Structural and Biophysical Analyses of Human N-Myc Downstream-Regulated Gene 3 (NDRG3) Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Cloning, Protein Expression, and Purification of NDRG3
2.2. Mutagenesis and Purification of NDRG3 ΔNC
2.3. Crystallization
2.4. X-ray Data Collection, Refinement and Structure Determination
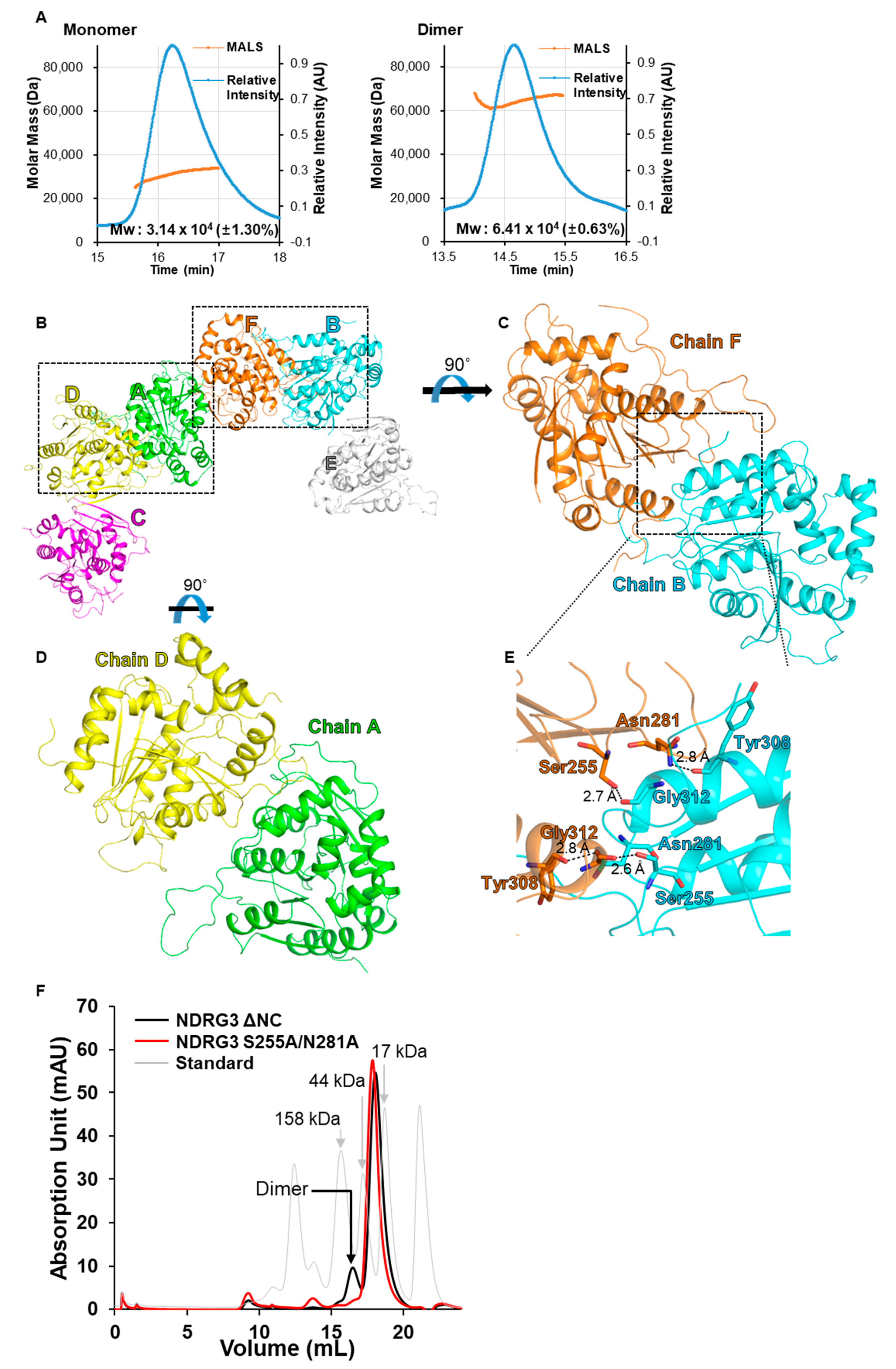
2.5. Size-Exclusion Chromatography with Multi-Angle Light Scattering (SEC-MALS) Analysis
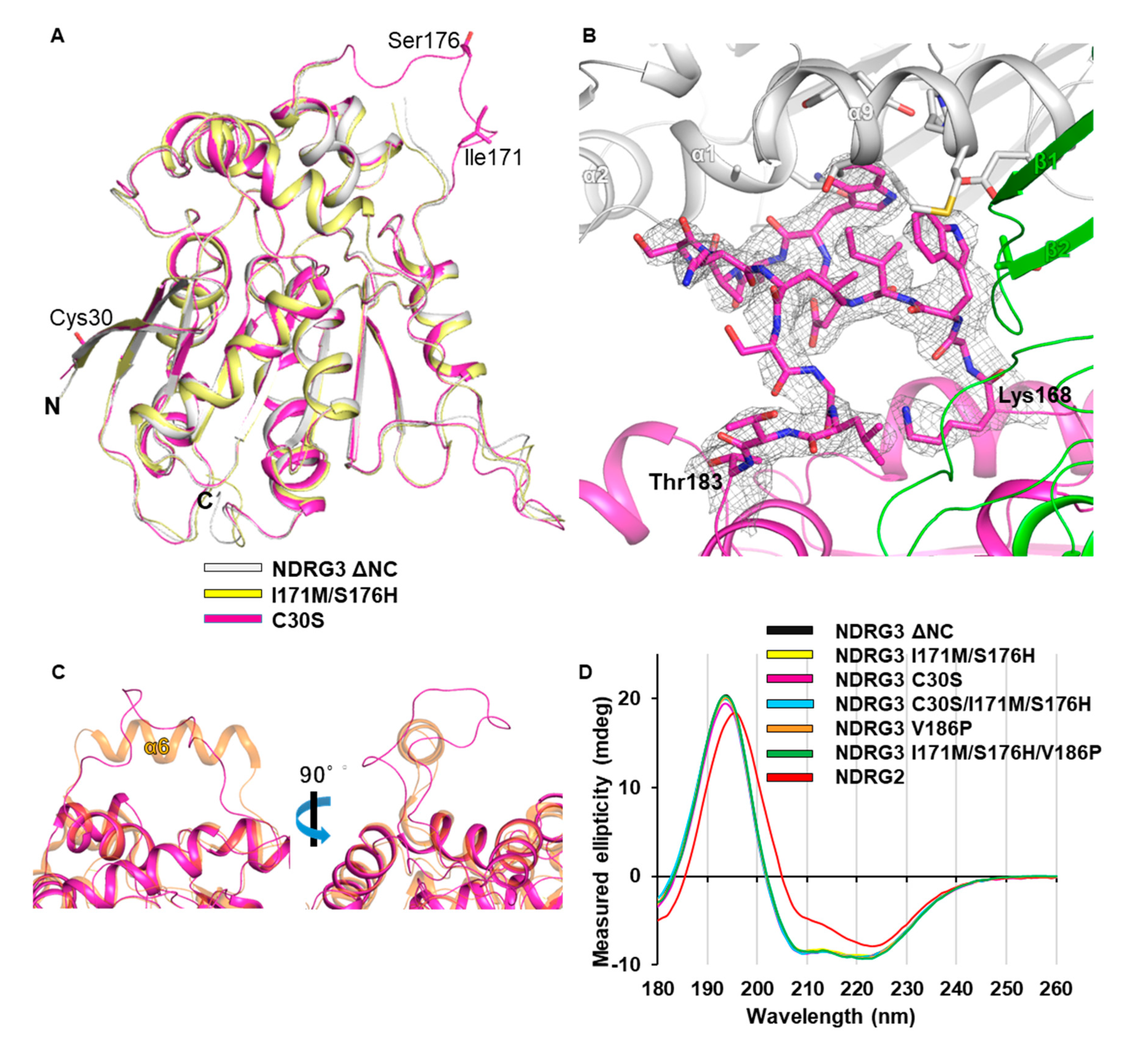
2.6. Circular Dichroism (CD)
3. Results
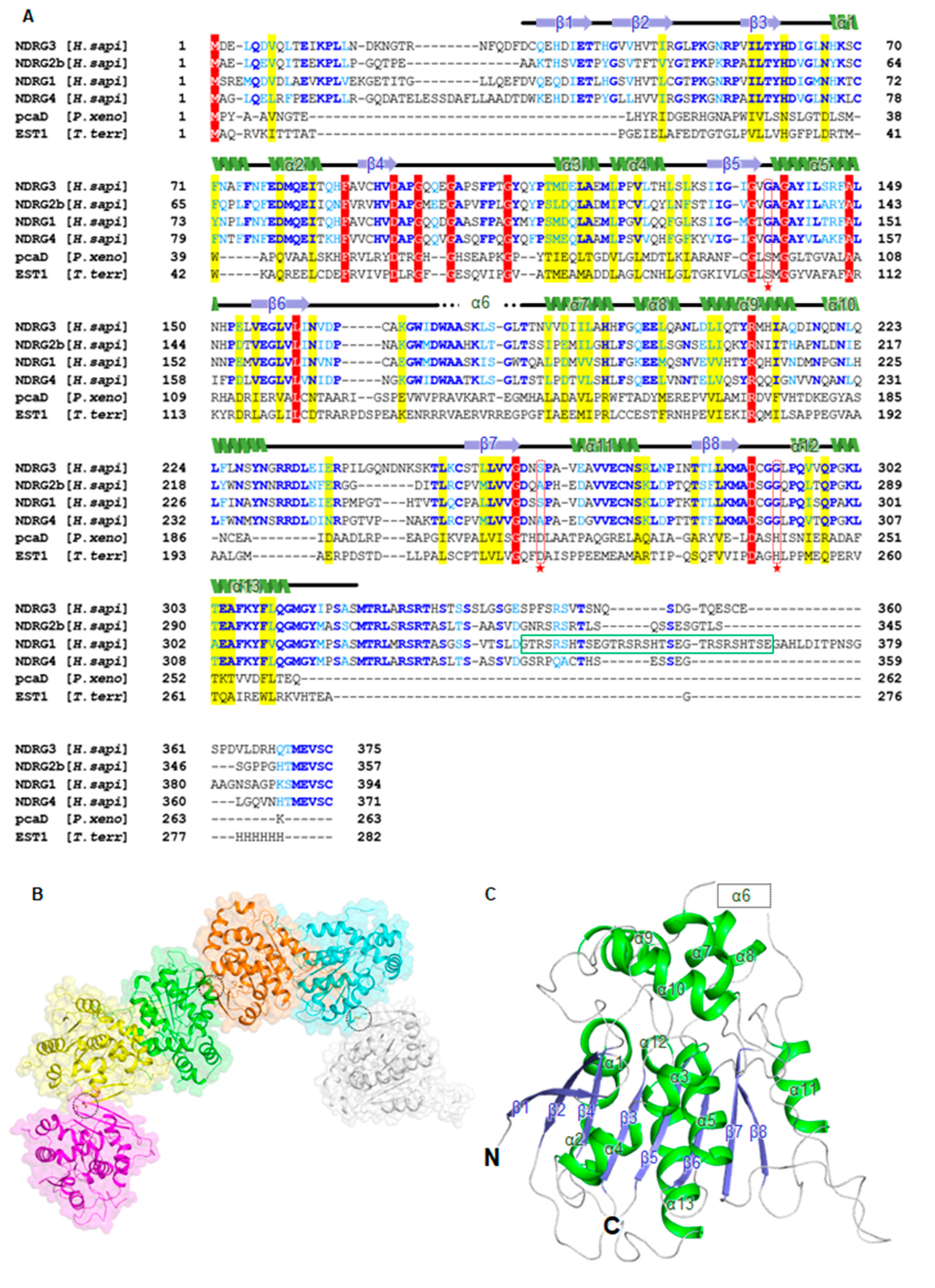
3.1. Overall Structure of Human NDRG3 Contains an α/β-Hydrolase Fold Domain and a Cap-Like Domain
3.2. Crystal Packing of NDRG3 Structure Indicates Dimeric Interactions
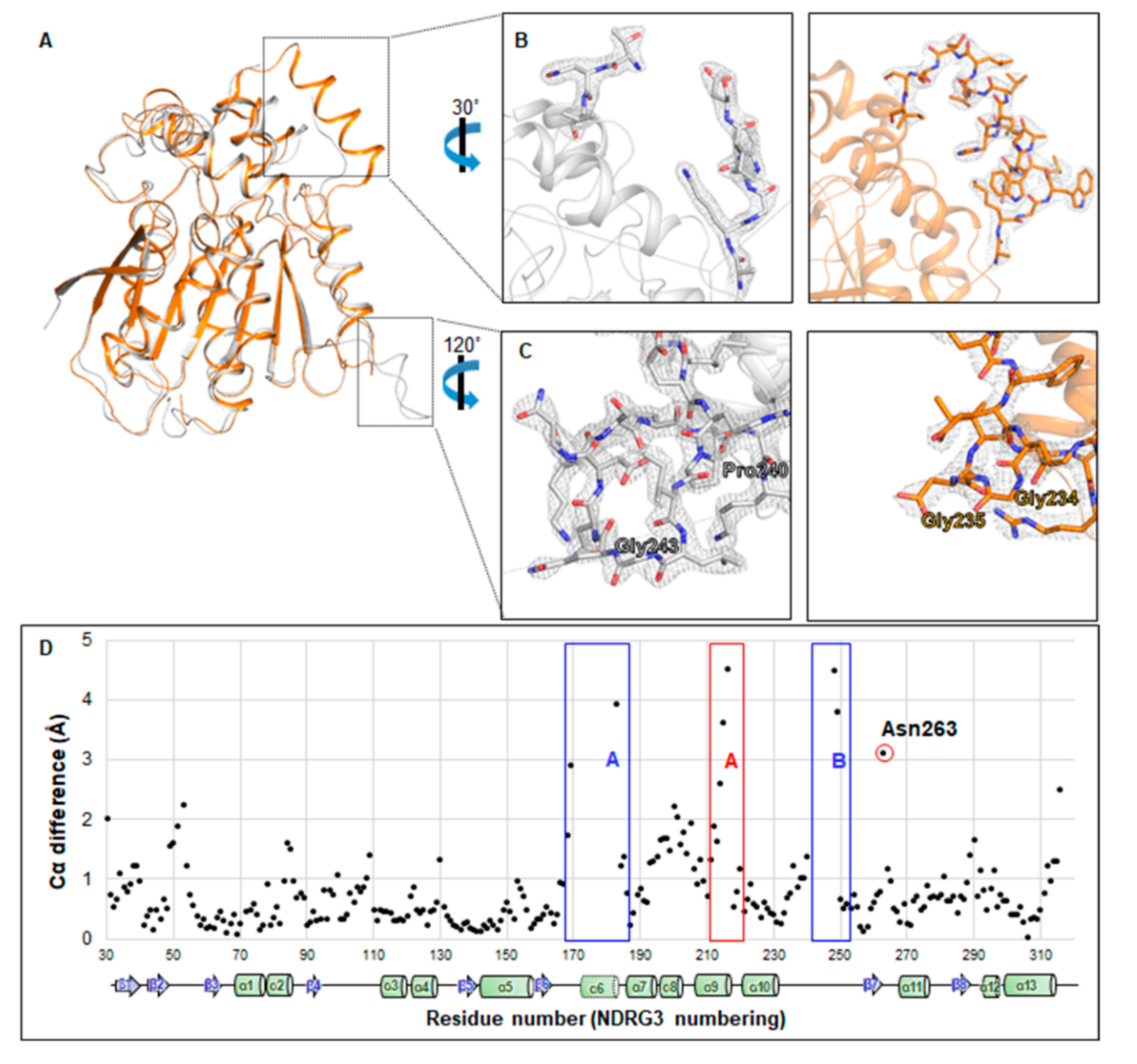
3.3. NDRG3 Shows a Structural Similarity to NDRG2 and Contains a Distinctive Disordered Region and a Solvent Accessible Cavity
3.4. Unfolded Helix α6 Region of NDRG3 Is a Flexible Loop
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, S.J.; Silman, I.; Schrag, J.; et al. The alpha/beta hydrolase fold. Protein Eng. 1992, 5, 197–211. [Google Scholar] [CrossRef] [PubMed]
- Botti, S.A.; Felder, C.E.; Sussman, J.L.; Silman, I. Electrotactins: A class of adhesion proteins with conserved electrostatic and structural motifs. Protein Eng. 1998, 11, 415–420. [Google Scholar] [CrossRef]
- De Jaco, A.; Comoletti, D.; Dubi, N.; Camp, S.; Taylor, P. Processing of cholinesterase-like alpha/beta-hydrolase fold proteins: Alterations associated with congenital disorders. Protein Pept. Lett. 2012, 19, 173–179. [Google Scholar] [CrossRef]
- Marchot, P.; Chatonnet, A. Enzymatic activity and protein interactions in alpha/beta hydrolase fold proteins: Moonlighting versus promiscuity. Protein Pept. Lett. 2012, 19, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Shimono, A.; Okuda, T.; Kondoh, H. N-myc-dependent repression of ndr1, a gene identified by direct subtraction of whole mouse embryo cDNAs between wild type and N-myc mutant. Mech. Dev. 1999, 83, 39–52. [Google Scholar] [CrossRef]
- Shaw, E.; McCue, L.A.; Lawrence, C.E.; Dordick, J.S. Identification of a novel class in the alpha/beta hydrolase fold superfamily: The N-myc differentiation-related proteins. Proteins 2002, 47, 163–168. [Google Scholar] [CrossRef]
- O’Connell, B.C.; Cheung, A.F.; Simkevich, C.P.; Tam, W.; Ren, X.; Mateyak, M.K.; Sedivy, J.M. A large scale genetic analysis of c-Myc-regulated gene expression patterns. J. Biol. Chem. 2003, 278, 12563–12573. [Google Scholar] [CrossRef]
- Vervoorts, J.; Luscher-Firzlaff, J.; Luscher, B. The ins and outs of MYC regulation by posttranslational mechanisms. J. Biol. Chem. 2006, 281, 34725–34729. [Google Scholar] [CrossRef]
- Qu, X.; Zhai, Y.; Wei, H.; Zhang, C.; Xing, G.; Yu, Y.; He, F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol. Cell Biochem. 2002, 229, 35–44. [Google Scholar] [CrossRef]
- Kurdistani, S.K.; Arizti, P.; Reimer, C.L.; Sugrue, M.M.; Aaronson, S.A.; Lee, S.W. Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res. 1998, 58, 4439–4444. [Google Scholar]
- Guan, R.J.; Ford, H.L.; Fu, Y.; Li, Y.; Shaw, L.M.; Pardee, A.B. Drg-1 as a differentiation-related, putative metastatic suppressor gene in human colon cancer. Cancer Res. 2000, 60, 749–755. [Google Scholar] [PubMed]
- Mao, Z.; Sun, J.; Feng, B.; Ma, J.; Zang, L.; Dong, F.; Zhang, D.; Zheng, M. The metastasis suppressor, N-myc downregulated gene 1 (NDRG1), is a prognostic biomarker for human colorectal cancer. PLoS ONE 2013, 8, e68206. [Google Scholar] [CrossRef]
- Deng, Y.; Yao, L.; Chau, L.; Ng, S.S.; Peng, Y.; Liu, X.; Au, W.S.; Wang, J.; Li, F.; Ji, S.; et al. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int. J. Cancer 2003, 106, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Yoon, S.Y.; Kim, J.T.; Choi, S.C.; Lim, J.S.; Kim, J.H.; Song, E.Y.; Lee, H.G.; Choi, I.; Kim, J.W. NDRG2 suppresses cell proliferation through down-regulation of AP-1 activity in human colon carcinoma cells. Int. J. Cancer 2009, 124, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Furuta, H.; Kondo, Y.; Nakahata, S.; Hamasaki, M.; Sakoda, S.; Morishita, K. NDRG2 is a candidate tumor-suppressor for oral squamous-cell carcinoma. Biochem. Biophys. Res. Commun. 2010, 391, 1785–1791. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.J.; Liao, C.G.; Jiang, X.; Zhao, H.D.; Yao, L.B.; Bao, T.Y. NDRG2 suppresses the proliferation of clear cell renal cell carcinoma cell A-498. J. Exp. Clin. Cancer Res. 2010, 29, 103. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Liu, W.; Yan, X.; Wang, Q.; Zhao, Q.; Xue, Y.; Ren, H.; Wu, L.; Cheng, Y.; Li, S.; et al. Inhibition of endothelial cell proliferation and tumor angiogenesis by up-regulating NDRG2 expression in breast cancer cells. PLoS ONE 2012, 7, e32368. [Google Scholar] [CrossRef]
- Huang, J.; Wu, Z.; Wang, G.; Cai, Y.; Cai, M.; Li, Y. N-Myc downstreamregulated gene 2 suppresses the proliferation of T24 human bladder cancer cells via induction of oncosis. Mol. Med. Rep. 2015, 12, 5730–5736. [Google Scholar] [CrossRef][Green Version]
- Hong, S.N.; Kim, S.J.; Kim, E.R.; Chang, D.K.; Kim, Y.H. Epigenetic silencing of NDRG2 promotes colorectal cancer proliferation and invasion. J. Gastroenterol. Hepatol. 2016, 31, 164–171. [Google Scholar] [CrossRef]
- Kim, Y.J.; Yoon, S.Y.; Kim, J.T.; Song, E.Y.; Lee, H.G.; Son, H.J.; Kim, S.Y.; Cho, D.; Choi, I.; Kim, J.H.; et al. NDRG2 expression decreases with tumor stages and regulates TCF/beta-catenin signaling in human colon carcinoma. Carcinogenesis 2009, 30, 598–605. [Google Scholar] [CrossRef]
- Kim, A.; Kim, M.J.; Yang, Y.; Kim, J.W.; Yeom, Y.I.; Lim, J.S. Suppression of NF-kappaB activity by NDRG2 expression attenuates the invasive potential of highly malignant tumor cells. Carcinogenesis 2009, 30, 927–936. [Google Scholar] [CrossRef] [PubMed]
- Takarada-Iemata, M.; Yoshikawa, A.; Ta, H.M.; Okitani, N.; Nishiuchi, T.; Aida, Y.; Kamide, T.; Hattori, T.; Ishii, H.; Tamatani, T.; et al. N-myc downstream-regulated gene 2 protects blood-brain barrier integrity following cerebral ischemia. Glia 2018, 66, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, Y.; Hong, A.; Wang, J.; Lin, B.; Li, R. NDRG3 is an androgen regulated and prostate enriched gene that promotes in vitro and in vivo prostate cancer cell growth. Int. J. Cancer 2009, 124, 521–530. [Google Scholar] [CrossRef]
- Lee, D.C.; Sohn, H.A.; Park, Z.Y.; Oh, S.; Kang, Y.K.; Lee, K.M.; Kang, M.; Jang, Y.J.; Yang, S.J.; Hong, Y.K.; et al. A lactate-induced response to hypoxia. Cell 2015, 161, 595–609. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Zhang, X.; Jiang, H.; Jiang, X.; Wang, L.; Qi, Q.; Bi, Y.; Wang, J.; Shi, Q.; Li, R. Ndrg3 gene regulates DSB repair during meiosis through modulation the ERK signal pathway in the male germ cells. Sci. Rep. 2017, 7, 44440. [Google Scholar] [CrossRef] [PubMed]
- Cui, C.; Lin, H.; Shi, Y.; Pan, R. Hypoxic postconditioning attenuates apoptosis via inactivation of adenosine A2a receptor through NDRG3-Raf-ERK pathway. Biochem. Biophys. Res. Commun. 2017, 491, 277–284. [Google Scholar] [CrossRef]
- Li, T.; Sun, R.; Lu, M.; Chang, J.; Meng, X.; Wu, H. NDRG3 facilitates colorectal cancer metastasis through activating Src phosphorylation. OncoTargets Ther. 2018, 11, 2843–2852. [Google Scholar] [CrossRef]
- Shi, J.; Zheng, H.; Yuan, L. High NDRG3 expression facilitates HCC metastasis by promoting nuclear translocation of beta-catenin. BMB Rep. 2019, 52, 451–456. [Google Scholar] [CrossRef]
- Lee, G.Y.; Shin, S.H.; Shin, H.W.; Chun, Y.S.; Park, J.W. NDRG3 lowers the metastatic potential in prostate cancer as a feedback controller of hypoxia-inducible factors. Exp. Mol. Med. 2018, 50, 61. [Google Scholar] [CrossRef]
- Di Tommaso, P.; Moretti, S.; Xenarios, I.; Orobitg, M.; Montanyola, A.; Chang, J.M.; Taly, J.F.; Notredame, C. T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011, 39, W13–W17. [Google Scholar] [CrossRef]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.; Kim, Y.; Kang, H.B.; Jaroszewski, L.; Deacon, A.M.; Lee, H.; Choi, W.C.; Kim, K.J.; Kim, C.H.; Kang, B.S.; et al. Crystal structure of the human N-Myc downstream-regulated gene 2 protein provides insight into its role as a tumor suppressor. J. Biol. Chem. 2011, 286, 12450–12460. [Google Scholar] [CrossRef] [PubMed]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Vagin, A.A.; Steiner, R.A.; Lebedev, A.A.; Potterton, L.; McNicholas, S.; Long, F.; Murshudov, G.N. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Winn, M.D.; Ballard, C.C.; Cowtan, K.D.; Dodson, E.J.; Emsley, P.; Evans, P.R.; Keegan, R.M.; Krissinel, E.B.; Leslie, A.G.; McCoy, A.; et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef]
- Joosten, R.P.; Long, F.; Murshudov, G.N.; Perrakis, A. The PDB_REDO server for macromolecular structure model optimization. IUCrJ 2014, 1, 213–220. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B., 3rd; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef]
- Schrodinger, L.L.C. The PyMOL Molecular Graphics System; Version 1.8; Schrodinger, L.L.C.: New York, NY, USA, 2015. [Google Scholar]
- Slabinski, L.; Jaroszewski, L.; Rychlewski, L.; Wilson, I.A.; Lesley, S.A.; Godzik, A. XtalPred: A web server for prediction of protein crystallizability. Bioinformatics 2007, 23, 3403–3405. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res. 2015, 43, D512–D520. [Google Scholar] [CrossRef]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef]
- Holm, L.; Laakso, L.M. Dali server update. Nucleic Acids Res. 2016, 44, W351–W355. [Google Scholar] [CrossRef] [PubMed]
- Pace, C.N.; Scholtz, J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998, 75, 422–427. [Google Scholar] [CrossRef]
- Manning, M.C.; Woody, R.W. Theoretical CD studies of polypeptide helices: Examination of important electronic and geometric factors. Biopolymers 1991, 31, 569–586. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.Y.; Taneja, A.K.; Hodges, R.S. Synthesis of a model protein of defined secondary and quaternary structure. Effect of chain length on the stabilization and formation of two-stranded alpha-helical coiled-coils. J. Biol. Chem. 1984, 259, 13253–13261. [Google Scholar] [PubMed]
- Choy, N.; Raussens, V.; Narayanaswami, V. Inter-molecular coiled-coil formation in human apolipoprotein E C-terminal domain. J. Mol. Biol. 2003, 334, 527–539. [Google Scholar] [CrossRef]
- Udeshi, N.D.; Svinkina, T.; Mertins, P.; Kuhn, E.; Mani, D.R.; Qiao, J.W.; Carr, S.A. Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell Proteom. 2013, 12, 825–831. [Google Scholar] [CrossRef]
- Appelhoff, R.J.; Tian, Y.M.; Raval, R.R.; Turley, H.; Harris, A.L.; Pugh, C.W.; Ratcliffe, P.J.; Gleadle, J.M. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J. Biol. Chem. 2004, 279, 38458–38465. [Google Scholar] [CrossRef]
| NDRG3 ΔNC | NDRG3 C30S | NDRG3 I171M/S176H | |
|---|---|---|---|
| Data Collection a | |||
| Beamline | PLS-7A | PLS-7A | PLS-11C |
| Space group | C2 | P3121 | P3221 |
| Cell dimensions | |||
| a, b, c (Å), α, β, γ (°) | 173.34, 100.15, 110.74, 90.00, 90.01, 90.00 | 99.76, 99.76, 332.71, 90.00, 90.00, 120.00 | 100.39, 100.39, 111.76, 90.00, 90.00, 120.00 |
| X-ray wavelength (Å) | 0.9793 | 0.9793 | 0.9794 |
| Resolution (Å) b | 50.0–2.2 (2.24–2.20) | 50.0–3.4 (3.46–3.40) | 50.0–3.3 (3.36–3.30) |
| <I/σ(I)> | 15.9 (2.5) | 16.6 (2.6) | 14.0 (3.0) |
| Unique reflections Redundancy | 95,796 (4,774) 5.2 (5.1) | 26,798 (1,311) 6.6 (6.7) | 10,073 (485) 10.5 (8.8) |
| Completeness (%) | 99.7 (99.3) | 97.7 (97.5) | 99.3 (95.8) |
| Rmerge (%) c Rp.i.m (%) d | 10.1 (64.1) 4.9 (31.1) | 10.3 (73.2) 3.9 (27.5) | 16.4 (62.9) 5.4 (21.5) |
| Refinement | |||
| No. of reflections | 88,791 | 25,238 | 9142 |
| Resolution (Å) | 50.0–2.2 (2.24–2.20) | 50.0–3.4 (3.46–3.40) | 50.0–3.3 (3.36–3.30) |
| Rework/Rfreef (%) | 16.8%/18.5% | 24.1%/27.7% | 19.6%/22.6% |
| Twin fraction | 0.172, 0.177, 0.195, 0.129, 0.131, 0.197 g | 0.502, 0.498 h | |
| No. of subunits | 6 | 4 | 2 |
| No. of protein atoms | 13,139 | 8673 | 4345 |
| No. of solvent atoms | 334 | 0 | 6 |
| Mean B value (Å2) | 31.59 | 125.18 | 33.22 |
| Ramachandran plot (%) | |||
| favored | 1629 (97.7%) | 1085 (98.4%) | 541 (98.0%) |
| allowed | 39 (2.3%) | 18 (1.6%) | 11 (2.0%) |
| outliers | 0 (0%) | 0 (0%) | 0 (0%) |
| Rotamer outliers (%) | 0 (0%) | 0 (0%) | 0 (0%) |
| r.m.s. deviations | |||
| bond lengths (Å) | 0.002 | 0.003 | 0.004 |
| bond angles (°) | 1.143 | 1.246 | 1.279 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.R.; Kim, K.A.; Park, J.S.; Jang, J.Y.; Choi, Y.; Lee, H.H.; Lee, D.C.; Park, K.C.; Yeom, Y.I.; Kim, H.-J.; et al. Structural and Biophysical Analyses of Human N-Myc Downstream-Regulated Gene 3 (NDRG3) Protein. Biomolecules 2020, 10, 90. https://doi.org/10.3390/biom10010090
Kim KR, Kim KA, Park JS, Jang JY, Choi Y, Lee HH, Lee DC, Park KC, Yeom YI, Kim H-J, et al. Structural and Biophysical Analyses of Human N-Myc Downstream-Regulated Gene 3 (NDRG3) Protein. Biomolecules. 2020; 10(1):90. https://doi.org/10.3390/biom10010090
Chicago/Turabian StyleKim, Kyung Rok, Kyung A. Kim, Joon Sung Park, Jun Young Jang, Yuri Choi, Hyung Ho Lee, Dong Chul Lee, Kyung Chan Park, Young Il Yeom, Hyun-Jung Kim, and et al. 2020. "Structural and Biophysical Analyses of Human N-Myc Downstream-Regulated Gene 3 (NDRG3) Protein" Biomolecules 10, no. 1: 90. https://doi.org/10.3390/biom10010090
APA StyleKim, K. R., Kim, K. A., Park, J. S., Jang, J. Y., Choi, Y., Lee, H. H., Lee, D. C., Park, K. C., Yeom, Y. I., Kim, H.-J., & Han, B. W. (2020). Structural and Biophysical Analyses of Human N-Myc Downstream-Regulated Gene 3 (NDRG3) Protein. Biomolecules, 10(1), 90. https://doi.org/10.3390/biom10010090






