Abstract
The biosynthesis of aromatic cytokinins in planta, unlike isoprenoid cytokinins, is still unknown. To compare the final steps of biosynthesis pathways of aromatic and isoprenoid cytokinins, we synthesized a series of nucleoside derivatives of natural cytokinins starting from acyl-protected ribofuranosyl-, 2′-deoxyribofuranosyl- and 5′-deoxyribofuranosyladenine derivatives using stereoselective alkylation with further deblocking. Their cytokinin activity was determined in two bioassays based on model plants Arabidopsis thaliana and Amaranthus caudatus. Unlike active cytokinins-bases, cytokinin nucleosides lack the hormonal activity until the ribose moiety is removed. According to our experiments, ribo-, 2′-deoxyribo- and 5′-deoxyribo-derivatives of isoprenoid cytokinin N6-isopentenyladenine turned in planta into active cytokinins with clear hormonal activity. As for aromatic cytokinins, both 2′-deoxyribo- and 5′-deoxyribo-derivatives did not exhibit analogous activity in Arabidopsis. The 5′-deoxyribo-derivatives cannot be phosphorylated enzymatically in vivo; therefore, they cannot be “activated” by the direct LOG-mediated cleavage, largely occurring with cytokinin ribonucleotides in plant cells. The contrasting effects exerted by deoxyribonucleosides of isoprenoid (true hormonal activity) and aromatic (almost no activity) cytokinins indicate a significant difference in the biosynthesis of these compounds.
1. Introduction
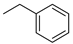
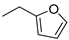
Cytokinins (CKs) are a group of phytohormones that play a crucial role in many processes of plant growth and development. One of the most important effects of CKs is the stimulation of plant cell division and growth. CKs promote the formation of shoots, control the root development, stimulate seed germination and the formation of pigments, activate the chloroplast formation, etc. [1,2]. Naturally occurring CKs are adenine derivatives with a hydrophobic substituent at the N6 position. CKs are divided into two groups depending on the structure of the N6 substituent: (i) aliphatic or isoprenoid, including N6-isopentenyladenine (iP) and zeatins (Figure 1A), and (ii) aromatic, including N6-benzyladenine (BA), topolins, and N6-furfuryladenine (kinetin) (Figure 1B) [1,2].
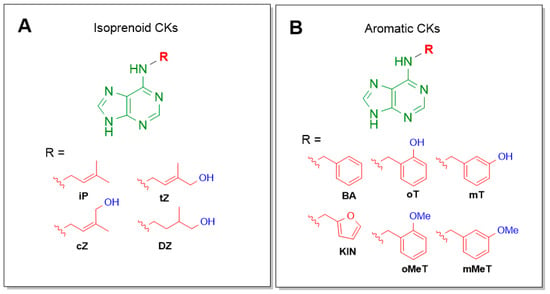
Figure 1.
Names and structures of the naturally occurring cytokinins. (A) Isoprenoid (aliphatic) CKs: iP—N6-isopentenyladenine; cZ—cis-zeatin (N6-(4-hydroxy-3-methyl-cis-2-buten-1-yl)adenine); tZ—trans-zeatin (N6-(4-hydroxy-3-methyl-trans-2-buten-1-yl)adenine); DZ—dihydrozeatin (N6-(4-hydroxy-3-methyl-2-butan-1-yl)adenine); (B) Aromatic CKs: oT—ortho-topolin (N6-(2-hydroxybenzyl)adenine); mT—meta-topolin (N6-(3-hydroxybenzyl)adenine); oMeT—ortho-methoxytopolin (N6-(2-methoxybenzyl)adenine); mMeT—meta-methoxytopolin (N6-(3-methoxybenzyl)adenine); BA—N6-benzyladenine; KIN—kinetin (N6-furfuryladenine).
Since the discovery of kinetin in 1955 by Miller, Skoog et al. [3], a large number of various CKs as well as corresponding nucleoside and nucleotide derivatives were synthesized or isolated from different plant sources [4,5]. Among these numerous molecular forms, only CKs as nucleobases possess hormonal activity, but not their nucleoside or nucleotide derivatives [6,7]. The structural diversity of CKs and CK derivatives in vivo is obviously due to complex pathways of their biosynthesis.
To date, the CK biosynthesis pathways are not completely deciphered. The biosynthesis of isoprenoid CKs, in particular iP, is most studied, while the biosynthesis pathway of aromatic CKs is practically unknown [5,8]. The key step of the iP biosynthesis is commonly assumed to be isopentenyl transferase-catalyzed transfer of isopentenyl moiety from dimethylallyl diphosphate (DMAPP) to the N6 position of adenosine-5′-monophosphate (AMP), adenosine-5′-diphosphate (ADP) or adenosine-5′-triphosphate (ATP), with the formation of the respective 5′-nucleotides (iPRMP, iPRDP, iPRTP) (Figure 2). iPRDP and iPRTP are able to dephosphorylate to iPRMP. At present, two pathways of iP formation are known: (i) 5′-ribonucleotide phosphohydrolase (5′-nucleotidase) catalyzed conversion of iPRm.p. into N6-isopentenyladenosine (iPR), followed by hydrolysis of the N-glycosidic bond catalyzed by adenosine nucleosidase. The reverse process of ribosylation with the formation of nucleosides is catalyzed by purine nucleoside phosphorylase, and the process of reverse phosphorylation of nucleosides with the formation of nucleotides is catalyzed by adenosine kinase; (ii) iPRm.p. can be directly cleaved to iP by specific enzyme phosphoribohydrolase (LOG). Importantly, LOG specifically binds only the 5′-monophosphates of cytokinin nucleosides, but not di- or triphosphates, Am.p. or cytokinin ribosides. The reverse process of iPRm.p. formation from iP is catalyzed by adenosine phosphoribosyltransferase [9].
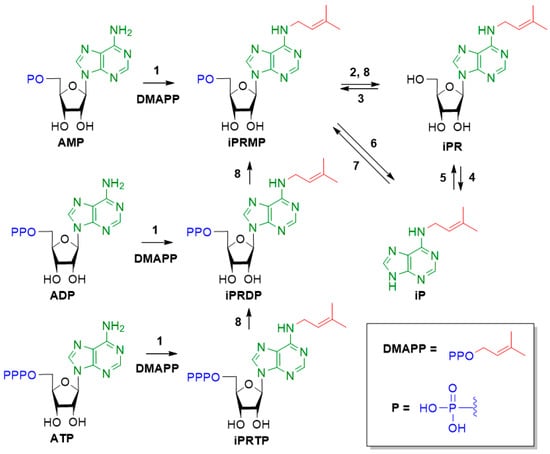
Figure 2.
Scheme of the iP biosynthesis. (1)—isopentenyltransferase; (2)—5′-ribonucleotide phosphohydrolase (5′-nucleotidase); (3)—adenosine kinase; (4)—adenosine nucleosidase; (5)—purine nucleoside phosphorylase; (6)—phosphorybohydrolase (LOG); (7)—adenosine phosphoribosyltransferase; (8)—phosphatase.
Notably, blocking the LOG-dependent pathway in complete knockout log mutants leads to severe cytokinin deficiency phenotype in Arabidopsis plants [10]. Therefore, although iPR is widely occurring in plants, it is considered to be mainly converted to iP via the second (i) LOG-dependent pathway (Figure 2, conversion 6) after reverse phosphorylation to 5′-monophosphate by adenosine kinase (Figure 2, conversion 3) [11].
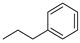
In the present study, a new approach was proposed for the comparative analysis of the biosynthesis pathway of aromatic and isoprenoid CKs. We synthesized a series of ribo-, 2′-deoxyribo- and 5′-deoxyribonucleoside derivatives of natural iP, BA, KIN, and also synthetic cytokinin analog N6-phenylethyladenine, which possesses a high hormonal activity [7]. To investigate the possibility of biochemical conversion in vivo into active CKs in comparison with corresponding ribonucleosides, a series of 2′-deoxyribonucleosides was obtained. It is well known that nucleosides are converted in vivo to 5′-mono-, di-, and triphosphates by cellular nucleoside kinases, and the biological activity of most nucleoside analogs is associated with this mechanism [12]. To study the role of 5′-phosphorylation of nucleosides in the mechanism of CK biosynthesis, a series of 5′-deoxyribonucleoside derivatives was obtained, which cannot be phosphorylated in vivo to form nucleotide 5′-monophosphates and converted into active CKs via LOG-mediated cleavage (Figure 2, conversion 6).
2. Materials and Methods
2.1. Synthesis
2.1.1. General
The reagents and solvents were reagent grade (Sigma Aldrich, St. Louis, MO, USA; Merck, Darmstadt, Germany; Alfa Aesar, Haverhill, MA, USA). Column chromatography was performed on silica gel (Kieselgel 60 Merck, 0.063–0.200 mm). Thin-layer chromatography (TLC) was performed on silica-coated aluminum plates with fluorescent indicator (Merck silica gel 60F254, Darmstadt, Germany or Alugram SIL G/UV254 Macherey-Nagel, Düren, Germany with UV visualization.
1H and 13C (with complete proton decoupling) NMR spectra were recorded on Bruker AMX 400 NMR instrument at 303 K. 1H-NMR spectra were recorded at 400 MHz and 13C-NMR spectra at 100 MHz. Chemical shifts in ppm were measured relative to the residual solvent signals as internal standards (CDCl3, 1H: 7.26 ppm, 13C: 77.1 ppm; DMSO-d6, 1H: 2.50 ppm, 13C: 39.5 ppm). Spin-spin coupling constants (J) are given in Hz. Hydroxyl protons in 1H-NMR were assigned by deuterium exchange on upon addition of D2O into DMSO-d6 solutions of nucleosides, which led to disappearance of hydroxyl signals and simplification of 1H-NMR spectra.
High-resolution mass spectra (HRMS) were registered on a Bruker Daltonics micrOTOF-Q II instrument (Bruker Daltonics, Billerica, MA, USA) using electrospray ionization (ESI). Samples were injected into the mass spectrometer chamber from the Agilent 1260 HPLC system (Santa Clara, CA, USA) equipped with an Agilent Poroshell 120 EC-C18 (3.0 × 50 mm; 2.7 µm) column (Santa Clara, CA, USA); flow rate 400 µL/min; samples were injected from the acetonitrile-water (1:1) solution and the column was eluted with a gradient of concentrations of acetonitrile (A) in water (B) in the following parameters: 0–15% A for 6.0 min, 15%–85% A for 1.5 min, 85%–0% A for 0.1 min, 0% A for 2.4 min. Retention times were as follows: 4—3.9 min; 5—3.9 min; 6—4.5 min; 7—3.7 min; 8—4.1 min; 9—4.1 min; 10—4.2 min; 11—3.8 min; 12—4.4 min; 13—4.3 min; 14—4.5 min; 15—4.0 min.
Melting points were measured with Electrothermal Melting Point Apparatus IA6301 (Camlab Ltd., Cambridge, UK) and are uncorrected.
The following nucleosides were prepared according to the methods reported earlier: N6-benzyladenosine (4), N6-isopentenyladenosine (5), N6-furfuryladenosine (7) [13], N6-(2-phenylethyl)-adenosine (6), N6-benzyl-2′-deoxyadenosine (8), N6-(2-phenylethyl)-2′-deoxyadenosine (10), N6-benzyl-5′-deoxyadenosine (12), N6-isopentenyl-5′-deoxyadenosine (13), N6-(2-phenylethyl)-5′-deoxyadenosine (14) [14].
NMR and HPLC-HRMS data are presented in full in Supplementary Information.
2.1.2. Typical Procedure for Preparation of Nucleosides by Alkylation with Alkyl Halides
A mixture of N6-acetyl-2′,3′,5′-tri-O-acetyladenosine (1) or N6-acetyl-3′,5′-di-O-acetyl-2′-deoxyadenosine (2) or N6-acetyl-2′,3′-di-O-acetyl-5′-deoxyadenosine (3) (0.5 mmol), 1,8-Diazabicyclo[5.4.0]undec-7-ene (DBU) (1 mmol), and corresponding bromide (1 mmol) in dry acetonitrile (5 mL) was kept at ambient temperature for 24 h. The reaction was monitored by TLC (silica gel, CH2Cl2-EtOH, 97:3). After 24 h the reaction mixture was concentrated in vacuo to dryness. The residue was diluted with ethyl acetate (20 mL) and washed successively with brine (2 × 20 mL), 10% aqueous sodium bicarbonate (20 mL) and water (2 × 20 mL). The organic layer was separated, dried over anhydrous sodium sulfate, filtered, and concentrated in vacuo. The residue was purified by column chromatography on silica gel. The column was washed with methylene chloride, the product was eluted with CH2Cl2-EtOH, 98:2. Purified acetyl-protected compound was dissolved in 4M n-PrNH2 in MeOH solution (50 mmol) and was left for 24 h, after which the mixture was concentrated in vacuo and the residue was purified by column chromatography on silica gel. The column was washed with CH2Cl2-EtOH, 95:5 and the product was eluted with CH2Cl2-EtOH, 90:10. The resulting product was dried for 24 h in a vacuum desiccator over phosphorous pentaoxide (P2O5).
N6-benzyladenosine (4). Yield for two steps was 72% as a powder. Rf = 0.32 (CHCl3-EtOH, 9:1 v/v). m.p. 164–165 °C. 1H-NMR (400 MHz, DMSO-d6): δ = 8.40 br s (1H, N6H), 8.37 s (1H, H-8), 8.20 br s (1H, H-2), 7.37–7.17 m (5H, Ph), 5.89 d (1H, J1′2′ = 6.1 Hz, H-1′), 5.41 d (1H, JOH,2′ = 6.2 Hz, 2-OH′), 5.34 dd (1H, JOH,5′b = 7.1 Hz, JOH,5′a = 4.6 Hz, 5-OH′), 5.15 d (1H, JOH,3′ = 4.7 Hz, 3-OH′), 4.72 br s (2H, N6HCH2), 4.61 ddd (1H, J2′3′ = 5.1 Hz, J2′1′ = 6.1 Hz, J2′OH = 6.2 Hz, H-2′), 4.15 ddd (1H, J3′4′ = 2.8 Hz, J3′2′ = 5.1 Hz, J3′OH = 4.7 Hz, H-3′), 3.97 ddd (1H, J4′5′b = 3.8 Hz, J4′5′a = 3.3 Hz, J4′3′ = 2.8 Hz, H-4′), 3.67 ddd (1H, J5′a5′b = −12.1 Hz, J5′a4′ = 3.3 Hz, J5′a,OH = 4.6 Hz, H-5′a), 3.56 ddd (1H, J5′b5′a = −12.1 Hz, J5′b4′ = 3.8 Hz, J5′b,OH = 7.1 Hz, H-5′b). 13C NMR (100 MHz, DMSO-d6): 154.62 (C-6), 152.39 (C-2), 148.48 (C-4), 139.96 (C-8), 128.26 (Ph), 127.16 (Ph), 126.68 (Ph), 119.78 (C-5), 88.05 (C-1′), 85.96 (C-4′), 73.58 (C-2′), 70.70 (C-3′), 61.72 (C-5′), 42.99 (NHCH2). HRMS: m/z [M + H]+ calculated C17H20N5O4+ 358.1510, found 358.1510.
N6-isopentenyladenosine (5). Yield for two steps was 91% as a powder. Rf = 0.45 (CH2Cl2-EtOH, 4:1 v/v). m.p. 144–146 °C. 1H-NMR (400 MHz, DMSO-d6): δ = 8.31 s (1H, H-8), 8.19 br s (1H, H-2), 7.82 br s (1H, N6H), 5.87 d (1H, J1′2′ = 6.0 Hz, H-1′), 5.40 d (1H, JOH,2′ = 6.2 Hz, 2-OH′), 5.39 dd (1H, JOH,5′b = 7.9 Hz, JOH,5′a = 4.6 Hz, 5-OH′), 5.33–5.27 m (1H, CCH = CMe2), 5.15 d (1H, JOH,3′ = 4.8 Hz, 3-OH′), 4.60 ddd (1H, J2′3′ = 5.3 Hz, J2′1′ = 6.0 Hz, J2′OH = 6.2 Hz, H-2′), 4.14 ddd (1H, J3′4′ = 2.8 Hz, J3′2′ = 5.3 Hz, J3′OH = 4.8 Hz, H-3′), 4.08 br s (2H, N6HCH2), 3.96 ddd (1H, J4′5′b = 3.0 Hz, J4′5′a = 3.6 Hz, J4′3′ = 2.8 Hz, H-4′), 3.67 ddd (1H, J5′a5′b = −12.1 Hz, J5′a4′ = 3.6 Hz, J5′a,OH = 4.6 Hz, H-5′a), 3.55 ddd (1H, J5′b5′a = −12.1 Hz, J5′b4′ = 3.0 Hz, J5′b,OH = 7.9 Hz, H-5′b). 13C NMR (100 MHz, DMSO-d6): 154.93 (C-6), 153.14 (C-2), 148.63 (C-4), 140.49 (C-8), 135.41 (=CMe2), 121.73 (CH=), 120.14 (C-5), 88.79 (C-1′), 86.57 (C-4′), 74.22 (C-2′), 71.20 (C-3′), 62.22 (C-5′), 38.57 (NHCH2), 26.02 (Me), 18.49 (Me). HRMS: m/z [M + H]+ calculated C15H22N5O4+ 336.1666, found 336.1666.
N6-benzyl-2′-deoxyadenosine (8). Yield for two steps was 55% as a foam. Rf 0.08 (CH2Cl2-EtOH, 97:3). 1H NMR (400 MHz, DMSO-d6): δ = 8.41 (br s, 1H, N6H), 8.35 (s, 1H, H2), 8.19 (s, 1H, H8), 7.37–7.16 (m, 5H, Ph), 6.35 (dd, 1H, J1′,2′b = 5.9 Hz, J1′,2′a = 7.8 Hz, H1′), 5.30 (d, 1H, JOH,3′ = 3.9 Hz, 3′OH), 5.20 (dd, 1H, JOH, 5′a = 5.2 Hz, JOH,5′b = 6.4 Hz, 5′OH), 4.70 (br s, 2H, NHCH2), 4.40 (dddd, 1H, J3′4′ = 2.5 Hz, J3′2′a = 5.7 Hz, J3′2′b = 2.3 Hz, J3′OH = 3.9 Hz, H3′), 3.88 (ddd, 1H, J4′,3′ = 2.3 Hz, J4′,5′a = 3.9 Hz, J4′,5′b = 6.2 Hz, H4′), 3.62 (ddd, 1H, J5′a,4′ = 3.9 Hz, J5′a,5′b = −12.0 Hz, J5′a,OH = 5.2 Hz, H5′a), 3.51 (ddd, 1H, J5′b,4′ = 6.2 Hz, J5′b,5′a = −12.0 Hz, J5′b,OH = 6.4 Hz, H5′b), 2.74 (ddd, 1H, J2′a,1′ = 7.8 Hz, J2′a,3′ = 5.7 Hz, J2′a,2′b = −13.1 Hz, H2′a), 2.25 (ddd, 1H, J2′b,1′ = 5.8 Hz, J2′b,3′ = 2.3 Hz, J2′b,2′a = −13.1 Hz, H2′b). 13C NMR (400 MHz, DMSO-d6): δ = 154.90 (C6), 152.86 (C2), 148.61 (C4), 140.10 (C8), 128.82 (Ph), 127.58 (Ph), 127.32 (Ph), 119.55 (C5), 88.28 (C1′), 84.62 (C4′), 71.34 (C3′), 62.20 (C5′), 43.38 (NHCH2), 39.49 (C2′ overlapping with DMSO). HRMS: m/z [M + H]+ calculated C17H20N5O3+ 342.1561, found 342.1562; m/z [M–deoxyribosyl]+ calculated C12H12N5+ 226.1087, found 226.1083.
N6-isopentenyl-2′-deoxyadenosine (9). Yield for two steps was 43% as a foam. Rf 0.33 (CH2Cl2-EtOH, 95:5). 1H NMR (400 MHz, DMSO-d6): δ = 8.30 s (1H, H-2), 8.18 br s (1H, H-8), 7.79 br s (1H, N6H), 6.34 dd (1H, J1′2′b = 6.0 Hz, J1′2′a = 7.5 Hz, H-1′), 5.30 dd (1H, JCHCH3 = 1.2 Hz, JCHCH2 = 6.5 Hz, CH = CMe2), 5.27 d (1H, JOH,3′ = 4.0 Hz, 3-OH′), 5.18 dd (1H, JOH,5′b = 6.4 Hz, JOH,5′a = 4.8 Hz, 5-OH′), 4.40 dddd (1H, J3′4′ = 2.7 Hz, J3′2′a = 5.9 Hz, J3′2′b = 2.9 Hz, J3′OH = 4.0 Hz, H-3′), 4.07 br s (2H, N6HCH2), 3.88 ddd (1H, J4′5′b = 4.2 Hz, J4′5′a = 4.4 Hz, J4′3′ = 2.7 Hz, H-4′), 3.62 ddd (1H, J5′a5′b = −12.0 Hz, J5′a4′ = 4.5 Hz, J5′a-OH = 4.8 Hz, H-5′a), 3.52 ddd (1H, J5′b5′a = −12.0 Hz, J5′b4′ = 4.2 Hz, J5′b,OH = 6.3 Hz, H-5′b), 2.71 (ddd, 1H, J2′a,1′ = 7.5 Hz, J2′a,3′ = 5.9 Hz, J2′a,2′b = −13.0 Hz, H2′a), 2.25 (ddd, 1H, J2′b,1′ = 6.0 Hz, J2′b,3′ = 2.8 Hz, J2′b,2′a = −13.0 Hz, H2′b), 1.69 (s, 3H, CH3-cis), 1.66 (s, 3H, CH3-trans). 13C NMR (100 MHz, DMSO-d6): δ = 154.32 (C6), 152.26 (C2), 148.09 (C4), 139.21 (C8), 133.14 (=CMe2), 122.07 (CH=), 119.57 (C5), 87.96 (C1′), 83.92 (C4′), 70.93 (C3′), 61.86 (C5′), 39.49 (C2′, overlapping with DMSO), 37.67 (NHCH2), 25.32 (CH3 isopentenyl), 17.77 (CH3 isopentenyl). HRMS: m/z [M + H]+ calculated C15H22N5O3+ 320.1717, found 320.1720; m/z [M–deoxyribosyl]+ calculated C12H12N5+ 204.1244, found 204.1236.
N6-(2-phenylethyl)-2′-deoxyadenosine (10). Yield for two steps was 58% as a foam. Rf 0.06 (CH2Cl2-EtOH, 97:3). 1H NMR (400 MHz, DMSO-d6): δ = 8.32 (s, 1H, H2), 8.22 (s, 1H, H8), 7.83 (br s, 1H, N6H), 7.35–7.15 (m, 5H, Ph), 6.35 (dd, 1H, J1′,2′b = 6.2 Hz, J1′,2′a = 7.3 Hz, H1′), 5.27 (d, 1H, JOH,3′ = 3.9 Hz, 3′OH), 5.18 (dd, 1H, JOH,5′b = 6.4 Hz, JOH,5′a = 5.1 Hz, 5′OH), 4.44–4.36 (m, 1H, H3′), 3.88 (ddd, 1H, J4′,3′ = 6.6 Hz, J4′,5′a = 4.3 Hz, J4′,5′b = 4.1 Hz, H4′), 3.82–3.60 (m, 2H, N6HCH2), 3.62 (ddd, 1H, J5′a,4′ = 4.3 Hz, J5′a,5′b = −12.0 Hz, JOH,5′a = 5.1 Hz, H5′a), 3.52 (ddd, 1H, J5′b,4′ = 4.1 Hz, J5′b,5′a = −12.0 Hz, JOH,5′b = 6.4 Hz, H5′b), 2.92 (dd, 2H, JCH2-CH2 = 7.8 Hz, J CH2-CH2 = 7.0 Hz, CH2Ph), 2.72 (ddd, 1H, J2′a,1′ = 7.3 Hz, J2′a,3′ = 5.6 Hz, J2′a,2′b = −13.1 Hz, H2′a), 2.72 (ddd, 1H, J2′b,1′ = 6.1 Hz, J2′b,3′ = 2.9 Hz, J2′b,2′a = −13.1 Hz, H2′a). 13C NMR (100 MHz, CD3OD): δ = 156.21 (C6), 153.51 (C2), 148.86 (C4), 140.93 (C8), 140.47 (Ph), 129.88 (Ph), 129.46 (Ph), 127.31 (Ph), 121.30 (C5), 89.91 (C1′), 87.15 (C4′), 73.07 (C3′), 63.67 (C5′), 43.19 (NHCH2), 41.58 (C2′), 36.68 (CH2Ph). HRMS: m/z [M + H]+ calculated C18H22N5O3+ 356.1717, found 356.1724; m/z [M–deoxyribosyl]+ calculated C13H14N5+ 240.1244, found 240.1241.
N6-benzyl-5′-deoxyadenosine (12). Yield for two steps was 77% as a powder. Rf 0.32 (CH2Cl2-EtOH, 95:5). m.p. 198–201 °C. 1H NMR (400 MHz, DMSO-d6): δ = 8.29–8.37 (m, 2H, H-2, N6H), 8.21 (s, 1H, H-8), 7.16–7.37 (m, 5H, Ph), 5.85 (dd, 1H, J1′-2′ = 4.77 Hz, J1′-2′OH = 2.54 Hz, H1′), 5.38 (d, 1H, J3′-OH = 5.25 Hz, 3′OH), 5.10 (d, 1H, J2′-OH = 4.61 Hz, 2′OH), 4.60–4.83 (m, 3H, H2′, NHCH2), 3.93–4.10 (m, 2H, H3′, H4′), 1.30 (dd, 3H, JCH3-4′= 4.77 Hz, JCH3-3′OH = 1.4 Hz, CH3). 13C NMR (100 MHz, DMSO-d6): δ = 154.47 (C6), 152.54 (C2), 148.79 (C4), 140.04 (Ph), 139.82 (C8), 128.14 (Ph), 127.08 (Ph), 126.54 (Ph), 119.51 (C5), 87.92 (C1′), 79.71 (C4′), 74.59 (C2′), 73.05 (C3′), 42.89 (CH2Ph), 16.89 (CH3). HRMS: m/z [M + H]+ calculated C17H20N5O3+ 342.1561, found 342.1566; m/z [M–deoxyribosyl]+ calculated C12H12N5+ 226.1087, found 226.1086.
N6-isopentenyl-5′-deoxyadenosine (13). Yield for two steps was 47% as a powder. Rf 0.31 (CH2Cl2-EtOH, 95:5). m.p. 117-120 °C. 1H NMR (400 MHz, DMSO-d6): δ = 8.29 (s, 1H, H2), 8.21 (s, 1H, H8), 7.77 (br s, 1H, N6H), 5.84 (d, 1H, J1′-2′ = 4.8 Hz, H1′), 5.38 (d, 1H, J2′-OH = 5.6 Hz, 2′OH), 5.30 (tq, 1H, JCH-CH2 = 6.6 Hz, JCH-CH3 = 1.3 Hz, CH=), 5.10 (d, 1H, J3′-OH = 5.3 Hz, 3′OH), 4.65 (ddd, 1H, J2′-3′=4.6 Hz, J2′-1′ = 4.8 Hz, J2′-OH = 5.6 Hz, H2′), 4.08 (br s, 2H, N6HCH2), 3.93–4.01 (m, 2H, H3′, H4′), 1.70 (d, 3H, JCH3-CH = 0.5 Hz, CH3-cis), 1.67 (d, 3H, JCH3-CH = 0.8 Hz, CH3-trans), 1.30 (d, 3H, JCH3-CH = 6.2 Hz, CH3). 13C NMR (100 MHz, DMSO-d6): δ = 154.30 (C6), 152.53 (C2), 148.43 (C4), 139.53 (C8), 133.11 (CMe2), 122.13 (CH=), 119.52 (C5), 87.84 (C1′), 79.63 (C4′), 74.57 (C2′), 73.05 (C3′), 37.67 (NHCH2), 25.33 (CH3), 18.86 (CH3 isopentenyl), 17.79 (CH3 isopentenyl). HRMS: m/z [M + H]+ calculated C15H22N5O3+ 320.1717, found 320.1723; m/z [M–deoxyribosyl]+ calculated C12H12N5+ 204.1244, found 204.1240.
2.1.3. Typical Procedure for Preparation of Nucleosides by Mitsunobu Reaction with Alcohols
To the solution of N6-acetyl-2′,3′,5′-tri-O-acetyladenosine (1) or N6-acetyl-3′,5′-di-O-acetyl-2′-deoxyadenosine (2) or N6-acetyl-2′,3′-di-O-acetyl-5′-deoxyadenosine (3) (1 mmol) with triphenylphosphine (Ph3P) (2 mmol) and corresponding alcohol (2 mmol) in tetrahydrofuran (THF) (5 mL) diethyl azodicarboxylate (DEAD) (2 mmol) was added in one portion and the solution was kept at r.t. for 48 h. The reaction was monitored by TLC (silica gel, CH2Cl2-EtOH, 97:3). After 48 h the reaction mixture was concentrated in vacuo and the residue was dissolved in CH2Cl2 and washed with brine (3 × 20 mL). The organic layer was separated, dried over anhydrous sodium sulfate, filtered, and concentrated in vacuo. The residue was purified by column chromatography (silica gel, CH2Cl2-EtOH, 97:3). Partially purified compound was dissolved in 4M n-PrNH2 in MeOH solution (50 mmol) and left at r.t. for 24 h, after which the mixture was concentrated in vacuo and the residue was purified by column chromatography on silica gel. The column was washed with CH2Cl2-EtOH, 95:5, the product was eluted with CH2Cl2-EtOH, 90:10. The resulting product was dried for 24 hrs in a vacuum desiccator over phosphorous pentaoxide (P2O5).
N6-(2-phenylethyl)-adenosine (6). Yield for two steps was 65% as a powder. Rf 0.28 (CH2Cl2-EtOH, 9:1 v/v). m.p. 169–171 °C. 1H NMR (400 MHz, DMSO-d6): δ = 8.34 (s, 1H, H8), 8.23 (br s, 1H, H2), 7.88 (br s, 1H, N6H), 7.16–7.33 (m, 5H, Ph), 5.89 (d, 1H, J1′,2′ = 6.1 Hz, H1′), 5.41 (d, 1H, JOH-2′ = 6.1 Hz, 2′OH), 5.36 (dd, 1H, JOH-5′b = 7.0 Hz, JOH-5′a = 4.6 Hz, 5′OH), 5.15 (d, 1H, JOH-3′ = 4.6 Hz, 3′OH), 4.60 (dd, 1H, J2′,3′ = 5.9 Hz, J2′,1′ = 6.1 Hz, H2′),4.15 (ddd, 1H, J3′,4′ = 3.2 Hz, J3′,2′ = 5.9 Hz, J3′,OH = 4.6 Hz, H3′), 3.97 (ddd, 1H, J4′,5′b = 3.0, J4′,5′a = 3.4, J4′,3′ = 3.2, H4′), 3.63–3.81 (m, 3H, N6HCH2, H5′a), 3.55 (ddd, 1H, J5′b,5′a = −12.0 Hz, J5′b,4′ = 3.4 Hz, J5′b,OH = 7.0 Hz, H5′b), 2.93 (t, 2H, JCH2-CH2 = 7.5 Hz, CH2Ph). 13C NMR (100 MHz, DMSO-d6): δ = 154.61 (C6), 152.35 (C2), 148.27 (C4), 139.70 (C8), 139.48 (Ph), 128.63 (Ph), 128.27 (Ph), 126.00 (Ph), 119.74 (C5), 87.94 (C1′), 85.88 (C4′), 73.49 (C2′), 70.63 (C3′), 61.66 (C5′), 41.23 (NCH2), 34.39 (CH2Ph). HRMS: m/z [M + H]+ calculated C18H22N5O4+ 372.1666, found 372.1666.
N6-furfuryladenosine (7). Yield for two steps was 64% as a powder. Rf = 0.33 (CH2Cl2-EtOH, 90:10). m.p. 150–155 °C. 1H NMR (400 MHz, DMSO-d6): δ = 8.36 s (1H, H-8), 8.23 br s (1H, N6H), 8.23 s (1H, H-2), 7.52 dd (1H, 3J=3.1 Hz, 4J=1.9 Hz, H5Fur), 6.35 dd (1H, 3J=3.1 Hz, 3J=1.9 Hz, H4Fur), 6.23 dd (1H, 3J = 3.1 Hz, 4J=0.7 Hz, H3Fur), 5.89 d (1H, J1′2′=6.2 Hz, H-1′), 5.41 d (1H, JOH,2′ = 6.2 Hz, 2′-OH), 5.34 dd (1H, JOH,5′b = 7.0 Hz, JOH,5′a = 4.6 Hz, 5′-OH), 5.15 d (1H, JOH,3′ = 4.8 Hz, 3′-OH), 4.71 br s (2H, N6HCH2), 4.61 ddd (1H, J2′3′ = 4.9 Hz, J2′1′ = 6.2 Hz, J2′OH = 6.2 Hz, H-2′), 4.15 ddd (1H, J3′4′ = 3.0 Hz, J3′2′ = 4.9 Hz, J3′OH = 4.8 Hz, H-3′), 3.97 ddd (1H, J4′5′b = 3.9 Hz, J4′5′a = 3.9 Hz, J4′3′ = 3.0 Hz, H-4′), 3.67 ddd (1H, J5′a5′b = −12.0 Hz, J5′a4′ = 3.9 Hz, J5′a,OH = 4.6 Hz, H-5′a), 3.56 ddd (1H, J5′b5′a = −12.0 Hz, J5′b4′ = 3.9 Hz, J5′b,OH = 7.0 Hz, H-5′b). 13C NMR (100 MHz, DMSO-d6): δ = 154.41 (C-6), 152.88 (Fur), 152.30 (C-2), 148.68 (C-4), 141.87 (Fur), 140.03 (C-8), 119.85 (C-5), 110.50 (Fur), 106.73 (Fur), 88.01 (C-1′), 85.93 (C-4′), 73.59 (C-2′), 70.66 (C-3′), 61.69 (C-5′), 36.62 (NHCH2). HRMS: m/z [M + H]‒ calculated C15H18N5O5+ 348.1302, found 348.1307.
N6-furfuryl-2′-deoxyadenosine (11). Yield for two steps was 58% as a foam. Rf 0.52 (CH2Cl2-EtOH, 95:5). 1H NMR (400 MHz, DMSO-d6): δ = 8.36 s (1H, H-2), 8.23 br s (2H, H-8, N6H), 7.53 dd (1H, JH2-H3 = 1.8 Hz, JH2-H4 = 0.9 Hz, H2-furan), 6.39-6.33 m (2H, H-1′, H3-furan), 6.23 dd (1H, JH4-H3 = 3.2 Hz, JH4-H2 = 0.9 Hz, H4-furan), 5.29 d (1H, JOH-3′ = 3.9 Hz, 3-OH′), 5.16 dd (1H, JOH-5′b = 6.2 Hz, JOH-5′a = 4.4 Hz, 5-OH′), 4.72 br s (2H, N6HCH2), 4.41 dddd (1H, J3′-4′ = 2.7 Hz, J3′-2′a = 5.8 Hz, J3′-2′b = 2.8 Hz, J3′-OH = 3.9 Hz, H-3′), 3.89 ddd (1H, J4′-5′b = 6.9 Hz, J4′-5′a = 4.6 Hz, J4′-3′ = 2.7 Hz, H-4′), 3.62 ddd (1H, J5′a-5′b = −11.7 Hz, J5′a-4′ = 4.6 Hz, J5′a-OH = 6.2 Hz, H-5′a), 3.52 ddd (1H, J5′b5′a = −11.7 Hz, J5′b4′ = 6.9 Hz, J5′b,OH = 4.4 Hz, H-5′b), 2.73 ddd (1H, J2′a-1′ = 7.8 Hz, J2′a-3′ = 5.8 Hz, J2′a-2′b = −13.2 Hz, H2′a), 2.25 ddd (1H, J2′b-1′ = 6.1 Hz, J2′b-3′ = 2.8 Hz, J2′b-2′a = −13.2 Hz, H2′b). 13C NMR (100 MHz, CDCl3): δ = 154.90 (C-6), 152.59 (C-2), 151.48 (C-4), 142.45 (C-8), 139.79 (Fur), 121.50 (C-5), 110.60 (Fur), 107.81 (Fur), 89.83 (C-1′), 87.86 (C-4′), 73.43 (C-3′), 63.56 (C-5′), 40.98 (C-2′), 37.72 (NHCH2). HRMS: m/z [M + H]+ calculated C15H18N5O4+ 332.1353, found 332.1350.
N6-(2-phenylethyl)-5′-deoxyadenosine (14). Yield for two steps was 54% as a powder; Rf 0.30 (CH2Cl2-EtOH, 95:5). m.p. 161–163 °C. 1H NMR (DMSO-d6): δ = 8.30 (s, 1H, H2), 8.24 (s, 1H, H8), 7.79 (br s, 1H, N6H), 7.15–7.40 (m, 5H, Ph), 5.85 (d, 1H, J1′-2′ = 4.9 Hz, H1′), 5.38 (d, 1H, J3′-OH = 5.57 Hz, 3′OH), 5.10 (d, 1H, J2′-OH = 5.1 Hz, 2′OH), 4.66 (ddd, 1H, J2′-1′ = 4.9 Hz, J2′-3′ = 4.5 Hz, J2′-OH′ = 5 Hz, H2′), 3.92–4.02 (m, 2H, H3′, H4′), 3.72 (br s, 2H, NHCH2), 2.93 (t, 2H, JCH2-CH2 = 7 Hz, CH2Ph), 1.31 (d, 3H, JCH3-4′ = 6.04 Hz, CH3). 13C NMR (DMSO-d6): δ = 154.50 (C6), 152.58 (C2), 148.61 (C4), 139.62 (C8, Ph), 128.62 (Ph), 128.25 (Ph), 125.98 (Ph), 119.54 (C5), 87.85 (C1′), 79.67 (C4′), 74.59 (C2′), 73.07 (C3′), 41.25 (CH2Ph), 35.01 (NHCH2), 18.88 (CH3). HRMS: m/z [M + H]+ calculated C18H22N5O3+ 356.1717, found 356.1727; m/z [M–deoxyribosyl]+ calculated C13H14N5+ 240.1244, found 240.1243.
N6-furfuryl-5′-deoxyadenosine (15). Yield for two steps was 73% as a powder; Rf 0.24 (CH2Cl2-EtOH, 97:3). m.p. 185–188 °C. 1H NMR (DMSO-d6): δ = 8.34 s (1H, H-2), 8.25 s (1H, H8), 8.22 br s (1H, N6H), 7.53 dd (1H, JH2-H3 = 1.7 Hz, JH2-H4 = 0.8 Hz, H2-furan), 6.36 dd (1H, JH3-H2 = 1.7 Hz, JH3-H4 = 3.1 Hz, H3-furan), 6.23 dd (1H, JH4-H3 = 3.1 Hz, JH4-H2 = 0.8 Hz, H4-furan), 5.86 d (J1′-2′ = 4.9 Hz, H-1′), 5.41 d (1H, JOH-3′ = 5.7 Hz, 3-OH′), 5.14 d (1H, JOH-2′ = 5.2 Hz, 2-OH′), 4.70 br s (2H, N6HCH2), 4.67 ddd (1H, J2′-1′ = 4.9 Hz, J2′-3′ = 4.5 Hz, J2′-OH = 5.2 Hz, H-2′), 4.05-3.90 m (H-3′, H-4′), 1.30 d (JCH3-CH = 6.1 Hz, CH3). 13C NMR (100 MHz, DMSO-d6): δ = 154.27 (C-6), 152.93 (C-4), 152.44 (C-2), 141.77 (C-8), 139.96 (Fur), 119.60 (C-5), 110.40 (Fur), 106.59 (Fur), 87.89 (C-1′), 79.72 (C-4′), 74.59 (C-2′), 73.05 (C-3′), 36.54 (NHCH2), 18.90 (CH3). HRMS: m/z [M + H]+ calculated C15H18N5O4+ 332.1353, found 332.1355.
2.2. Cytokinin Activity Assays
Investigated compounds were dissolved in 100% DMSO to a concentration of 0.1 M. Then these solutions were diluted with distilled water to concentrations of 1–10 μM. Thus, the content of DMSO in 1 μM ligand solutions was only 0.001%.
CK activities were measured in two bioassays based on seedlings of Arabidopsis thaliana or Amaranthus caudatus. In the first assay, we used double mutants of Arabidopsis, in which only one of the three CK receptors (AHK2, AHK3, or CRE1/AHK4) was kept active, as well as wild type (WT) Arabidopsis seedlings with all three functioning receptors [15]. All Arabidopsis plants used, including WT control, carried the reporter GUS gene under control of CK-dependent promoter PARR5 [16]. 4- to 5-day-old Arabidopsis seedlings were incubated for 16 h in aqueous solutions of tested compounds [17]. The level of CK activity of each compound was determined through the level of GUS activity, since it reflects the intensity of PARR5:GUS expression [18]. Each sample contained 10 aligned seedlings; experiments were performed in two biological replicates.
In the case of Amaranthus bioassay, 3- to 4-day-old etiolated seedlings with removed roots were incubated for 16 h in solutions of tested compounds in the dark. Each sample contained 10 aligned seedlings; all samples were in triplicate. The CK activity of compounds corresponds to the level of the pigment amaranthin accumulation in cotyledons, which was determined with spectrophotometer [19,20].
The experiments were carried out in 2–3 biological replications. In all our experiments, probes with BA were included as positive control and its activity was set as 100%. CK activity of all tested compounds was evaluated as percentage relative to the BA activity (after background subtraction) at the same concentration in the same experiment.
3. Results and Discussion
3.1. Chemistry
To synthesize desired nucleosides, we used our mild and efficient approach based on regioselective N6-alkylation of acetyl-protected adenosine, 2′-deoxyadenosine and 5′-deoxyadenosine derivatives 1-3 either with alcohols under Mitsunobu reaction conditions (Scheme 1, conditions (i)) or with alkyl halides in the presence of the base (Scheme 1, conditions (ii)) [13,14]. The main advantage of these approaches is the possibility to use both alkyl halides and alcohols for N6-modification. The following deacetylation with n-PrNH2 in MeOH (Scheme 1, conditions (iii)) yielded the final nucleosides with high overall yields. The initial triacetyl-5′-deoxyadenosine 3 was synthesized by radical reduction of corresponding 5′-chloro-5′-deoxyadenosine derivative in the presence of Bu3SnH according to the literature [21].
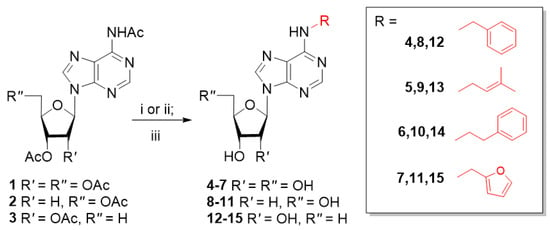
Scheme 1.
Synthesis of ribo- (4–7), 2′-deoxyribo- (8–11) and 5′-deoxyribonucleoside (12–15) derivatives of CKs. Reagents and conditions: (i) ROH, Ph3P, DEAD, THF, 20 °C, 24–48 h; (ii) RBr, DBU, CH3CN, r.t., 1–3 days; (iii) 4M n-PrNH2 in MeOH, 20 °C, 24 h.
The structure of the compounds was confirmed by 1H and 13C NMR spectroscopy. In addition, all compounds were characterized for purity and homogeneity by high-resolution mass spectrometry (HRMS). The obtained M/z values of the compounds correspond to the calculated M/z values. According to HPLC, all the obtained products are individual compounds. All spectral data are presented in full in the Supplementary Information.
3.2. Cytokinin Activity
The CK activity of all synthesized compounds was studied in two plant bioassay systems. First bioassay was based on seedlings of a model plant Arabidopsis thaliana [17]. To study the effects of CK nucleosides (as CK precursors) on different CK receptors in this assay we used double insertion mutants, in which only one isoform of the CK receptors (AHK2, AHK3 or CRE1/AHK4/WOL) was active in each mutant clone, as well as the wild type (WT) plants with all three functioning receptors (Table 1).

Table 1.
Cytokinin activity (in %) of nucleoside derivatives (at two concentrations) in bioassay based on Arabidopsis thaliana double mutants. BA activity was taken as 100% 1.
All plants were stably transformed with the GUS gene fused to CK-dependent promoter of the ARR5 gene. The hormonal activity of the compounds was determined by the level of GUS activity reflecting the intensity of the PARR5:GUS expression [17]. Based on the results of the Arabidopsis bioassay (Table 1), all tested compounds were subdivided into three conventional groups, of low, medium, and high activity. The first group included compounds whose CK activity did not exceed 30% (colored blue), the second group included derivatives with medium activity from 30% to 80% (colored black), and the third highly active group exhibited activity above 80% (colored red) of the BA activity.
As an additional bioassay we used Amaranthus seedlings which quickly responded to CK by accumulation of the red pigment amaranthin in the dark (Table 2) [19]. This assay is rather specific and fast and considered to be a classical cytokinin bioassay [20]. As in the case of Arabidopsis bioassay compounds with activity values of less than 30% of BA activity were considered to be low active or inactive. Compounds with activity no less than 80% of BA activity were considered to be highly active.

Table 2.
Cytokinin activity (in %) of nucleoside derivatives in bioassay based on Amaranthus caudatus seedlings. BA activity was taken as 100% *.
3.3. Analysis of Results with Arabidopsis Bioassay
Ribosides of natural CKs 4, 5, and 7 exhibited CK effect in all experimental variants of Arabidopsis bioassay. Compounds 4, 5, and 7 exhibited the highest activity at a concentration of 10−5 M for the WT, leading to nearly 100% activation of CK signaling system. 10-Fold decrease in CK concentration led to approx. twofold decrease in GUS activity activation. On the other hand, compounds 4 and 5 were more active at the more physiological concentration of 10−6 M for the mutant clones (nearly 100% receptor activation for AHK2, 80% for AHK3 and approximately 50% for AHK4) [22]. Compound 7, a kinetin derivative, was more active at 10−5 M for AHK2 and AHK4 and manifested comparable activity at both 10−5 M and 10−6 M for AHK3. N6-Phenylethyladenosine (6), an artificial CK derivative, demonstrated some, though weak activity at a concentration of 10−6 M only with the AHK2 receptor (Table 1). Two other receptors were inactive or very weakly active with 6 at 10−6 M.
Generally, as regards double mutant clones, our Arabidopsis assay makes it possible to perform a unique task, i.e. to analyze the response to CKs of individual CK receptors. In our study, the clones reacted quite uniformly, there were no case when any of CK derivatives exerted high effect with one receptor and simultaneously weak effect with another one. If WT plants or any of mutant clones demonstrated high CK activity (red color) with a defined compound, all other mutant clones showed with this compound the same (red) or at least medium (black) activity. Conversely, if WT plants or any of mutant clones demonstrated low/no CK activity (blue color) with some other compound, all other mutant clones with different receptors showed the same (blue) or medium (black) but never high (red) activity (Table 1). This may be a consequence of the use of the limited set of different CKs. In any case, such regularity can serve as an argument for the validity of the assay results.
2′-Deoxyribo-derivative of iP (9) was active at higher concentrations (10−5 M) with all individual receptors and manifested weaker activity at more physiological concentration. 2′-Deoxyribo-derivatives of aromatic CKs (8, 10) were virtually inactive even at a concentration of 10−5 M except for derivative 11 which showed some activity at both concentrations, but only with AHK2.
5′-Deoxyribo-derivative of iP (13) manifested high activity for WT, medium activity for AHK2 and AHK4 and weak activity for AHK3 at 10−5 M and manifested weak activity for all individual receptors at 10−6 M. On the other hand, 5′-deoxyribo-derivatives of aromatic CKs (12, 14, 15) were inactive even at a concentration of 10−5 M.
Taking into account these results, several inferences can be made. 5′-Deoxyribo-derivative of iP (13) cannot be phosphorylated enzymatically in vivo, thereby the LOG-mediated one-step formation of iP is not possible here. However, since this compound exerted the CK effect, the only way for this derivative conversion into active CK is the cleavage by adenosine nucleosidase (Figure 3A). In turn, the 5′-deoxyribo-derivatives of all aromatic CKs were inactive, and since the pathway associated with the action of LOG enzyme is blocked, this LOG-dependent mechanism may be suggested to play a major role in the formation of aromatic CKs (Figure 3B). For the iP nucleoside derivatives, there were no such restrictions as deoxyribo-derivatives may be able to turn in planta into free bases, at least at 10−5 M. Therefore, the results showed that the conversion of 5′-monophosphates into active CKs via LOG-mediated cleavage in Arabidopsis strongly depends on the nature of the side group (aliphatic or aromatic) of the CK ribosides.
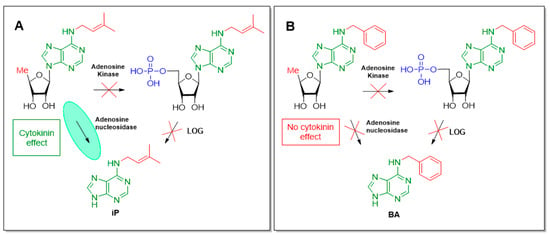
Figure 3.
The effect of structural modification of cytokinin nucleosides in Arabidopsis on the biosynthesis pathways of iP (A) and BA (B).
3.4. Analysis of Results with Amaranthus Bioassay
In the case of Amaranthus bioassay, N6-benzyladenosine (4) and N6-isopentenyladenosine (5) showed pronounced CK activity, while N6-phenylethyladenosine (6) demonstrated weak activity at 10−6 M (Table 2). Contrary to Arabidopsis, in Amaranthus there were no advantage of 5 over 4 in CK activity. Besides, another aromatic CK (ribo)nucleoside N6-furfuryladenosine (7) also showed weak activity in contrast to its medium activity in Arabidopsis bioassay. It is interesting that all 5′-deoxyribo-derivatives (12–15) as well as 2′-deoxyribo-derivatives (8–11) of both aromatic and isoprenoid CKs turned out to be essentially inactive in the Amaranthus bioassay. This peculiarity is indicative of a species specificity in the final step of active CK biosynthesis in plants.
It is known that adenosine nucleosidase, isolated from various sources, has a broad substrate specificity. Substrates for this enzyme may be adenosine, guanosine, inosine, xanthosine [23], N6-benzyladenosine [24], N6-isopentenyladenosine [25], as well as 5′-deoxyadenosine, 2′-deoxyadenosine [24] and several other purine nucleosides. Despite this generally accepted view, it cannot be excluded that the lack of activity of the deoxyribo-derivatives of aromatic CKs may be related to the substrate specificity of adenosine nucleosidase in this plant species. Another possible pathway for the formation of active CKs from 2′-deoxyribo-derivatives associated with the enzymatic phosphorylation followed by cleavage of 5′-monophosphates by the action of LOG also turned out to be unrealized. In that case, it may be due to the lack of 5′-phosphorylation of 2′-deoxyribo-derivatives by adenosine kinase, or due to the inability of cleavage of the corresponding 2′-deoxyribo-5′-monophosphates by LOG.
4. Conclusions
A series of nucleoside derivatives of isoprenoid CK N6-isopentenyladenine and various aromatic CKs was synthesized starting from acyl-protected ribofuranosyl-, 2′-deoxyribofuranosyl- and 5′-deoxyribofuranosyladenine derivatives using mild and efficient stereoselective N6-alkylation with further acyl deblocking, and their hormonal (cytokinin) activity was determined in two bioassays based on seedlings of Arabidopsis thaliana and Amaranthus caudatus. We demonstrated that ribo-, 2′-deoxyribo-, 5′-deoxyribo-derivatives of N6-isopentenyladenine, as well as ribosides of aromatic CKs were able to transform into active CKs and exhibited hormonal activity in Arabidopsis, while 5′-deoxyribo- and 2′-deoxyribo-derivatives of aromatic CKs were inactive. Therefore, not only ribo- but also 5′-deoxyribo- and 2′-deoxyribonucleosides may serve as precursors of active CKs, at least in Arabidopsis. However, in the latter case, the only way of biosynthesis of aromatic CKs seems to be the direct cleavage of 5′-monophosphates to active nucleobases catalyzed by phosphoribohydrolase LOG, whereas the biosynthesis of N6-isopentenyladenine may evidently proceed also by hydrolysis of CK nucleosides catalyzed by adenosine nucleosidase. As a result, it can be concluded that the biosynthesis pathways of isoprenoid (exemplified by N6-isopentenyladenine) and aromatic CKs may be different, at least in Arabidopsis, and that further research is needed to shed light on the mechanisms of biosynthesis of this class of compounds. Of course, at present it cannot be excluded that in other plant species the CK biosynthesis pathways are different. For example, in Amaranthus deoxyribo-derivatives of both N6-isopentenyladenine and aromatic CKs were equally low active. Taking into consideration the variety of biological effects of CKs on the growth, development, resistance and productivity of plants, the knowledge gained may contribute to the development of new approaches to handling the growth and development of plant species valuable to humans.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/10/1/86/s1, 1H and 13C NMR spectra; High resolution mass spectra (HRMS).
Author Contributions
Conceptualization, S.N.M. and G.A.R.; experimental work, V.E.O., E.M.S. and M.S.D.; data analysis V.E.O., E.M.S., M.S.D., G.A.R. and S.N.M.; writing—original draft preparation, V.E.O.; writing—review and editing, S.N.M. and G.A.R.; supervision, S.N.M. and G.A.R.; funding acquisition, V.E.O., G.A.R. All authors have read and agreed to the published version of the manuscript.
Funding
The research was supported by the Russian Foundation for Basic Research (grants No. 18-34-00084 and 17-04-00969).
Acknowledgments
The authors thank Pavel N. Solyev (EIMB RAS) for HPLC-HRMS spectra registration. The double mutants’ seeds of Arabidopsis thaliana were kindly provided by T. Schmülling and M. Riefler.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A. How do cytokinins affect the cell? Rus. J. Plant Physiol. 2009, 56, 268–290. [Google Scholar] [CrossRef]
- Miller, C.; Skoog, F.; Saltza, M.; Strong, M. Kinetin, a cell division factor from deoxyribonucleic acid. J. Am. Chem. Soc. 1955, 77, 1329–1334. [Google Scholar] [CrossRef]
- Plihalova, L.; Vylíčilová, H.; Doležal, K.; Zahajska, L.; Zatloukal, M.; Strnad, M. Synthesis of aromatic cytokinins for plant biotechnology. New Biotech. 2016, 33, 614–624. [Google Scholar] [CrossRef]
- Kamínek, M. Tracking the story of cytokinin research. J. Plant Growth Regul. 2015, 34, 723–739. [Google Scholar] [CrossRef]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Arkhipov, D.V.; Osolodkin, D.I.; Schmülling, T.; Romanov, G.A. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 2015, 66, 1851–1863. [Google Scholar] [CrossRef]
- Savelieva, E.M.; Oslovsky, V.E.; Karlov, D.S.; Kurochkin, N.N.; Getman, I.A.; Lomin, S.N.; Sidorov, G.V.; Mikhailov, S.N.; Osolodkin, D.I.; Romanov, G.A. Cytokinin activity of N6-benzyladenine derivatives assayed by interaction with the receptors in planta, in vitro, and in silico. Phytochemistry 2018, 149, 161–177. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinin biosynthesis and metabolism. In Plant Hormones, 3rd ed.; Davies, P.J., Ed.; Springer: Dordrecht, The Netherlands, 2010; pp. 95–114. [Google Scholar]
- Komada-Nobusada, T.; Sakakibara, H. Molecular basis for cytokinin biosynthesis. Phytochemistry 2009, 70, 444–449. [Google Scholar] [CrossRef]
- Tokunaga, H.; Kojima, M.; Kuroha, T.; Ishida, T.; Sugimoto, K.; Kiba, T.; Sakakibara, H. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012, 69, 355–365. [Google Scholar] [CrossRef]
- Osugi, A.; Sakakibara, H. Q&A: How do plants respond to cytokinins and what is their importance? BMC Biol. 2015, 13, 102. [Google Scholar]
- Jordheim, L.P.; Durantel, D.; Zoulim, F.; Dumontet, C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat. Rev. Drug Discov. 2013, 12, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Drenichev, M.S.; Oslovsky, V.E.; Tararov, V.I.; Mikhailov, S.N. Synthesis of N6-substituted adenosines as cytokinin nucleosides. Curr. Protoc. Nucleic Acid Chem. 2018, 72, 14.15.1–14.15.16. [Google Scholar] [CrossRef] [PubMed]
- Drenichev, M.S.; Oslovsky, V.E.; Sun, L.; Tijsma, A.; Kurochkin, N.N.; Tararov, V.I.; Chizhov, A.O.; Neyts, J.; Pannecouque, C.; Leyssen, P.; et al. Modification of the length and structure of the linker of N6-benzyladenosine modulates its selective antiviral activity against enterovirus 71. Eur. J. Med. Chem. 2016, 111, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Riefler, M.; Novak, O.; Strnad, M.; Schmülling, T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell 2006, 18, 40–54. [Google Scholar] [CrossRef] [PubMed]
- Stolz, A.; Riefler, M.; Lomin, S.N.; Achazi, K.; Romanov, G.A.; Schmülling, T. The specificity of cytokinin signalling in Arabidopsis thaliana is mediated by differing ligand affinities and expression profiles of the receptors. Plant J. 2011, 67, 157–168. [Google Scholar] [CrossRef] [PubMed]
- Romanov, G.A.; Kieber, J.J.; Schmülling, T. A rapid cytokinin response assay in Arabidopsis indicates a role for phospholipase D in cytokinin signalling. FEBS Lett. 2002, 515, 39–43. [Google Scholar] [CrossRef]
- Zvereva, S.D.; Romanov, G.A. Reporter genes for plant genetic engineering: Characteristics and detection. Russ. J. Plant Physiol. 2000, 47, 424–432. [Google Scholar]
- Romanov, G.A.; Getman, I.A.; Schmülling, T. Investigation of early cytokinin effects in a rapid Amaranthus seedling test. Plant Growth Regul. 2000, 34, 337–344. [Google Scholar] [CrossRef]
- Biddington, N.L.; Thomas, T.H. A modified Amaranthus betacyanin bioassay for the rapid determination of cytokinins in plant extracts. Planta 1973, 111, 183–186. [Google Scholar] [CrossRef]
- Wang, Y.; Hogenkamp, H.P.; Long, R.A.; Revankar, G.R.; Robins, R.K. A convenient synthesis of 5′-deoxyribonucleosides. Carbohydr. Res. 1977, 59, 449–457. [Google Scholar] [CrossRef]
- Oslovsky, V.E.; Savelieva, E.M.; Drenichev, M.S.; Romanov, G.A.; Mikhailov, S.N. Comparative analysis of the biosynthesis of isoprenoid and aromatic cytokinins. Dokl. Biochem. Biophys. 2019, 488, 346–349. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, J.; Takeda, S.; Xie, S.X.; Hatanaka, H.; Ashikari, T.; Amachi, T.; Shimizu, S. Purification, characterization, and gene cloning of purine nucleosidase from Ochrobactrum anthropi. Appl. Environ. Microbiol. 2001, 67, 1783–1787. [Google Scholar] [CrossRef] [PubMed]
- Abusamhadneh, E.; McDonald, N.E.; Kline, P.C. Isolation and characterization of adenosine nucleosidase from yellow lupin (Lupinus luteus). Plant Sci. 2000, 153, 25–32. [Google Scholar] [CrossRef]
- Chen, C.M.; Kristopeit, S.M. Metabolism of cytokinin: Deribosylation of cytokinin ribonucleoside by adenosine nucleosidase from wheat germ cells. Plant Physiol. 1981, 68, 1020–1023. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).