Omics-Based Platforms: Current Status and Potential Use for Cholangiocarcinoma
Abstract
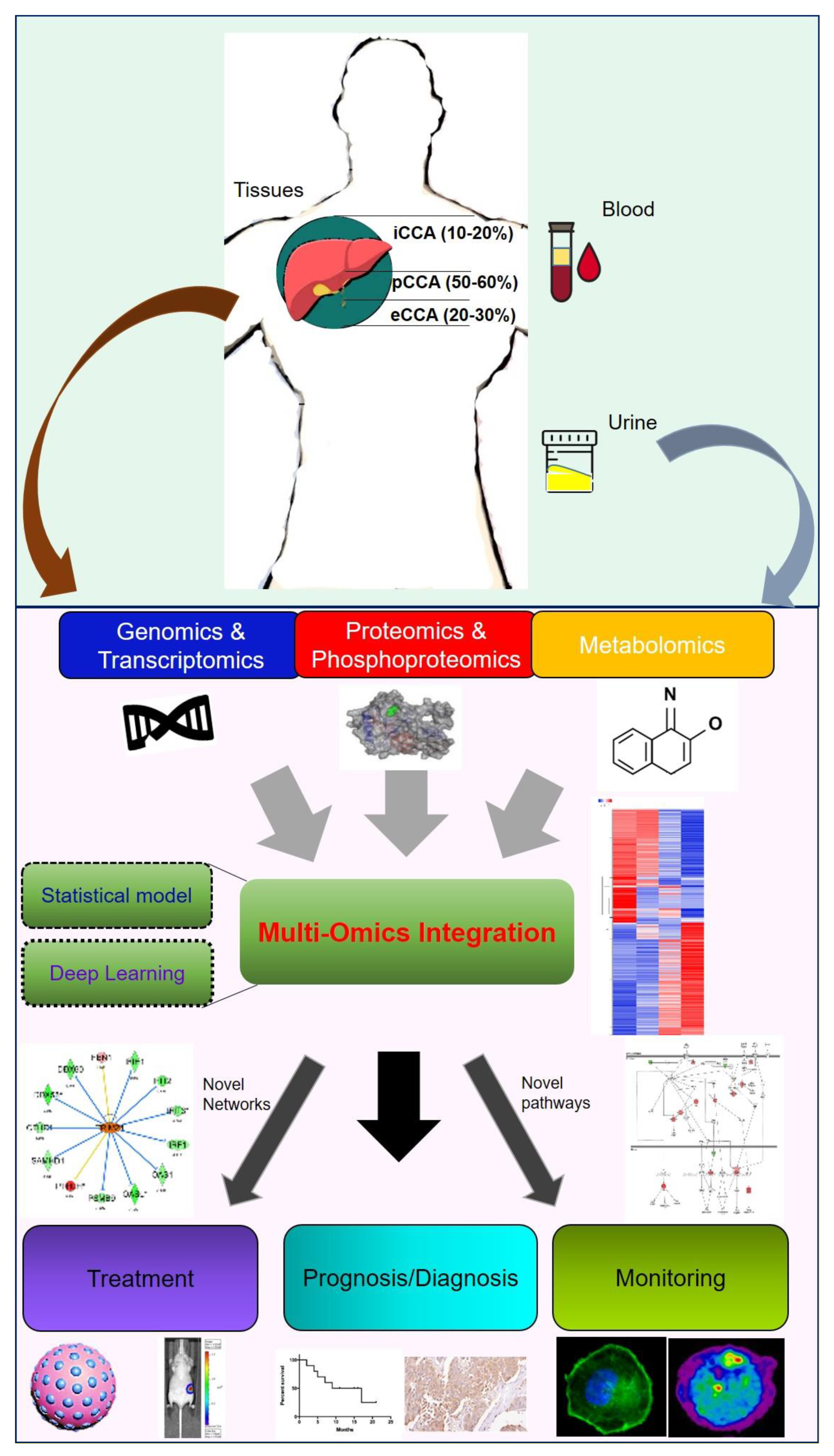
:1. Introduction to CCA
2. Genomics Alterations and Epigenetic Changes in CCA
2.1. Genomic Alterations
2.2. Epigenetic Events
3. Canonical Pathways and Key Drivers of the Transcriptome
4. Protein Levels and Post-Translational Modifications in CCA
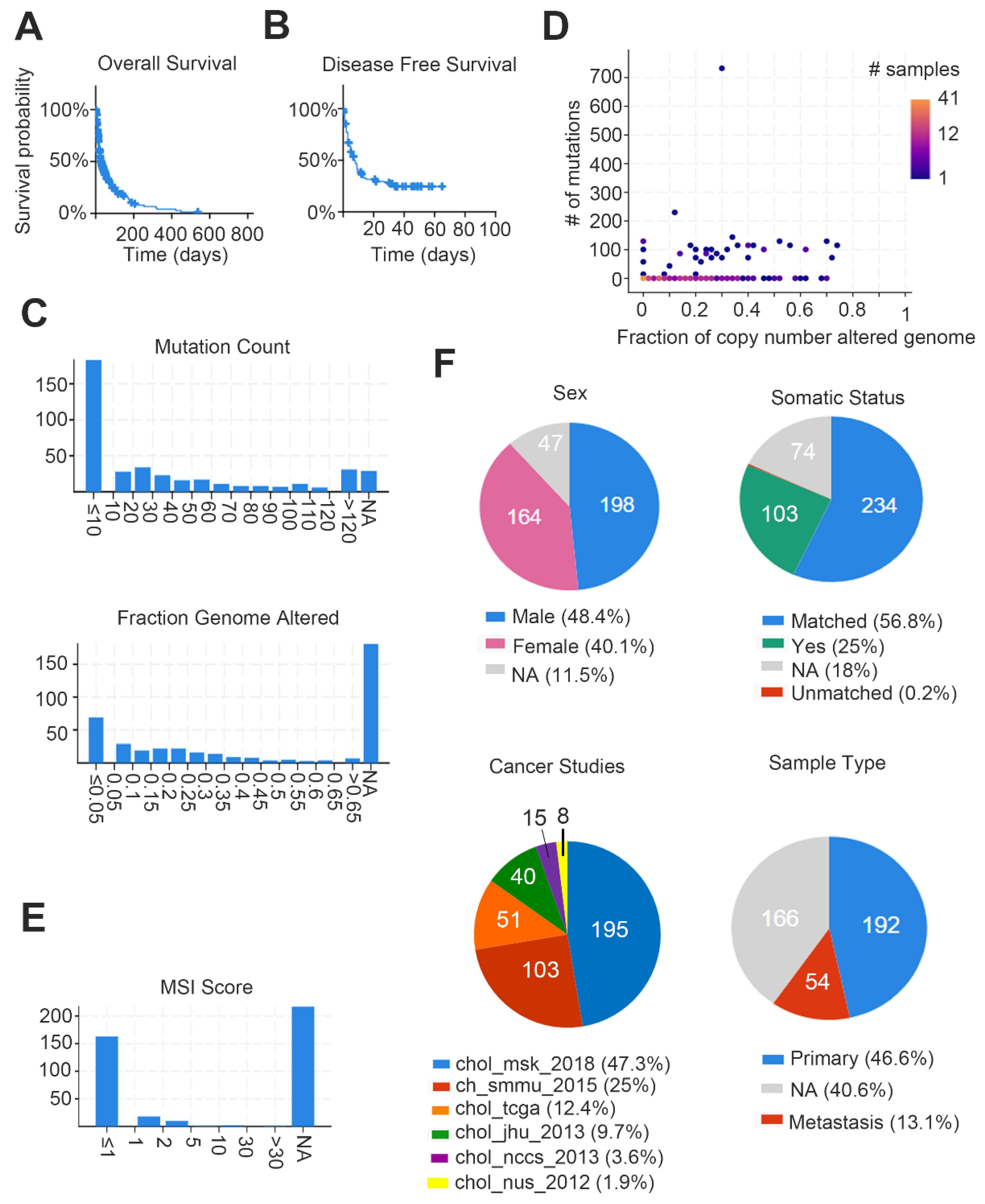
5. Available Omics Datasets for CCA
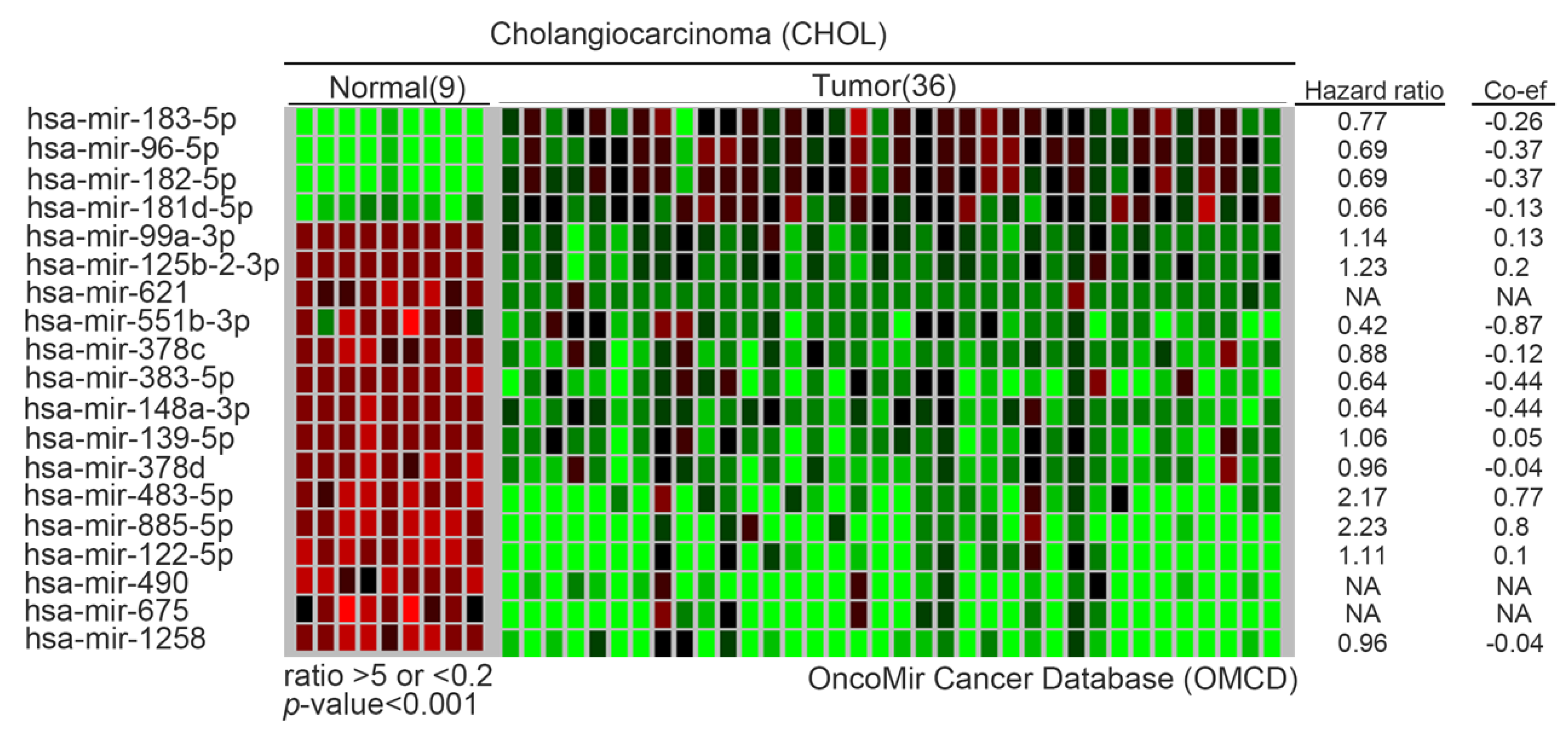
6. Metabolomics Analysis of Oncometabolites and Metabolic Reprogramming in CCA
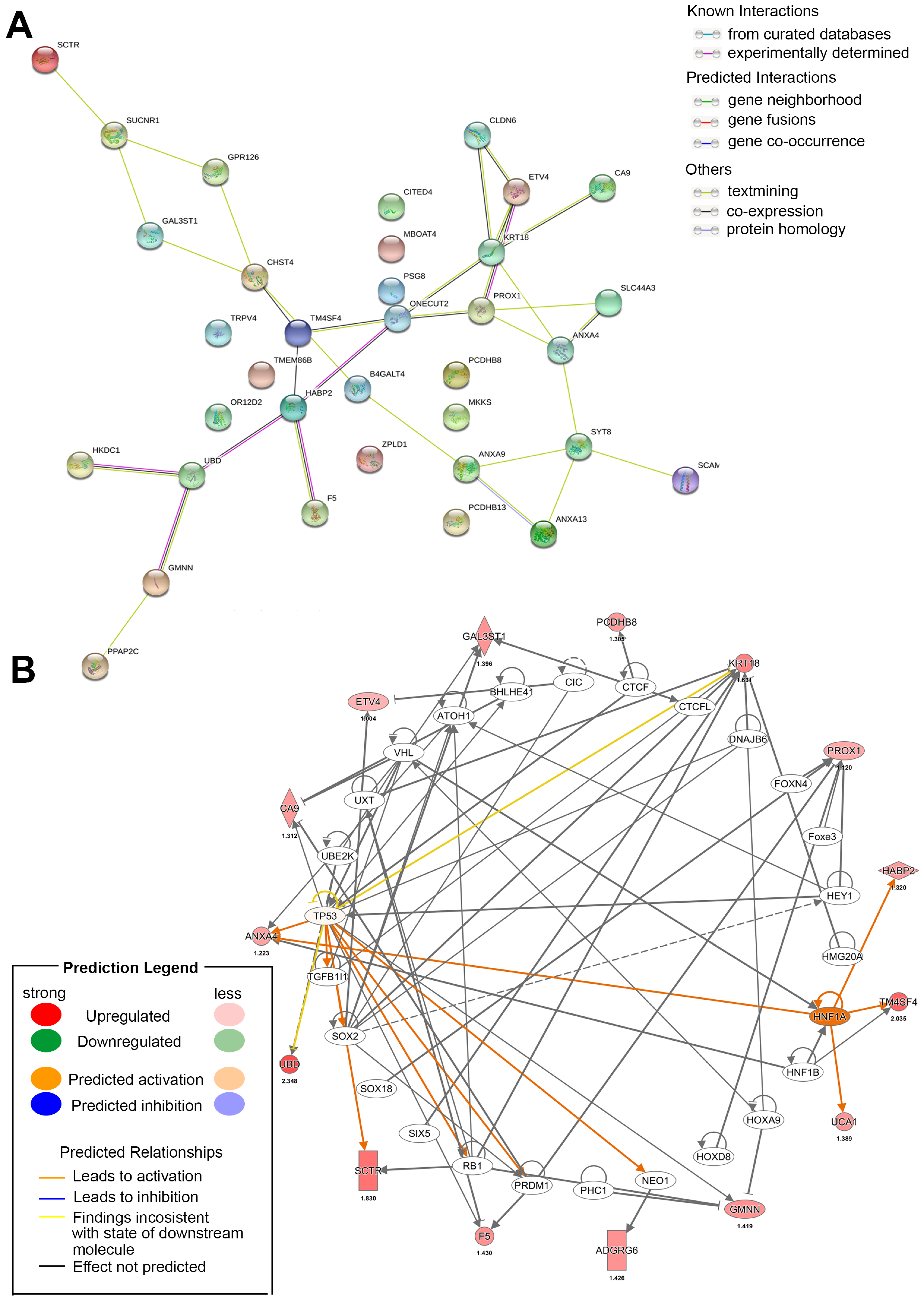
7. Using Multiple-Omics Approaches to Determine Prognostic Factors
8. Potential Small Molecule Compounds and Clinical Drugs from Omics Datasets
8.1. FGFR Family
8.2. IDH Mutants
8.3. Others
8.4. Immune Checkpoint Blockage
9. Future Prospects
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Banales, J.M.; Cardinale, V.; Carpino, G.; Marzioni, M.; Andersen, J.B.; Invernizzi, P.; Lind, G.E.; Folseraas, T.; Forbes, S.J.; Fouassier, L.; et al. Expert consensus document: Cholangiocarcinoma: Current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 261–280. [Google Scholar] [CrossRef]
- Rizvi, S.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J. Cholangiocarcinoma—Evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018, 15, 95–111. [Google Scholar] [CrossRef] [Green Version]
- Nakeeb, A.; Pitt, H.A.; Sohn, T.A.; Coleman, J.; Abrams, R.A.; Piantadosi, S.; Hruban, R.H.; Lillemoe, K.D.; Yeo, C.J.; Cameron, J.L. Cholangiocarcinoma. A spectrum of intrahepatic, perihilar, and distal tumors. Ann. Surg. 1996, 224, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Akita, M.; Sofue, K.; Fujikura, K.; Otani, K.; Itoh, T.; Ajiki, T.; Fukumoto, T.; Zen, Y. Histological and molecular characterization of intrahepatic bile duct cancers suggests an expanded definition of perihilar cholangiocarcinoma. HPB 2019, 21, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Banales, J.M.; Marin, J.J.G.; Lamarca, A.; Rodrigues, P.M.; Khan, S.A.; Roberts, L.R.; Cardinale, V.; Carpino, G.; Andersen, J.B.; Braconi, C.; et al. Cholangiocarcinoma 2020: The next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Bertuccio, P.; Malvezzi, M.; Carioli, G.; Hashim, D.; Boffetta, P.; El-Serag, H.B.; La Vecchia, C.; Negri, E. Global trends in mortality from intrahepatic and extrahepatic cholangiocarcinoma. J. Hepatol. 2019, 71, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Hu, W.; Shi, X.; Liu, P.; Ma, X.; Zhao, W.; Qu, L.; Zhang, S.; Shi, W.; Liu, A.; et al. Comprehensive genomic profile of cholangiocarcinomas in China. Oncol. Lett. 2020, 19, 3101–3110. [Google Scholar] [CrossRef] [Green Version]
- Utada, M.; Ohno, Y.; Tamaki, T.; Sobue, T.; Endo, G. Long-term trends in incidence and mortality of intrahepatic and extrahepatic bile duct cancer in Japan. J. Epidemiol. 2014, 24, 193–199. [Google Scholar] [CrossRef] [Green Version]
- Kim, B.W.; Oh, C.M.; Choi, H.Y.; Park, J.W.; Cho, H.; Ki, M. Incidence and Overall Survival of Biliary Tract Cancers in South Korea from 2006 to 2015: Using the National Health Information Database. Gut Liver 2019, 13, 104–113. [Google Scholar] [CrossRef]
- Chaiteerakij, R.; Pan-Ngum, W.; Poovorawan, K.; Soonthornworasiri, N.; Treeprasertsuk, S.; Phaosawasdi, K. Characteristics and outcomes of cholangiocarcinoma by region in Thailand: A nationwide study. World J. Gastroenterol. 2017, 23, 7160–7167. [Google Scholar] [CrossRef]
- Chen, M.F.; Jan, Y.Y.; Jeng, L.B.; Hwang, T.L.; Wang, C.S.; Chen, S.C.; Chao, T.C.; Chen, H.M.; Lee, W.C.; Yeh, T.S.; et al. Intrahepatic cholangiocarcinoma in Taiwan. J. Hepato-Biliary-Pancreat. Surg. 1999, 6, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Pinter, M.; Hucke, F.; Zielonke, N.; Waldhör, T.; Trauner, M.; Peck-Radosavljevic, M.; Sieghart, W. Incidence and mortality trends for biliary tract cancers in Austria. Liver Int. Off. J. Int. Assoc. Study Liver 2014, 34, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.J.; Wu, A.T.; Chiou, H.Y.; Chuang, M.T.; Meng, T.C.; Chien, L.N.; Yen, Y. Interactive role of diabetes mellitus and female sex in the risk of cholangiocarcinoma: A population-based nested case-control study. Oncotarget 2017, 8, 6642–6651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alabraba, E.; Joshi, H.; Bird, N.; Griffin, R.; Sturgess, R.; Stern, N.; Sieberhagen, C.; Cross, T.; Camenzuli, A.; Davis, R.; et al. Increased multimodality treatment options has improved survival for Hepatocellular carcinoma but poor survival for biliary tract cancers remains unchanged. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2019, 45, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Cambridge, W.A.; Fairfield, C.; Powell, J.J.; Harrison, E.M.; Søreide, K.; Wigmore, S.J.; Guest, R.V. Meta-analysis and Meta-regression of Survival After Liver Transplantation for Unresectable Perihilar Cholangiocarcinoma. Ann. Surg. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kamsa-Ard, S.; Luvira, V.; Suwanrungruang, K.; Kamsa-Ard, S.; Luvira, V.; Santong, C.; Srisuk, T.; Pugkhem, A.; Bhudhisawasdi, V.; Pairojkul, C. Cholangiocarcinoma Trends, Incidence, and Relative Survival in Khon Kaen, Thailand from 1989 Through 2013: A Population-Based Cancer Registry Study. J. Epidemiol. 2019, 29, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Komaya, K.; Ebata, T.; Yokoyama, Y.; Igami, T.; Sugawara, G.; Mizuno, T.; Yamaguchi, J.; Nagino, M. Recurrence after curative-intent resection of perihilar cholangiocarcinoma: Analysis of a large cohort with a close postoperative follow-up approach. Surgery 2018, 163, 732–738. [Google Scholar] [CrossRef]
- Strijker, M.; Belkouz, A.; van der Geest, L.G.; van Gulik, T.M.; van Hooft, J.E.; de Meijer, V.E.; Haj Mohammad, N.; de Reuver, P.R.; Verheij, J.; de Vos-Geelen, J.; et al. Treatment and survival of resected and unresected distal cholangiocarcinoma: A nationwide study. Acta Oncol. 2019, 58, 1048–1055. [Google Scholar] [CrossRef] [PubMed]
- Sripa, B.; Pairojkul, C. Cholangiocarcinoma: Lessons from Thailand. Curr. Opin. Gastroenterol. 2008, 24, 349–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigopoulou, E.I.; Zachou, K.; Gatselis, N.K.; Papadamou, G.; Koukoulis, G.K.; Dalekos, G.N. Primary biliary cirrhosis in HBV and HCV patients: Clinical characteristics and outcome. World J. Hepatol. 2013, 5, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zong, C.; Zhang, L.; Garner, E.; Sugie, S.; Huang, C.; Wu, W.; Chang, J.; Sakurai, T.; Kato, M.; et al. Exposure of Mice to 1,2-Dichloropropane Induces CYP450-Dependent Proliferation and Apoptosis of Cholangiocytes. Toxicol. Sci. Off. J. Soc. Toxicol. 2018, 162, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varamo, C.; Peraldo-Neia, C.; Ostano, P.; Basiricò, M.; Raggi, C.; Bernabei, P.; Venesio, T.; Berrino, E.; Aglietta, M.; Leone, F.; et al. Establishment and Characterization of a New Intrahepatic Cholangiocarcinoma Cell Line Resistant to Gemcitabine. Cancers 2019, 11, 519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.J.; Chun, Y.S. Intrahepatic cholangiocarcinoma: The AJCC/UICC 8th edition updates. Chin. Clin. Oncol. 2018, 7, 52. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.C.; Korkut, A.; Kanchi, R.S.; Hegde, A.M.; Lenoir, W.; Liu, W.; Liu, Y.; Fan, H.; Shen, H.; Ravikumar, V.; et al. A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell 2018, 33, 690–705. [Google Scholar] [CrossRef] [Green Version]
- Koplev, S.; Lin, K.; Dohlman, A.B.; Ma’ayan, A. Integration of pan-cancer transcriptomics with RPPA proteomics reveals mechanisms of epithelial-mesenchymal transition. PLoS Comput. Biol. 2018, 14, e1005911. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, H.; Arai, Y.; Totoki, Y.; Shirota, T.; Elzawahry, A.; Kato, M.; Hama, N.; Hosoda, F.; Urushidate, T.; Ohashi, S.; et al. Genomic spectra of biliary tract cancer. Nat. Genet. 2015, 47, 1003–1010. [Google Scholar] [CrossRef]
- Arai, Y.; Totoki, Y.; Hosoda, F.; Shirota, T.; Hama, N.; Nakamura, H.; Ojima, H.; Furuta, K.; Shimada, K.; Okusaka, T.; et al. Fibroblast growth factor receptor 2 tyrosine kinase fusions define a unique molecular subtype of cholangiocarcinoma. Hepatology 2014, 59, 1427–1434. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [Green Version]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.F.; Jiang, H.; Zhang, C.S.; Yu, S.P.; Wang, Z.Q.; Su, H.L. Targeted drug regulation on methylation of p53-BAX mitochondrial apoptosis pathway affects the growth of cholangiocarcinoma cells. J. Int. Med Res. 2012, 40, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Kong, F.M.; Xu, Z.; Yu, S.P.; Sun, F.B.; Zhang, C.S.; Huang, Q.X.; Zhou, X.T.; Song, Z.W. Promoter hypermethylation of death-associated protein kinase gene in cholangiocarcinoma. Hepatobiliary Pancreat. Dis. Int. 2007, 6, 407–411. [Google Scholar] [PubMed]
- Shin, S.H.; Lee, K.; Kim, B.H.; Cho, N.Y.; Jang, J.Y.; Kim, Y.T.; Kim, D.; Jang, J.J.; Kang, G.H. Bile-based detection of extrahepatic cholangiocarcinoma with quantitative DNA methylation markers and its high sensitivity. J. Mol. Diagn. JMD 2012, 14, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Taniai, M.; Higuchi, H.; Burgart, L.J.; Gores, G.J. p16INK4a promoter mutations are frequent in primary sclerosing cholangitis (PSC) and PSC-associated cholangiocarcinoma. Gastroenterology 2002, 123, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Tannapfel, A.; Busse, C.; Geissler, F.; Witzigmann, H.; Hauss, J.; Wittekind, C. INK4a-ARF alterations in liver cell adenoma. Gut 2002, 51, 253–258. [Google Scholar] [CrossRef] [Green Version]
- Vedeld, H.M.; Andresen, K.; Eilertsen, I.A.; Nesbakken, A.; Seruca, R.; Gladhaug, I.P.; Thiis-Evensen, E.; Rognum, T.O.; Boberg, K.M.; Lind, G.E. The novel colorectal cancer biomarkers CDO1, ZSCAN18 and ZNF331 are frequently methylated across gastrointestinal cancers. Int. J. Cancer 2015, 136, 844–853. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, K.; Wang, J.; Wu, X.; Liu, X.; Li, B.; Zhu, Y.; Yu, Y.; Cheng, Q.; Hu, Z.; et al. Underexpression of LKB1 tumor suppressor is associated with enhanced Wnt signaling and malignant characteristics of human intrahepatic cholangiocarcinoma. Oncotarget 2015, 6, 18905–18920. [Google Scholar] [CrossRef] [Green Version]
- Wehbe, H.; Henson, R.; Meng, F.; Mize-Berge, J.; Patel, T. Interleukin-6 contributes to growth in cholangiocarcinoma cells by aberrant promoter methylation and gene expression. Cancer Res. 2006, 66, 10517–10524. [Google Scholar] [CrossRef] [Green Version]
- Xiang, J.; Luo, F.; Chen, Y.; Zhu, F.; Wang, J. si-DNMT1 restore tumor suppressor genes expression through the reversal of DNA hypermethylation in cholangiocarcinoma. Clin. Res. Hepatol. Gastroenterol. 2014, 38, 181–189. [Google Scholar] [CrossRef]
- Xiaofang, L.; Kun, T.; Shaoping, Y.; Zaiqiu, W.; Hailong, S. Correlation between promoter methylation of p14(ARF), TMS1/ASC, and DAPK, and p53 mutation with prognosis in cholangiocarcinoma. World J. Surg. Oncol. 2012, 10, 5. [Google Scholar] [CrossRef] [Green Version]
- Tian, X.; Zhang, S.; Liu, H.M.; Zhang, Y.B.; Blair, C.A.; Mercola, D.; Sassone-Corsi, P.; Zi, X. Histone lysine-specific methyltransferases and demethylases in carcinogenesis: New targets for cancer therapy and prevention. Curr. Cancer Drug Targets 2013, 13, 558–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biegel, J.A.; Busse, T.M.; Weissman, B.E. SWI/SNF chromatin remodeling complexes and cancer. Am. J. Med. Genet. Part C Semin. Med. Genet. 2014, 166c, 350–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, R.C.; Wang, T.L.; Shih Ie, M. The emerging roles of ARID1A in tumor suppression. Cancer Biol. Ther. 2014, 15, 655–664. [Google Scholar] [CrossRef] [Green Version]
- Braconi, C.; Huang, N.; Patel, T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology 2010, 51, 881–890. [Google Scholar] [CrossRef] [Green Version]
- Nakaoka, T.; Saito, Y.; Saito, H. Aberrant DNA Methylation as a Biomarker and a Therapeutic Target of Cholangiocarcinoma. Int. J. Mol. Sci. 2017, 18, 1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, F.; Wehbe-Janek, H.; Henson, R.; Smith, H.; Patel, T. Epigenetic regulation of microRNA-370 by interleukin-6 in malignant human cholangiocytes. Oncogene 2008, 27, 378–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaki, J.; Kikuchi, K.; Mizuguchi, Y.; Kawahigashi, Y.; Yoshida, H.; Uchida, E.; Takizawa, T. MiR-376c down-regulation accelerates EGF-dependent migration by targeting GRB2 in the HuCCT1 human intrahepatic cholangiocarcinoma cell line. PLoS ONE 2013, 8, e69496. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Chen, Y.; Tang, C.; Li, H.; Wang, B.; Yan, Q.; Hu, J.; Zou, S. MicroRNA-144 suppresses cholangiocarcinoma cell proliferation and invasion through targeting platelet activating factor acetylhydrolase isoform 1b. BMC Cancer 2014, 14, 917. [Google Scholar] [CrossRef] [Green Version]
- Sarver, A.L.; Sarver, A.E.; Yuan, C.; Subramanian, S. OMCD: OncomiR Cancer Database. BMC Cancer 2018, 18, 1223. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Xiao, J.; Chai, Y.; Du, Y.Y.; Liu, Z.; Huang, K.; Zhou, X.; Zhou, W. LncRNA-CCAT1 Promotes Migration, Invasion, and EMT in Intrahepatic Cholangiocarcinoma Through Suppressing miR-152. Dig. Dis. Sci. 2017, 62, 3050–3058. [Google Scholar] [CrossRef]
- Xu, Y.; Yao, Y.; Qin, W.; Zhong, X.; Jiang, X.; Cui, Y. Long non-coding RNA CCAT2 promotes cholangiocarcinoma cells migration and invasion by induction of epithelial-to-mesenchymal transition. Biomed. Pharmacother. 2018, 99, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Zu, C.; Liu, S.; Cao, W.; Liu, Z.; Qiang, H.; Li, Y.; Cheng, C.; Ji, L.; Li, J.; Li, J. MiR-590-3p suppresses epithelial-mesenchymal transition in intrahepatic cholangiocarcinoma by inhibiting SIP1 expression. Oncotarget 2017, 8, 34698–34708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Wang, Z.; Jiang, X.; Cui, Y. Overexpression of long noncoding RNA H19 indicates a poor prognosis for cholangiocarcinoma and promotes cell migration and invasion by affecting epithelial-mesenchymal transition. Biomed. Pharmacother. 2017, 92, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Li, J.; Ji, D.; Leng, K.; Xu, Y.; Huang, L.; Jiang, X.; Cui, Y. Overexpressed long noncoding RNA Sox2ot predicts poor prognosis for cholangiocarcinoma and promotes cell proliferation and invasion. Gene 2018, 645, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yang, X.; Zhang, H.; Yu, M.; Long, J.; Yang, T. Inhibition of miR-10a-5p suppresses cholangiocarcinoma cell growth through downregulation of Akt pathway. Oncotargets Ther. 2018, 11, 6981–6994. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Lu, L.; Tu, J.; Zhang, J.; Xiong, T.; Fan, W.; Wang, J.; Li, M.; Chen, Y.; Steggerda, J.; et al. Reciprocal Regulation Between Forkhead Box M1/NF-kB and Methionine Adenosyltransferase 1A Drives Liver Cancer. Hepatology 2020. [Google Scholar] [CrossRef]
- Fan, W.; Yang, H.; Liu, T.; Wang, J.; Li, T.W.; Mavila, N.; Tang, Y.; Yang, J.; Peng, H.; Tu, J.; et al. Prohibitin 1 suppresses liver cancer tumorigenesis in mice and human hepatocellular and cholangiocarcinoma cells. Hepatology 2017, 65, 1249–1266. [Google Scholar] [CrossRef] [Green Version]
- Srijiwangsa, P.; Ponnikorn, S.; Na-Bangchang, K. Effect of β-Eudesmol on NQO1 suppression-enhanced sensitivity of cholangiocarcinoma cells to chemotherapeutic agents. BMC Pharmacol. Toxicol. 2018, 19, 32. [Google Scholar] [CrossRef]
- Yeh, C.N.; Chen, M.H.; Chang, Y.C.; Wu, R.C.; Tsao, L.C.; Wang, S.Y.; Cheng, C.T.; Chiang, K.C.; Chen, T.W.; Hsiao, M.; et al. Over-expression of TNNI3K is associated with early-stage carcinogenesis of cholangiocarcinoma. Mol. Carcinog. 2019, 58, 270–278. [Google Scholar] [CrossRef]
- Hatzaras, I.; Schmidt, C.; Muscarella, P.; Melvin, W.S.; Ellison, E.C.; Bloomston, M. Elevated CA 19-9 portends poor prognosis in patients undergoing resection of biliary malignancies. HPB Off. J. Int. Hepato Pancreato Biliary Assoc. 2010, 12, 134–138. [Google Scholar] [CrossRef] [Green Version]
- Aksorn, N.; Roytrakul, S.; Kittisenachai, S.; Leelawat, K.; Chanvorachote, P.; Topanurak, S.; Hamano, S.; Lek-Uthai, U. Novel Potential Biomarkers for Opisthorchis viverrini Infection and Associated Cholangiocarcinoma. In Vivo 2018, 32, 871–878. [Google Scholar] [CrossRef] [PubMed]
- Boonla, C.; Sripa, B.; Thuwajit, P.; Cha-On, U.; Puapairoj, A.; Miwa, M.; Wongkham, S. MUC1 and MUC5AC mucin expression in liver fluke-associated intrahepatic cholangiocarcinoma. World J. Gastroenterol. 2005, 11, 4939–4946. [Google Scholar] [CrossRef] [PubMed]
- Gardini, A.; Corti, B.; Fiorentino, M.; Altimari, A.; Ercolani, G.; Grazi, G.L.; Pinna, A.D.; Grigioni, W.F.; D’Errico Grigioni, A. Expression of connective tissue growth factor is a prognostic marker for patients with intrahepatic cholangiocarcinoma. Dig. Liver Dis. Off. J. Ital. Soc. Gastroenterol. Ital. Assoc. Study Liver 2005, 37, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.Y.; Ke, A.W.; Shi, G.M.; Ding, Z.B.; Devbhandari, R.P.; Gu, F.M.; Li, Q.L.; Dai, Z.; Zhou, J.; Fan, J. Overexpression of CD151 as an adverse marker for intrahepatic cholangiocarcinoma patients. Cancer 2010, 116, 5440–5451. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.F.; Zhao, R.; Guo, S.; Wang, X.Q.; Lian, P.L.; Chen, Y.G.; Xu, K.S. Expression and clinical significance of hepatoma-derived growth factor as a prognostic factor in human hilar cholangiocarcinoma. Ann. Surg. Oncol. 2011, 18, 872–879. [Google Scholar] [CrossRef]
- Mao, X.; Chen, D.; Wu, J.; Li, J.; Zhou, H.; Wu, Y.; Duan, X. Differential expression of fascin, E-cadherin and vimentin: Proteins associated with survival of cholangiocarcinoma patients. Am. J. Med. Sci. 2013, 346, 261–268. [Google Scholar] [CrossRef]
- Merino-Azpitarte, M.; Lozano, E.; Perugorria, M.J.; Esparza-Baquer, A.; Erice, O.; Santos-Laso, Á.; O’Rourke, C.J.; Andersen, J.B.; Jiménez-Agüero, R.; Lacasta, A.; et al. SOX17 regulates cholangiocyte differentiation and acts as a tumor suppressor in cholangiocarcinoma. J. Hepatol. 2017, 67, 72–83. [Google Scholar] [CrossRef]
- Morine, Y.; Imura, S.; Ikemoto, T.; Iwahashi, S.; Saito, Y.U.; Shimada, M. CD44 Expression Is a Prognostic Factor in Patients with Intrahepatic Cholangiocarcinoma after Surgical Resection. Anticancer Res. 2017, 37, 5701–5705. [Google Scholar] [CrossRef]
- Nepal, C.; O’Rourke, C.J.; Oliveira, D.; Taranta, A.; Shema, S.; Gautam, P.; Calderaro, J.; Barbour, A.; Raggi, C.; Wennerberg, K.; et al. Genomic perturbations reveal distinct regulatory networks in intrahepatic cholangiocarcinoma. Hepatology 2018, 68, 949–963. [Google Scholar] [CrossRef] [Green Version]
- Shi, R.Y.; Yang, X.R.; Shen, Q.J.; Yang, L.X.; Xu, Y.; Qiu, S.J.; Sun, Y.F.; Zhang, X.; Wang, Z.; Zhu, K.; et al. High expression of Dickkopf-related protein 1 is related to lymphatic metastasis and indicates poor prognosis in intrahepatic cholangiocarcinoma patients after surgery. Cancer 2013, 119, 993–1003. [Google Scholar] [CrossRef]
- Shimada, M.; Sugimoto, K.; Iwahashi, S.; Utsunomiya, T.; Morine, Y.; Imura, S.; Ikemoto, T. CD133 expression is a potential prognostic indicator in intrahepatic cholangiocarcinoma. J. Gastroenterol. 2010, 45, 896–902. [Google Scholar] [CrossRef]
- Yoshikawa, D.; Ojima, H.; Iwasaki, M.; Hiraoka, N.; Kosuge, T.; Kasai, S.; Hirohashi, S.; Shibata, T. Clinicopathological and prognostic significance of EGFR, VEGF, and HER2 expression in cholangiocarcinoma. Br. J. Cancer 2008, 98, 418–425. [Google Scholar] [CrossRef]
- Navaneethan, U.; Lourdusamy, V.; Gk Venkatesh, P.; Willard, B.; Sanaka, M.R.; Parsi, M.A. Bile proteomics for differentiation of malignant from benign biliary strictures: A pilot study. Gastroenterol. Rep. 2015, 3, 136–143. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Morikawa, K.; Mori, Y.; Zong, C.; Zhang, L.; Garner, E.; Huang, C.; Wu, W.; Chang, J.; Nagashima, D.; et al. Proteomic analysis of liver proteins of mice exposed to 1,2-dichloropropane. Arch. Toxicol. 2020. [Google Scholar] [CrossRef]
- Son, K.H.; Ahn, C.B.; Kim, H.J.; Kim, J.S. Quantitative proteomic analysis of bile in extrahepatic cholangiocarcinoma patients. J. Cancer 2020, 11, 4073–4080. [Google Scholar] [CrossRef]
- Le Large, T.Y.S.; Meijer, L.L.; Paleckyte, R.; Boyd, L.N.C.; Kok, B.; Wurdinger, T.; Schelfhorst, T.; Piersma, S.R.; Pham, T.V.; van Grieken, N.C.T.; et al. Combined Expression of Plasma Thrombospondin-2 and CA19-9 for Diagnosis of Pancreatic Cancer and Distal Cholangiocarcinoma: A Proteome Approach. Oncologist 2020, 25, 634–643. [Google Scholar] [CrossRef] [Green Version]
- Cheon, Y.K.; Cho, Y.D.; Moon, J.H.; Jang, J.Y.; Kim, Y.S.; Kim, Y.S.; Lee, M.S.; Lee, J.S.; Shim, C.S. Diagnostic utility of interleukin-6 (IL-6) for primary bile duct cancer and changes in serum IL-6 levels following photodynamic therapy. Am. J. Gastroenterol. 2007, 102, 2164–2170. [Google Scholar] [CrossRef]
- Leelawat, K.; Sakchinabut, S.; Narong, S.; Wannaprasert, J. Detection of serum MMP-7 and MMP-9 in cholangiocarcinoma patients: Evaluation of diagnostic accuracy. BMC Gastroenterol. 2009, 9, 30. [Google Scholar] [CrossRef] [Green Version]
- Onsurathum, S.; Haonon, O.; Pinlaor, P.; Pairojkul, C.; Khuntikeo, N.; Thanan, R.; Roytrakul, S.; Pinlaor, S. Proteomics detection of S100A6 in tumor tissue interstitial fluid and evaluation of its potential as a biomarker of cholangiocarcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2018, 40. [Google Scholar] [CrossRef] [Green Version]
- Shen, J.; Wang, W.; Wu, J.; Feng, B.; Chen, W.; Wang, M.; Tang, J.; Wang, F.; Cheng, F.; Pu, L.; et al. Comparative proteomic profiling of human bile reveals SSP411 as a novel biomarker of cholangiocarcinoma. PLoS ONE 2012, 7, e47476. [Google Scholar] [CrossRef]
- Wang, M.; Fang, M.; Zhu, J.; Feng, H.; Warner, E.; Yi, C.; Ji, J.; Gu, X.; Gao, C. Serum N-glycans outperform CA19-9 in diagnosis of extrahepatic cholangiocarcinoma. Electrophoresis 2017, 38, 2749–2756. [Google Scholar] [CrossRef]
- Da, Z.; Gao, L.; Su, G.; Yao, J.; Fu, W.; Zhang, J.; Zhang, X.; Pei, Z.; Yue, P.; Bai, B.; et al. Bioinformatics combined with quantitative proteomics analyses and identification of potential biomarkers in cholangiocarcinoma. Cancer Cell Int. 2020, 20, 130. [Google Scholar] [CrossRef]
- Sulpice, L.; Desille, M.; Turlin, B.; Fautrel, A.; Boudjema, K.; Clément, B.; Coulouarn, C. Gene expression profiling of the tumor microenvironment in human intrahepatic cholangiocarcinoma. Genom. Data 2016, 7, 229–232. [Google Scholar] [CrossRef]
- Sulpice, L.; Rayar, M.; Desille, M.; Turlin, B.; Fautrel, A.; Boucher, E.; Llamas-Gutierrez, F.; Meunier, B.; Boudjema, K.; Clément, B.; et al. Molecular profiling of stroma identifies osteopontin as an independent predictor of poor prognosis in intrahepatic cholangiocarcinoma. Hepatology 2013, 58, 1992–2000. [Google Scholar] [CrossRef] [PubMed]
- Peraldo Neia, C.; Cavalloni, G.; Chiorino, G.; Ostano, P.; Aglietta, M.; Leone, F. Gene and microRNA modulation upon trabectedin treatment in a human intrahepatic cholangiocarcinoma paired patient derived xenograft and cell line. Oncotarget 2016, 7, 86766–86780. [Google Scholar] [CrossRef] [Green Version]
- Meijer, L.L.; Puik, J.R.; Le Large, T.Y.S.; Heger, M.; Dijk, F.; Funel, N.; Wurdinger, T.; Garajová, I.; van Grieken, N.C.T.; van de Wiel, M.A.; et al. Unravelling the Diagnostic Dilemma: A MicroRNA Panel of Circulating MiR-16 and MiR-877 as A Diagnostic Classifier for Distal Bile Duct Tumors. Cancers 2019, 11, 1181. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Yang, H.; Wan, L.; Wang, Z.; Wang, H.; Ge, C.; Liu, Y.; Hao, Y.; Zhang, D.; Shi, G.; et al. Single cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020. [Google Scholar] [CrossRef]
- Saengboonmee, C.; Seubwai, W.; Pairojkul, C.; Wongkham, S. High glucose enhances progression of cholangiocarcinoma cells via STAT3 activation. Sci. Rep. 2016, 6, 18995. [Google Scholar] [CrossRef]
- Ma, L.; Hernandez, M.O.; Zhao, Y.; Mehta, M.; Tran, B.; Kelly, M.; Rae, Z.; Hernandez, J.M.; Davis, J.L.; Martin, S.P.; et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell 2019, 36, 418–430. [Google Scholar] [CrossRef] [Green Version]
- Lapitz, A.; Arbelaiz, A.; O’Rourke, C.J.; Lavin, J.L.; Casta, A.; Ibarra, C.; Jimeno, J.P.; Santos-Laso, A.; Izquierdo-Sanchez, L.; Krawczyk, M.; et al. Patients with Cholangiocarcinoma Present Specific RNA Profiles in Serum and Urine Extracellular Vesicles Mirroring the Tumor Expression: Novel Liquid Biopsy Biomarkers for Disease Diagnosis. Cells 2020, 9, 721. [Google Scholar] [CrossRef] [Green Version]
- Bledea, R.; Vasudevaraja, V.; Patel, S.; Stafford, J.; Serrano, J.; Esposito, G.; Tredwin, L.M.; Goodman, N.; Kloetgen, A.; Golfinos, J.G.; et al. Functional and topographic effects on DNA methylation in IDH1/2 mutant cancers. Sci. Rep. 2019, 9, 16830. [Google Scholar] [CrossRef] [PubMed]
- Montal, R.; Sia, D.; Montironi, C.; Leow, W.Q.; Esteban-Fabró, R.; Pinyol, R.; Torres-Martin, M.; Bassaganyas, L.; Moeini, A.; Peix, J.; et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J. Hepatol. 2020. [Google Scholar] [CrossRef]
- Mondesir, J.; Willekens, C.; Touat, M.; de Botton, S. IDH1 and IDH2 mutations as novel therapeutic targets: Current perspectives. J. Blood Med. 2016, 7, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Raineri, S.; Mellor, J. IDH1: Linking Metabolism and Epigenetics. Front. Genet. 2018, 9, 493. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef]
- Liang, Q.; Liu, H.; Zhang, T.; Jiang, Y.; Xing, H.; Zhang, H. Serum metabolomics uncovering specific metabolite signatures of intra- and extrahepatic cholangiocarcinoma. Mol. Biosyst. 2016, 12, 334–340. [Google Scholar] [CrossRef]
- Alsaleh, M.; Leftley, Z.; Barbera, T.A.; Koomson, L.K.; Zabron, A.; Crossey, M.M.E.; Reeves, H.L.; Cramp, M.; Ryder, S.; Greer, S.; et al. Characterisation of the Serum Metabolic Signature of Cholangiocarcinoma in a United Kingdom Cohort. J. Clin. Exp. Hepatol. 2020, 10, 17–29. [Google Scholar] [CrossRef]
- Alsaleh, M.; Sithithaworn, P.; Khuntikeo, N.; Loilome, W.; Yongvanit, P.; Chamadol, N.; Hughes, T.; O’Connor, T.; Andrews, R.H.; Holmes, E.; et al. Characterisation of the Urinary Metabolic Profile of Liver Fluke-Associated Cholangiocarcinoma. J. Clin. Exp. Hepatol. 2019, 9, 657–675. [Google Scholar] [CrossRef] [Green Version]
- Prasopdee, S.; Thitapakorn, V.; Sathavornmanee, T.; Tesana, S. A comprehensive review of omics and host-parasite interplays studies, towards control of Opisthorchis viverrini infection for prevention of cholangiocarcinoma. Acta Trop. 2019, 196, 76–82. [Google Scholar] [CrossRef]
- Winter, H.; Kaisaki, P.J.; Harvey, J.; Giacopuzzi, E.; Ferla, M.P.; Pentony, M.M.; Knight, S.J.L.; Sharma, R.A.; Taylor, J.C.; McCullagh, J.S.O. Identification of Circulating Genomic and Metabolic Biomarkers in Intrahepatic Cholangiocarcinoma. Cancers 2019, 11, 1895. [Google Scholar] [CrossRef] [Green Version]
- Banales, J.M.; Iñarrairaegui, M.; Arbelaiz, A.; Milkiewicz, P.; Muntané, J.; Muñoz-Bellvis, L.; La Casta, A.; Gonzalez, L.M.; Arretxe, E.; Alonso, C.; et al. Serum Metabolites as Diagnostic Biomarkers for Cholangiocarcinoma, Hepatocellular Carcinoma, and Primary Sclerosing Cholangitis. Hepatology 2019, 70, 547–562. [Google Scholar] [CrossRef] [Green Version]
- Kotawong, K.; Chaijaroenkul, W.; Roytrakul, S.; Phaonakrop, N.; Na-Bangchang, K. Screening of Molecular Targets of Action of Atractylodin in Cholangiocarcinoma by Applying Proteomic and Metabolomic Approaches. Metabolites 2019, 9, 260. [Google Scholar] [CrossRef] [Green Version]
- Murakami, Y.; Kubo, S.; Tamori, A.; Itami, S.; Kawamura, E.; Iwaisako, K.; Ikeda, K.; Kawada, N.; Ochiya, T.; Taguchi, Y.H. Comprehensive analysis of transcriptome and metabolome analysis in Intrahepatic Cholangiocarcinoma and Hepatocellular Carcinoma. Sci. Rep. 2015, 5, 16294. [Google Scholar] [CrossRef] [Green Version]
- Urman, J.M.; Herranz, J.M.; Uriarte, I.; Rullán, M.; Oyón, D.; González, B.; Fernandez-Urién, I.; Carrascosa, J.; Bolado, F.; Zabalza, L.; et al. Pilot Multi-Omic Analysis of Human Bile from Benign and Malignant Biliary Strictures: A Machine-Learning Approach. Cancers 2020, 12, 1644. [Google Scholar] [CrossRef]
- Zhou, Z.; Ma, J. miR-378 serves as a prognostic biomarker in cholangiocarcinoma and promotes tumor proliferation, migration, and invasion. Cancer Biomark. Sect. A Dis. Markers 2019, 24, 173–181. [Google Scholar] [CrossRef]
- Liu, C.H.; Huang, Q.; Jin, Z.Y.; Xie, F.; Zhu, C.L.; Liu, Z.; Wang, C. Circulating microRNA-21 as a prognostic, biological marker in cholangiocarcinoma. J. Cancer Res. Ther. 2018, 14, 220–225. [Google Scholar] [CrossRef]
- Silakit, R.; Loilome, W.; Yongvanit, P.; Chusorn, P.; Techasen, A.; Boonmars, T.; Khuntikeo, N.; Chamadol, N.; Pairojkul, C.; Namwat, N. Circulating miR-192 in liver fluke-associated cholangiocarcinoma patients: A prospective prognostic indicator. J. Hepato-Biliary-Pancreat. Sci. 2014, 21, 864–872. [Google Scholar] [CrossRef]
- Wang, L.J.; Zhang, K.L.; Zhang, N.; Ma, X.W.; Yan, S.W.; Cao, D.H.; Shi, S.J. Serum miR-26a as a diagnostic and prognostic biomarker in cholangiocarcinoma. Oncotarget 2015, 6, 18631–18640. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wan, M.; Zhou, Q.; Wang, H.; Wang, Z.; Zhong, X.; Zhang, L.; Tai, S.; Cui, Y. The Prognostic Role of SOCS3 and A20 in Human Cholangiocarcinoma. PLoS ONE 2015, 10, e0141165. [Google Scholar] [CrossRef]
- Thongsom, S.; Chaocharoen, W.; Silsirivanit, A.; Wongkham, S.; Sripa, B.; Choe, H.; Suginta, W.; Talabnin, C. YKL-40/chitinase-3-like protein 1 is associated with poor prognosis and promotes cell growth and migration of cholangiocarcinoma. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2016, 37, 9451–9463. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, 98–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Ma, S. A selective review of robust variable selection with applications in bioinformatics. Brief. Bioinform. 2015, 16, 873–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, C.; Zhou, F.; Ren, J.; Li, X.; Jiang, Y.; Ma, S. A Selective Review of Multi-Level Omics Data Integration Using Variable Selection. High-Throughput 2019, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Abou-Alfa, G.K.; Sahai, V.; Hollebecque, A.; Vaccaro, G.; Melisi, D.; Al-Rajabi, R.; Paulson, A.S.; Borad, M.J.; Gallinson, D.; Murphy, A.G.; et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: A multicentre, open-label, phase 2 study. Lancet Oncol. 2020, 21, 671–684. [Google Scholar] [CrossRef]
- Javle, M.; Lowery, M.; Shroff, R.T.; Weiss, K.H.; Springfeld, C.; Borad, M.J.; Ramanathan, R.K.; Goyal, L.; Sadeghi, S.; Macarulla, T.; et al. Phase II Study of BGJ398 in Patients With FGFR-Altered Advanced Cholangiocarcinoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2018, 36, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.G.; Yu, Y.; Eathiraj, S.; Wang, Y.; Savage, R.E.; Lapierre, J.M.; Schwartz, B.; Abbadessa, G. Preclinical Activity of ARQ 087, a Novel Inhibitor Targeting FGFR Dysregulation. PLoS ONE 2016, 11, e0162594. [Google Scholar] [CrossRef] [Green Version]
- Goyal, L.; Shi, L.; Liu, L.Y.; Fece de la Cruz, F.; Lennerz, J.K.; Raghavan, S.; Leschiner, I.; Elagina, L.; Siravegna, G.; Ng, R.W.S.; et al. TAS-120 Overcomes Resistance to ATP-Competitive FGFR Inhibitors in Patients with FGFR2 Fusion-Positive Intrahepatic Cholangiocarcinoma. Cancer Discov. 2019, 9, 1064–1079. [Google Scholar] [CrossRef] [Green Version]
- Chae, Y.K.; Hong, F.; Vaklavas, C.; Cheng, H.H.; Hammerman, P.; Mitchell, E.P.; Zwiebel, J.A.; Ivy, S.P.; Gray, R.J.; Li, S.; et al. Phase II Study of AZD4547 in Patients With Tumors Harboring Aberrations in the FGFR Pathway: Results From the NCI-MATCH Trial (EAY131) Subprotocol W. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2020, 38, 2407–2417. [Google Scholar] [CrossRef]
- Voss, M.H.; Hierro, C.; Heist, R.S.; Cleary, J.M.; Meric-Bernstam, F.; Tabernero, J.; Janku, F.; Gandhi, L.; Iafrate, A.J.; Borger, D.R.; et al. A Phase I, Open-Label, Multicenter, Dose-escalation Study of the Oral Selective FGFR Inhibitor Debio 1347 in Patients with Advanced Solid Tumors Harboring FGFR Gene Alterations. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 2699–2707. [Google Scholar] [CrossRef] [Green Version]
- Borad, M.J.; Champion, M.D.; Egan, J.B.; Liang, W.S.; Fonseca, R.; Bryce, A.H.; McCullough, A.E.; Barrett, M.T.; Hunt, K.; Patel, M.D.; et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014, 10, e1004135. [Google Scholar] [CrossRef]
- Chong, D.Q.; Zhu, A.X. The landscape of targeted therapies for cholangiocarcinoma: Current status and emerging targets. Oncotarget 2016, 7, 46750–46767. [Google Scholar] [CrossRef] [Green Version]
- Hedvat, M.; Huszar, D.; Herrmann, A.; Gozgit, J.M.; Schroeder, A.; Sheehy, A.; Buettner, R.; Proia, D.; Kowolik, C.M.; Xin, H.; et al. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell 2009, 16, 487–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotschy, A.; Szlavik, Z.; Murray, J.; Davidson, J.; Maragno, A.L.; Le Toumelin-Braizat, G.; Chanrion, M.; Kelly, G.L.; Gong, J.N.; Moujalled, D.M.; et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016, 538, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Bekaii-Saab, T.; Phelps, M.A.; Li, X.; Saji, M.; Goff, L.; Kauh, J.S.; O’Neil, B.H.; Balsom, S.; Balint, C.; Liersemann, R.; et al. Multi-institutional phase II study of selumetinib in patients with metastatic biliary cancers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 2357–2363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padthaisong, S.; Dokduang, H.; Yothaisong, S.; Techasen, A.; Namwat, N.; Yongvanit, P.; Khuntikeo, N.; Titapun, A.; Sangkhamanon, S.; Loilome, W. Inhibitory effect of NVP-BKM120 on cholangiocarcinoma cell growth. Oncol. Lett. 2018, 16, 1627–1633. [Google Scholar] [CrossRef]
- Fujisaka, Y.; Kurata, T.; Tanaka, K.; Kudo, T.; Okamoto, K.; Tsurutani, J.; Kaneda, H.; Okamoto, I.; Namiki, M.; Kitamura, C.; et al. Phase I study of amatuximab, a novel monoclonal antibody to mesothelin, in Japanese patients with advanced solid tumors. Investig. New Drugs 2015, 33, 380–388. [Google Scholar] [CrossRef] [Green Version]
- Kwak, T.W.; Kim, D.H.; Jeong, Y.I.; Kang, D.H. Antitumor activity of vorinostat-incorporated nanoparticles against human cholangiocarcinoma cells. J. Nanobiotechnol. 2015, 13, 60. [Google Scholar] [CrossRef] [Green Version]
- Wang, B.; Li, H.; Yang, R.; Zhou, S.; Zou, S. Decitabine inhibits the cell growth of cholangiocarcinoma in cultured cell lines and mouse xenografts. Oncol. Lett. 2014, 8, 1919–1924. [Google Scholar] [CrossRef]
- Zhang, M.; Pan, Y.; Tang, D.; Dorfman, R.G.; Xu, L.; Zhou, Q.; Zhou, L.; Wang, Y.; Li, Y.; Yin, Y.; et al. Low levels of pyruvate induced by a positive feedback loop protects cholangiocarcinoma cells from apoptosis. Cell Commun. Signal. CCS 2019, 17, 23. [Google Scholar] [CrossRef] [Green Version]
- Pant, S.; Saleh, M.; Bendell, J.; Infante, J.R.; Jones, S.; Kurkjian, C.D.; Moore, K.M.; Kazakin, J.; Abbadessa, G.; Wang, Y.; et al. A phase I dose escalation study of oral c-MET inhibitor tivantinib (ARQ 197) in combination with gemcitabine in patients with solid tumors. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 1416–1421. [Google Scholar] [CrossRef]
- Demols, A.; Borbath, I.; Van den Eynde, M.; Houbiers, G.; Peeters, M.; Marechal, R.; Delaunoit, T.; Goemine, J.C.; Laurent, S.; Holbrechts, S.; et al. Regorafenib after failure of gemcitabine and platinum-based chemotherapy for locally advanced/metastatic biliary tumors: REACHIN, a randomized, double-blind, phase II trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020. [Google Scholar] [CrossRef]
- Sun, W.; Patel, A.; Normolle, D.; Patel, K.; Ohr, J.; Lee, J.J.; Bahary, N.; Chu, E.; Streeter, N.; Drummond, S. A phase 2 trial of regorafenib as a single agent in patients with chemotherapy-refractory, advanced, and metastatic biliary tract adenocarcinoma. Cancer 2019, 125, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.N.; Chang, Y.C.; Su, Y.; Shin-Shian Hsu, D.; Cheng, C.T.; Wu, R.C.; Chung, Y.H.; Chiang, K.C.; Yeh, T.S.; Lu, M.L.; et al. Identification of MALT1 as both a prognostic factor and a potential therapeutic target of regorafenib in cholangiocarcinoma patients. Oncotarget 2017, 8, 113444–113459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontugne, J.; Augustin, J.; Pujals, A.; Compagnon, P.; Rousseau, B.; Luciani, A.; Tournigand, C.; Cherqui, D.; Azoulay, D.; Pawlotsky, J.M.; et al. PD-L1 expression in perihilar and intrahepatic cholangiocarcinoma. Oncotarget 2017, 8, 24644–24651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Sun, L.; Li, Y.; Xie, F.; Zhou, X.; Yang, H.; Du, S.; Xu, H.; Mao, Y. The Clinicopathological and Prognostic Value of PD-L1 Expression in Cholangiocarcinoma: A Meta-Analysis. Front. Oncol. 2019, 9, 897. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, X.Y.; Zhang, Y.; Xu, D.; Dong, J.; Zhang, Z.; Yi, C.H.; Jia, H.L.; Yang, X. Programmed death ligand 1 expression in human intrahepatic cholangiocarcinoma and its association with prognosis and CD8(+) T-cell immune responses. Cancer Manag. Res. 2018, 10, 4113–4123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, K.T.; Yeh, C.N.; Chang, Y.C.; Cheng, J.H.; Fang, W.L.; Yeh, Y.C.; Wang, Y.C.; Hsu, D.S.; Wu, C.E.; Lai, J.I.; et al. PRKDC: New biomarker and drug target for checkpoint blockade immunotherapy. J. Immunother. Cancer 2020, 8. [Google Scholar] [CrossRef] [Green Version]

| Copy-Number Alteration (CNA) | ||||
| Gene | Cytoband | CAN | # | Frequency (%) |
| CDKN2A | 9p21.3 | HOMDEL | 21 | 9.1 |
| CDKN2B | 9p21.3 | HOMDEL | 16 | 6.9 |
| MCL1 | 1q21.2 | AMP | 11 | 4.8 |
| CCND1 | 11q13.3 | AMP | 8 | 3.5 |
| FDF4 | 11q13.3 | AMP | 8 | 3.5 |
| MDM2 | 12q15 | AMP | 8 | 3.5 |
| RIT1 | 1q22 | AMP | 8 | 3.5 |
| FDF19 | 11q13.3 | AMP | 8 | 3.5 |
| HIST2H3C | 1q21.2 | AMP | 6 | 3.4 |
| HIST2H3D | 1q21.2 | AMP | 6 | 3.4 |
| ERBB2 | 17q12 | AMP | 7 | 3 |
| FGF3 | 11q13.3 | AMP | 7 | 3 |
| MUC1 | 1q22 | AMP | 7 | 3 |
| MYC | 8q24.21 | AMP | 7 | 3 |
| NOTCH2 | 1p12 | AMP | 7 | 3 |
| Mutated Genes | ||||
| Gene | #Mut | Frequency (%) | ||
| TP53 | 114 | 27.3 | ||
| IDH1 | 64 | 16.2 | ||
| ARID1A | 69 | 16.2 | ||
| KRAS | 49 | 12.4 | ||
| BAP1 | 49 | 11.9 | ||
| PBRM1 | 37 | 9.3 | ||
| KMT2C | 28 | 7.1 | ||
| HLA-A | 37 | 6.7 | ||
| SMAD4 | 27 | 6.3 | ||
| ATM | 27 | 6.1 | ||
| Fusion Genes | ||||
| Gene | #Mut | Frequency (%) | ||
| FDFR2 | 19 | 4.8 | ||
| RAD21 | 1 | 0.3 | ||
| BRCA1 | 1 | 0.3 | ||
| BRAF | 1 | 0.3 | ||
| ESR1 | 1 | 0.3 | ||
| MAP3K1 | 1 | 0.3 | ||
| NOTCH2 | 1 | 0.3 | ||
| PIK3C2G | 1 | 0.3 | ||
| MAP2K1 | 1 | 0.3 | ||
| RB1 | 1 | 0.3 | ||
| miRNAs Significantly Associated with Survival in CHOL | |||
| miRNA Name | Z-Score | Log Rank p-Value | Upregulated in: |
| hsa-miR-802 | 0.000 | 3.02 × 10−3 | Living |
| hsa-miR-500b-5p | 2.718 | 3.57 × 10−3 | Living |
| hsa-miR-500a-5p | 2.674 | 4.27 × 10−3 | Living |
| hsa-miR-202-5p | 0.000 | 4.91 × 10−3 | Living |
| hsa-miR-551b-3p | 1.746 | 5.57 × 10−3 | Living |
| hsa-miR-129-5p | 2.166 | 5.79 × 10−3 | Living |
| hsa-miR-3161 | 3.025 | 9.51 × 10−3 | Decreased |
| hsa-miR-3199 | 0.173 | 1.50 × 10−2 | Living |
| hsa-miR-1228-5p | 0.000 | 2.03 × 10−2 | Living |
| hsa-miR-10b-3p | 1.970 | 2.40 × 10−2 | Living |
| miRNAs Associated with Tumorigenesis of CHOL | |||
| miRNA Name | t-Test p-Value | t-Test FDR | Upregulated in: |
| hsa-miR-183-5p | 3.84 × 10−9 | 2.16 × 10−6 | Tumor |
| hsa-miR-182-5p | 1.15 × 10−8 | 3.25 × 10−6 | Tumor |
| hsa-miR-194-3p | 2.88 × 10−8 | 5.41 × 10−6 | Normal |
| hsa-miR-125b-2-3p | 1.35 × 10−7 | 1.85 × 10−5 | Normal |
| hsa-let-7c-5p | 1.71 × 10−7 | 1.85 × 10−5 | Normal |
| hsa-miR-378a-3p | 1.97 × 10−7 | 1.85 × 10−5 | Normal |
| hsa-miR-92b-3p | 2.60 × 10−7 | 2.09 × 10−5 | Tumor |
| hsa-miR-1258 | 3.43 × 10−7 | 2.41 × 10−5 | Normal |
| hsa-miR-378c | 4.13 × 10−7 | 2.59 × 10−5 | Normal |
| hsa-miR-23c | 8.50 × 10−7 | 4.36 × 10−5 | Normal |
| LncRNAs/CircRNAs Dependency of CCA | |||
|---|---|---|---|
| LncRNAs Symbol | Disease Name | Confidence Score | Database ID |
| SPRY4-IT1 | CCA | 0.9820 | LDA0003741 |
| CPS1-IT1 | iCCA | 0.9820 | LDA0004726 |
| MALAT1 | CCA | 0.9526 | LDA0003742 |
| NEAT1 | CCA | 0.9526 | LDA0003743 |
| SOX2-OT | CCA | 0.9462 | LDA0003746 |
| AFAP1-AS1 | CCA | 0.8808 | LDA0003736 |
| UCA1 | CCA | 0.8808 | LDA0003749 |
| CCAT1 | iCCA | 0.8808 | LDA0004724 |
| H19 | CCA | 0.8785 | LDA0003740 |
| HULC | CCA | 0.8785 | LDA0003741 |
| PCAT1 | CCA | 0.8785 | LDA0003745 |
| CCAT2 | CCA | 0.7311 | LDA0003737 |
| PANDAR | CCA | 0.7311 | LDA0003744 |
| TUG1 | CCA | 0.7311 | LDA0003748 |
| PCAT1 | eCCA | 0.7311 | LDA0004199 |
| TLINC | iCCA | 0.7311 | LDA0004727 |
| TUG1 | iCCA | 0.7311 | LDA0004728 |
| CDR1-AS | CCA | 0.7311 | LDA0178708 |
| hsa-circ_0001649 | CCA | 0.7311 | LDA0178709 |
| CCAT2 | iCCA | 0.6606 | LDA0004725 |
| ENST00000517758.1 | CCA | 0.5483 | LDA0003738 |
| ENST00000588480.1 | CCA | 0.5483 | LDA0003739 |
| Overall Survival | ||
| Gene Symbol | Gene ID | pValue |
| CTD-2033C11.1 | ENSG00000269961.1 | 4.36 × 10−5 |
| AD001527.7 | ENSG00000270760.1 | 1.46 × 10−4 |
| FAM86KP | ENSG00000163612.10 | 4.47 × 10−4 |
| RP11-209M4.1 | ENSG00000267253.1 | 9.00 × 10−4 |
| PTGER3 | ENSG00000050628.20 | 9.41 × 10−4 |
| AP003774.6 | ENSG00000231680.1 | 9.95 × 10−4 |
| FAM177A1 | ENSG00000151327.12 | 1.00 × 10−3 |
| RP11-574F21.2 | ENSG00000228606.1 | 1.05 × 10−3 |
| CXCL17 | ENSG00000189377.8 | 1.14 × 10−3 |
| FUT4 | ENSG00000196371.3 | 1.27 × 10−3 |
| Disease-Free Survival | ||
| Gene Symbol | Gene ID | pValue |
| AC104654.2 | ENSG00000234362.5 | 1.31 × 10−6 |
| AP003774.6 | ENSG00000231680.1 | 5.59 × 10−6 |
| NADK | ENSG00000008130.15 | 3.05 × 10−5 |
| AGAP2 | ENSG00000135439.11 | 3.59 × 10−5 |
| NRXN2 | ENSG00000110076.18 | 4.80 × 10−5 |
| RP1-28O10.1 | ENSG00000227591.5 | 4.81 × 10−5 |
| CPSF3L | ENSG00000127054.18 | 7.45 × 10−5 |
| TPRG1L | ENSG00000158109.14 | 8.32 × 10−5 |
| CDC40 | ENSG00000168438.14 | 8.54 × 10−5 |
| KLHL34 | ENSG00000185915.5 | 8.86 × 10−5 |
| Compound | Target | Clinical Trial Number | Phase | Status |
|---|---|---|---|---|
| NVP-BGJ398 (infigratinib) | FGFR | NCT02150967 | II | Recruiting |
| JNJ-42756493 (Erdafitinib) | FGFR | NCT02699606 | II | Recruiting |
| ARQ 087 (derazantinib) | FGFR | NCT017529520 | I + II | Active |
| TAS-120 | FGFR | NCT02052778 | I + II | Active |
| CH5183284 (debio 1347) | FGFR | NCT03834220 | II | Active |
| Ponatinib | FGFR2 | NCT02265341 | II | Completed |
| FPA144 (bemarituzumab) | FGFR2 | N/A | ||
| Pemazyre (pemigatinib) | FGFR2 | NCT03656536 | III | Recruiting |
| AG-120 (Ivosidenib) | IDH1 | NCT02073994 | I | Active |
| Trametinib | MEK | NCT02070549/NCT02042443 | I/II | Recruiting/Completed |
| Pazopanib | MEK/VEGFR/PDGFR/REF | NCT01855724 | II | Terminated |
| Pembrolizumab | PD-1 | NCT03111732 | II | Recruiting |
| Regorafenib | MEK/VEGFR/PDGFR/REF | NCT02115542 | II | Active |
| AGI-5198 | IDH1 | N/A | ||
| AGI-6780 | IDH2 | N/A | ||
| AG-221 (enasidenib) | IDH2 | NCT02273739 | I + II | Completed |
| Isoquinoline | PKA | N/A | ||
| S63845 | MCL1 | N/A | ||
| AZD1480 | JAK | N/A | ||
| AZD6244 (selumetinib) | MEK1/2 | NCT00553332 | II | Completed |
| BKM120 (buparlisib) | PI3K | NCT01501604 | II | Withdrawn |
| Amatuximab | Mesothelin | NCT01766219 | I + II | Completed |
| Azacitidine | DNMT | N/A | ||
| Decitabine | DNMT | N/A | ||
| Vorinostat | HDAC | N/A | ||
| Romidepsin | HDAC | N/A | ||
| Tivantinib | MET | N/A | ||
| Cabozantinib | MET | NCT01954745 | II | Completed |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, Y.-C.; Chen, M.-H.; Yeh, C.-N.; Hsiao, M. Omics-Based Platforms: Current Status and Potential Use for Cholangiocarcinoma. Biomolecules 2020, 10, 1377. https://doi.org/10.3390/biom10101377
Chang Y-C, Chen M-H, Yeh C-N, Hsiao M. Omics-Based Platforms: Current Status and Potential Use for Cholangiocarcinoma. Biomolecules. 2020; 10(10):1377. https://doi.org/10.3390/biom10101377
Chicago/Turabian StyleChang, Yu-Chan, Ming-Huang Chen, Chun-Nan Yeh, and Michael Hsiao. 2020. "Omics-Based Platforms: Current Status and Potential Use for Cholangiocarcinoma" Biomolecules 10, no. 10: 1377. https://doi.org/10.3390/biom10101377
APA StyleChang, Y.-C., Chen, M.-H., Yeh, C.-N., & Hsiao, M. (2020). Omics-Based Platforms: Current Status and Potential Use for Cholangiocarcinoma. Biomolecules, 10(10), 1377. https://doi.org/10.3390/biom10101377







