. [Google Scholar] [CrossRef] [Green Version]
- Luque, A.; Farwati, A.; Krupinski, J.; Aran, J.M. Association between low levels of serum miR-638 and atherosclerotic plaque vulnerability in patients with high-grade carotid stenosis. J. Neurosurg. 2019, 131, 72–79. [Google Scholar] [CrossRef]
- Aryal, B.; Singh, A.K.; Rotllan, N.; Price, N.D.; Fernández-Hernando, C. MicroRNAs and lipid metabolism. Curr. Opin. Lipidol. 2017, 28, 273–280. [Google Scholar] [CrossRef]
- Ensembl.org. Available online: https://www.ensembl.org/Multi/Search/Results (accessed on 3 April 2020).
- Lisker, R.; Granados, J.; Babinsky, V.; De Rubens, J.; Armendares, S.; Buentello, L.; Perez-Briceño, R. Gene frequencies and admixture estimates in a Mexico City population. Am. J. Phys. Anthr. 1986, 71, 203–207. [Google Scholar] [CrossRef]
- Lisker, R.; Ramirez, E.; Briceño, R.P.; Granados, J.; Babinsky, V. Gene frequencies and admixture estimates in four Mexican urban centers. Hum. Biol. 1990, 62, 791–801. [Google Scholar] [PubMed]
- Juárez-Cedillo, T.; Zúñiga, J.; Acuña-Alonzo, V.; Pérez-Hernández, N.; Rodriguez-Perez, J.M.; Barquera, R.; Gallardo, G.J.; Arenas, R.S.; García-Peña, M.D.C.; Granados, J.; et al. Genetic admixture and diversity estimations in the Mexican Mestizo population from Mexico City using 15 STR polymorphic markers. Forensic Sci. Int. Genet. 2008, 2, e37–e39. [Google Scholar] [CrossRef] [PubMed]
| Characteristic | ACS Patients (n = 618) | Healthy Controls (n = 666) | p-Value | |
|---|---|---|---|---|
| Median (percentile 25–75) | Median (percentile 25–75) | |||
| Age (years) | 58 (51–65) | 54 (49–59) | 0.001 | |
| Gender n (%) | Male | 505 (82) | 453 (68) | <0.001 |
| Female | 113 (18) | 213 (32) | ||
| BMI (kg/m2) | 27 (25–29) | 28 (26–31) | 0.521 | |
| Blood pressure (mmHg) | Systolic | 132 (114–144) | 117 (106–126) | <0.001 |
| Diastolic | 80 (70–90) | 73 (67–78) | <0.001 | |
| Glucose (mg/dL) | 159 (102–188) | 98 (84–99) | <0.001 | |
| Total cholesterol (mg/dL) | 164(128–199) | 191 (165–210) | <0.001 | |
| HDL-C (mg/dL) | 39 (34–45) | 44 (35–53) | 0.017 | |
| LDL-C (mg/dL) | 106 (75–132) | 116 (94–134) | <0.001 | |
| Triglycerides (mg/dL) | 169 (109–201) | 176 (113–208) | 0.301 | |
| Hypertension n (%) | Yes | 350 (57) | 201 (30) | <0.001 |
| Type II diabetes mellitus n (%) | Yes | 216 (35) | 63 (9) | <0.001 |
| Dyslipidemia n (%) | Yes | 528 (85) | 479 (72) | <0.001 |
| Smoking n (%) | Yes | 222 (36) | 147 (22) | <0.001 |
| Polymorphic Site | n (Genotype Frequency) | Model | OR (95%CI) | pC | n (Allele Frequency) | OR | 95%CI | pC | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| LDLR UTR′3 | c.*52 A/G (rs14158) | *Risk allele | |||||||||
| Control | GG | AG | AA | G | *A | A vs. G | |||||
| (n = 666) | 385 (0.578) | 252 (0.378) | 29 (0.043) | Co-dominant | 2.02 (1.18–3.46) | 0.033 | 1022 (0.766) | 310 (0.232) | |||
| Dominant | 1.13 (0.88–1.45) | 0.35 | 1.20 | 1.00–1.44 | 0.02 | ||||||
| ACS | 337 (0.545) | 231 (0.374) | 50 (0.081) | Recessive | 2.00 (1.18–3.39) | 0.009 | 905 (0.732) | 331 (0.267) | |||
| (n = 618) | Over-dominant | 0.96 (0.74–1.24) | 0.73 | ||||||||
| Additive | 1.20 (0.98–1.45) | 0.075 | |||||||||
| LDLR UTR′3 | c.*504 A/G (rs2738465) | ||||||||||
| Control | GG | AG | AA | G | *A | A vs. G | |||||
| (n = 666) | 323 (0.485) | 283 (0.425) | 60 (0.090) | Co-dominant | 1.50 (0.99–2.26) | 0.16 | 929 (0.696) | 403 (0.302) | |||
| Dominant | 1.15 (0.90–1.47) | 0.27 | 1.18 | 1.00–1.40 | 0.02 | ||||||
| ACS | 280 (0.453) | 256 (0.414) | 82 (0.133) | Recessive | 1.45 (0.98–2.16) | 0.06 | 816 (0.660) | 420 (0.339) | |||
| (n = 618) | Over-dominant | 0.99 (0.77–1.27) | 0.94 | ||||||||
| Additive | 1.17 (0.97–1.41) | 0.09 | |||||||||
| LDLR UTR′3 | c.*773 A/G (rs2738466) | ||||||||||
| Control | AA | AG | GG | A | *G | G vs. A | |||||
| (n = 666) | 386 (0.580) | 250 (0.375) | 30 (0.045) | Co-dominant | 2.04 (1.35–3.45) | 0.027 | 1022 (0.766) | 310 (0.232) | |||
| Dominant | 1.14 (0.88–1.46) | 0.32 | 1.22 | 1.02–1.146 | 0.01 | ||||||
| ACS | 335 (0.542) | 231 (0.374) | 52 (0.084) | Recessive | 2.01 (1.20–3.38) | 0.007 | 901 (0.728) | 335 (0.271) | |||
| (n = 618) | Over-dominant | 0.96 (0.74–1.24) | 0.73 | ||||||||
| Additive | 1.21 (0.99–1.48) | 0.062 | |||||||||
| c.*52 A/G | c.*504 A/G | c.*773 A/G | ACS (n = 618) | Controls (n = 666) | OR | 95%CI | pC |
|---|---|---|---|---|---|---|---|
| Haplotype | Hf | Hf | |||||
| G | G | A | 0.658 | 0.695 | 0.84 | 0.71–0.99 | 0.023 |
| A | A | G | 0.267 | 0.230 | 1.22 | 1.02–1.46 | 0.016 |
| A | G | A | 0.070 | 0.069 | 1.03 | 0.76–1.39 | 0.446 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vargas-Alarcon, G.; Perez-Mendez, O.; Ramirez-Bello, J.; Posadas-Sanchez, R.; Gonzalez-Pacheco, H.; Escobedo, G.; Nieto-Lima, B.; Carreon-Torres, E.; Fragoso, J.M. The c.*52 A/G and c.*773 A/G Genetic Variants in the UTR′3 of the LDLR Gene Are Associated with the Risk of Acute Coronary Syndrome and Lower Plasma HDL-Cholesterol Concentration. Biomolecules 2020, 10, 1381. https://doi.org/10.3390/biom10101381
Vargas-Alarcon G, Perez-Mendez O, Ramirez-Bello J, Posadas-Sanchez R, Gonzalez-Pacheco H, Escobedo G, Nieto-Lima B, Carreon-Torres E, Fragoso JM. The c.*52 A/G and c.*773 A/G Genetic Variants in the UTR′3 of the LDLR Gene Are Associated with the Risk of Acute Coronary Syndrome and Lower Plasma HDL-Cholesterol Concentration. Biomolecules. 2020; 10(10):1381. https://doi.org/10.3390/biom10101381
Chicago/Turabian StyleVargas-Alarcon, Gilberto, Oscar Perez-Mendez, Julian Ramirez-Bello, Rosalinda Posadas-Sanchez, Hector Gonzalez-Pacheco, Galileo Escobedo, Betzabe Nieto-Lima, Elizabeth Carreon-Torres, and Jose Manuel Fragoso. 2020. "The c.*52 A/G and c.*773 A/G Genetic Variants in the UTR′3 of the LDLR Gene Are Associated with the Risk of Acute Coronary Syndrome and Lower Plasma HDL-Cholesterol Concentration" Biomolecules 10, no. 10: 1381. https://doi.org/10.3390/biom10101381






