Mediator of DNA Damage Checkpoint Protein 1 Facilitates V(D)J Recombination in Cells Lacking DNA Repair Factor XLF
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Abelson Murine Leukemia Virus-Transformed (vAbl) Cell Lines
2.2. Antibodies
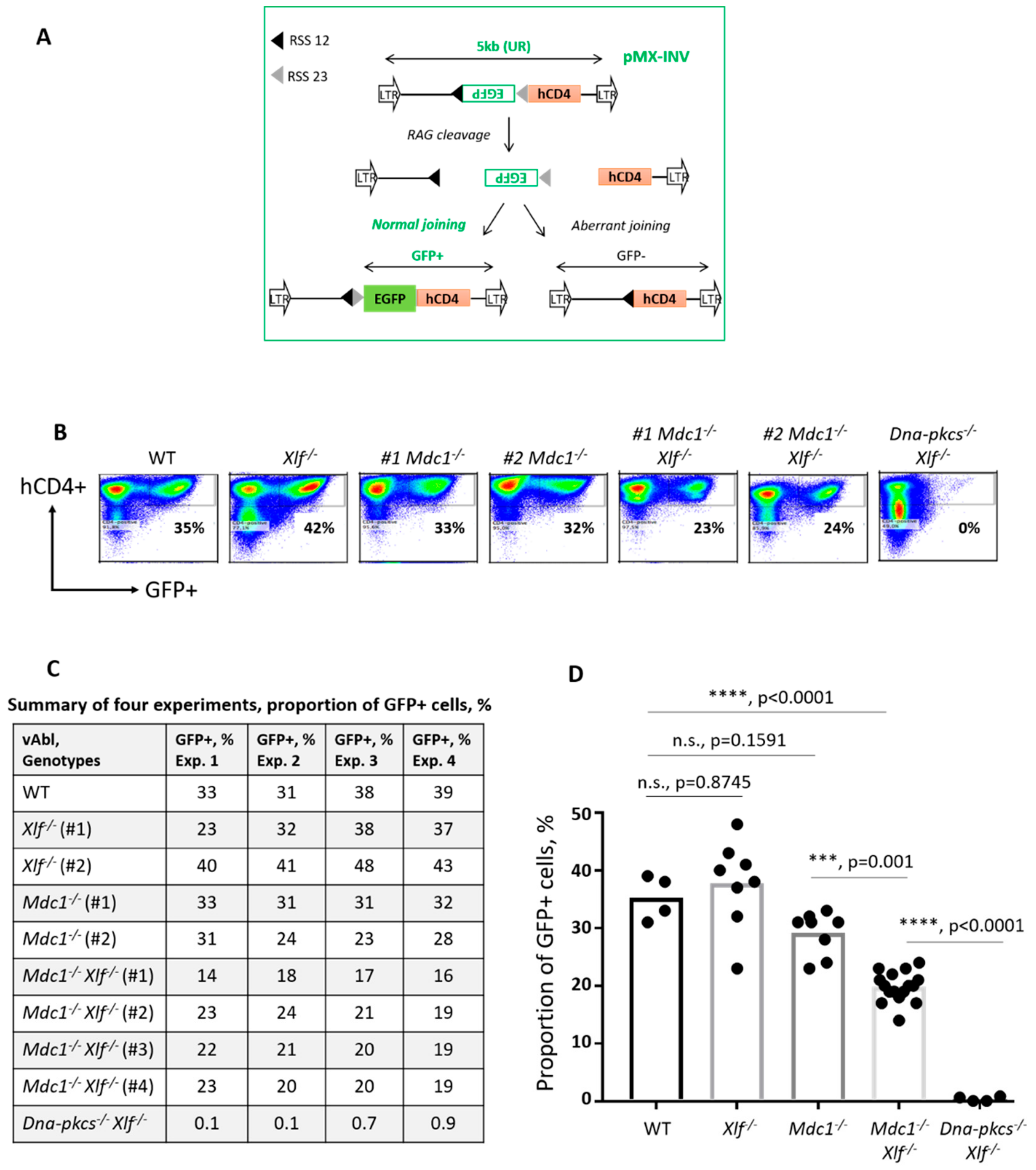
2.3. Variable (V), Diversity (D) and Joining (J) Gene Segments Recombination (V(D)J Recombination) Assays Based on Chromosomally Integrated pMX Cassettes
2.4. Mice
2.5. Proliferation Assay
3. Results
3.1. Robust V(D)J Recombination in Progenitor-B Cells Lacking Mediator of DNA Damage Checkpoint Protein 1 (MDC1)
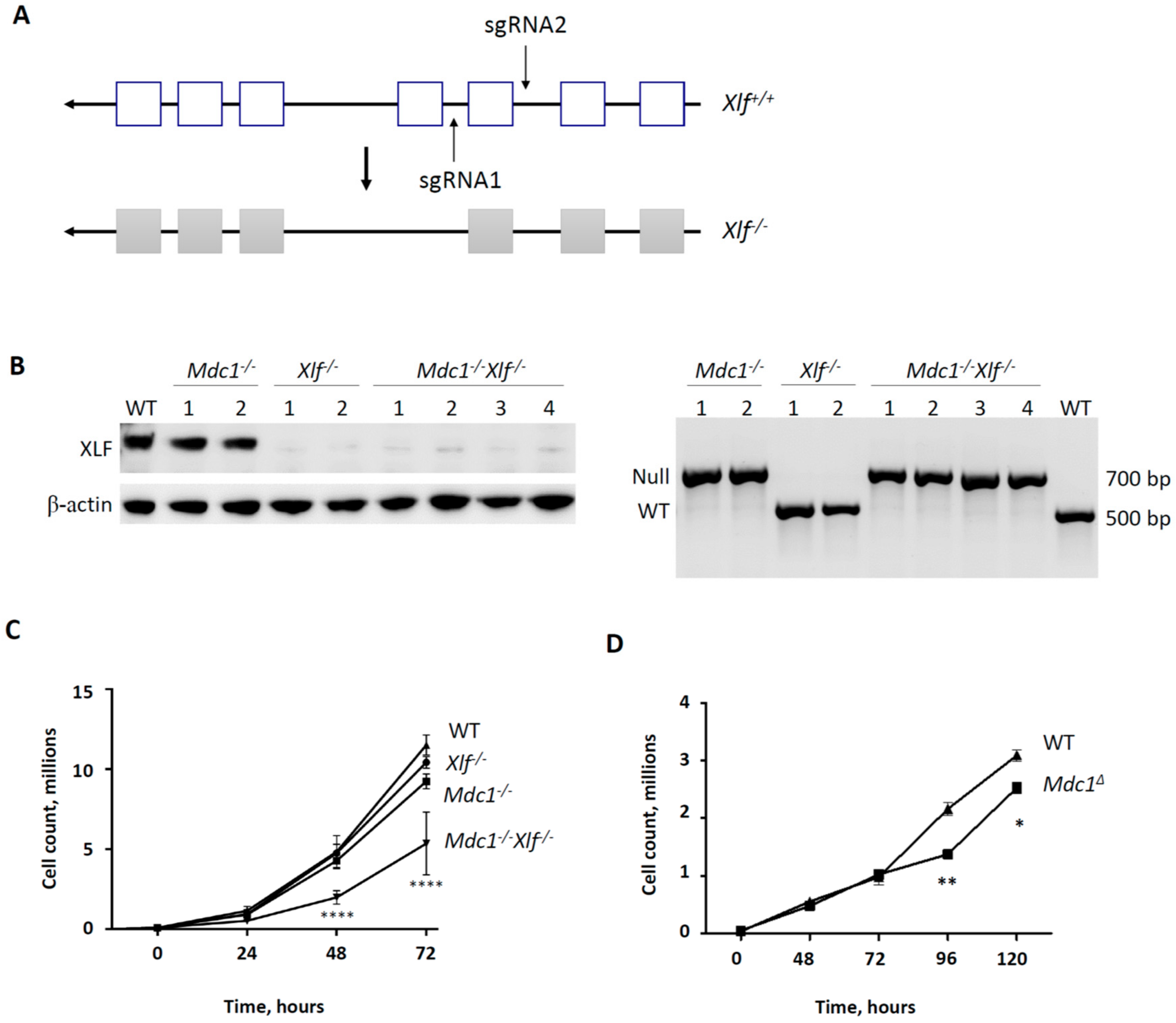
3.2. Synthetic Lethality Between Mediator of DNA Damage Checkpoint Protein 1 (MDC1) and XRCC4-Like Factor (XLF) in Mouse
3.3. Generation of XLF−/− Knockout and MDC1−/−XLF−/− Double Knockout vAbl Cell Lines
3.4. Reduced V(D)J Recombination Efficiency in vAbl Pro-B Cells Lacking both MDC1 and XLF
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, V.; Alt, F.W.; Oksenych, V. Functional overlaps between XLF and the ATM-dependent DNA double strand break response. DNA Repair 2014, 16, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Oksenych, V.; Zhovmer, A.; Ziani, S.; Mari, P.O.; Eberova, J.; Nardo, T.; Stefanini, M.; Giglia-Mari, G.; Egly, J.M.; Coin, F. Histone methyltransferase DOT1L drives recovery of gene expression after a genotoxic attack. PLoS Genet. 2013, 9, e1003611. [Google Scholar] [CrossRef] [PubMed]
- Botuyan, M.V.; Lee, J.; Ward, I.M.; Kim, J.E.; Thompson, J.R.; Chen, J.; Mer, G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 2006, 127, 1361–1373. [Google Scholar] [CrossRef] [PubMed]
- Huyen, Y.; Zgheib, O.; Ditullio, R.A., Jr.; Gorgoulis, V.G.; Zacharatos, P.; Petty, T.J.; Sheston, E.A.; Mellert, H.S.; Stavridi, E.S.; Halazonetis, T.D. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature 2004, 432, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.S.; Park, J.H.; Kim, S.J.; Kwon, S.J.; Kwon, J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010, 29, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pannicke, U.; Schwarz, K.; Lieber, M.R. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell 2002, 108, 781–794. [Google Scholar] [CrossRef]
- Ahnesorg, P.; Smith, P.; Jackson, S.P. XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining. Cell 2006, 124, 301–313. [Google Scholar] [CrossRef]
- Buck, D.; Malivert, L.; de Chasseval, R.; Barraud, A.; Fondaneche, M.C.; Sanal, O.; Plebani, A.; Stephan, J.L.; Hufnagel, M.; le Deist, F.; et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell 2006, 124, 287–299. [Google Scholar] [CrossRef]
- Craxton, A.; Somers, J.; Munnur, D.; Jukes-Jones, R.; Cain, K.; Malewicz, M. XLS (c9orf142) is a new component of mammalian DNA double-stranded break repair. Cell Death Differ. 2015, 22, 890–897. [Google Scholar] [CrossRef]
- Ochi, T.; Blackford, A.N.; Coates, J.; Jhujh, S.; Mehmood, S.; Tamura, N.; Travers, J.; Wu, Q.; Draviam, V.M.; Robinson, C.V.; et al. DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku to promote DNA double-strand break repair. Science 2015, 347, 185–188. [Google Scholar] [CrossRef]
- Xing, M.; Yang, M.; Huo, W.; Feng, F.; Wei, L.; Jiang, W.; Ning, S.; Yan, Z.; Li, W.; Wang, Q.; et al. Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 2015, 6, 6233. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.J.; Johnson, B.; Chen, B.R.; Byrum, A.K.; Bredemeyer, A.L.; Yewdell, W.T.; Johnson, T.E.; Lee, B.J.; Deivasigamani, S.; Hindi, I.; et al. MRI Is a DNA Damage Response Adaptor during Classical Non-homologous End Joining. Mol. Cell 2018, 71, 332–342.e8. [Google Scholar] [CrossRef]
- Castaneda-Zegarra, S.; Huse, C.; Røsand, Ø.; Sarno, A.; Xing, M.; Gago-Fuentes, R.; Zhang, Q.; Alirezaylavasani, A.; Werner, J.; Ji, P.; et al. Generation of a Mouse Model Lacking the Non-Homologous End-Joining Factor Mri/Cyren. Biomolecules 2019, 9, 798. [Google Scholar] [CrossRef] [PubMed]
- Alt, F.W.; Zhang, Y.; Meng, F.L.; Guo, C.; Schwer, B. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 2013, 152, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Seidl, K.J.; Rathbun, G.A.; Zhu, C.; Manis, J.P.; van der Stoep, N.; Davidson, L.; Cheng, H.L.; Sekiguchi, J.M.; Frank, K.; et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity 1997, 7, 653–665. [Google Scholar] [CrossRef]
- Nussenzweig, A.; Chen, C.; da Costa Soares, V.; Sanchez, M.; Sokol, K.; Nussenzweig, M.C.; Li, G.C. Requirement for Ku80 in growth and immunoglobulin V(D)J recombination. Nature 1996, 382, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.T.; Kaushal, D.; Murphy, M.; Zhang, Y.; Datta, A.; Chen, C.; Monroe, B.; Mostoslavsky, G.; Coakley, K.; Gao, Y.; et al. XRCC4 suppresses medulloblastomas with recurrent translocations in p53-deficient mice. Proc. Natl. Acad. Sci. USA 2006, 103, 7378–7383. [Google Scholar] [CrossRef]
- Boboila, C.; Oksenych, V.; Gostissa, M.; Wang, J.H.; Zha, S.; Zhang, Y.; Chai, H.; Lee, C.S.; Jankovic, M.; Saez, L.M.; et al. Robust chromosomal DNA repair via alternative end-joining in the absence of X-ray repair cross-complementing protein 1 (XRCC1). Proc. Natl. Acad. Sci. USA 2012, 109, 2473–2478. [Google Scholar] [CrossRef]
- Bosma, G.C.; Custer, R.P.; Bosma, M.J. A severe combined immunodeficiency mutation in the mouse. Nature 1983, 301, 527–530. [Google Scholar] [CrossRef]
- Rooney, S.; Sekiguchi, J.; Zhu, C.; Cheng, H.L.; Manis, J.; Whitlow, S.; DeVido, J.; Foy, D.; Chaudhuri, J.; Lombard, D.; et al. Leaky Scid phenotype associated with defective V(D)J coding end processing in Artemis-deficient mice. Mol. Cell 2002, 10, 1379–1390. [Google Scholar] [CrossRef]
- Zha, S.; Jiang, W.; Fujiwara, Y.; Patel, H.; Goff, P.H.; Brush, J.W.; Dubois, R.L.; Alt, F.W. Ataxia telangiectasia-mutated protein and DNA-dependent protein kinase have complementary V(D)J recombination functions. Proc. Natl. Acad. Sci. USA 2011, 108, 2028–2033. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Crowe, J.L.; Liu, X.; Nakajima, S.; Wang, Y.; Li, C.; Lee, B.J.; Dubois, R.L.; Liu, C.; Yu, X.; et al. Differential phosphorylation of DNA-PKcs regulates the interplay between end-processing and end-ligation during nonhomologous end-joining. Mol. Cell 2015, 58, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Chaudhuri, J.; Zhu, C.; Davidson, L.; Weaver, D.T.; Alt, F.W. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J recombination. Immunity 1998, 9, 367–376. [Google Scholar] [CrossRef]
- Li, G.; Alt, F.W.; Cheng, H.L.; Brush, J.W.; Goff, P.H.; Murphy, M.M.; Franco, S.; Zhang, Y.; Zha, S. Lymphocyte-specific compensation for XLF/cernunnos end-joining functions in V(D)J recombination. Mol. Cell 2008, 31, 631–640. [Google Scholar] [CrossRef]
- Vera, G.; Rivera-Munoz, P.; Abramowski, V.; Malivert, L.; Lim, A.; Bole-Feysot, C.; Martin, C.; Florkin, B.; Latour, S.; Revy, P.; et al. Cernunnos deficiency reduces thymocyte life span and alters the T cell repertoire in mice and humans. Mol. Cell Biol. 2013, 33, 701–711. [Google Scholar] [CrossRef]
- Balmus, G.; Barros, A.C.; Wijnhoven, P.W.; Lescale, C.; Hasse, H.L.; Boroviak, K.; le Sage, C.; Doe, B.; Speak, A.O.; Galli, A.; et al. Synthetic lethality between PAXX and XLF in mammalian development. Genes Dev. 2016, 30, 2152–2157. [Google Scholar] [CrossRef]
- Gago-Fuentes, R.; Xing, M.; Saeterstad, S.; Sarno, A.; Dewan, A.; Beck, C.; Bradamante, S.; Bjoras, M.; Oksenych, V. Normal development of mice lacking PAXX, the paralogue of XRCC4 and XLF. FEBS Open Bio 2018, 8, 426–434. [Google Scholar] [CrossRef]
- Liu, X.; Shao, Z.; Jiang, W.; Lee, B.J.; Zha, S. PAXX promotes KU accumulation at DNA breaks and is essential for end-joining in XLF-deficient mice. Nat. Commun. 2017, 8, 13816. [Google Scholar] [CrossRef]
- Abramowski, V.; Etienne, O.; Elsaid, R.; Yang, J.; Berland, A.; Kermasson, L.; Roch, B.; Musilli, S.; Moussu, J.P.; Lipson-Ruffert, K.; et al. PAXX and Xlf interplay revealed by impaired CNS development and immunodeficiency of double KO mice. Cell Death Differ. 2018, 25, 444–452. [Google Scholar] [CrossRef]
- Kumar, V.; Alt, F.W.; Frock, R.L. PAXX and XLF DNA repair factors are functionally redundant in joining DNA breaks in a G1-arrested progenitor B-cell line. Proc. Natl. Acad. Sci. USA 2016, 113, 10619–10624. [Google Scholar] [CrossRef]
- Hung, P.J.; Chen, B.R.; George, R.; Liberman, C.; Morales, A.J.; Colon-Ortiz, P.; Tyler, J.K.; Sleckman, B.P.; Bredemeyer, A.L. Deficiency of XLF and PAXX prevents DNA double-strand break repair by non-homologous end joining in lymphocytes. Cell Cycle 2017, 16, 286–295. [Google Scholar] [CrossRef]
- Lescale, C.; Lenden Hasse, H.; Blackford, A.N.; Balmus, G.; Bianchi, J.J.; Yu, W.; Bacoccina, L.; Jarade, A.; Clouin, C.; Sivapalan, R.; et al. Specific Roles of XRCC4 Paralogs PAXX and XLF during V(D)J Recombination. Cell Rep. 2016, 16, 2967–2979. [Google Scholar] [CrossRef]
- Castaneda-Zegarra, S.; Xing, M.; Gago-Fuentes, R.; Saeterstad, S.; Oksenych, V. Synthetic lethality between DNA repair factors Xlf and Paxx is rescued by inactivation of Trp53. DNA Repair 2019, 73, 164–169. [Google Scholar] [CrossRef]
- Oksenych, V.; Kumar, V.; Liu, X.; Guo, C.; Schwer, B.; Zha, S.; Alt, F.W. Functional redundancy between the XLF and DNA-PKcs DNA repair factors in V(D)J recombination and nonhomologous DNA end joining. Proc. Natl. Acad. Sci. USA 2013, 110, 2234–2239. [Google Scholar] [CrossRef] [PubMed]
- Xing, M.; Bjoras, M.; Daniel, J.A.; Alt, F.W.; Oksenych, V. Synthetic lethality between murine DNA repair factors XLF and DNA-PKcs is rescued by inactivation of Ku70. DNA Repair 2017, 57, 133–138. [Google Scholar] [CrossRef] [PubMed]
- Lescale, C.; Abramowski, V.; Bedora-Faure, M.; Murigneux, V.; Vera, G.; Roth, D.B.; Revy, P.; de Villartay, J.P.; Deriano, L. RAG2 and XLF/Cernunnos interplay reveals a novel role for the RAG complex in DNA repair. Nat. Commun. 2016, 7, 10529. [Google Scholar] [CrossRef]
- Zha, S.; Sekiguchi, J.; Brush, J.W.; Bassing, C.H.; Alt, F.W. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc. Natl. Acad. Sci. USA 2008, 105, 9302–9306. [Google Scholar] [CrossRef]
- Bassing, C.H.; Chua, K.F.; Sekiguchi, J.; Suh, H.; Whitlow, S.R.; Fleming, J.C.; Monroe, B.C.; Ciccone, D.N.; Yan, C.; Vlasakova, K.; et al. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. USA 2002, 99, 8173–8178. [Google Scholar] [CrossRef]
- Bassing, C.H.; Suh, H.; Ferguson, D.O.; Chua, K.F.; Manis, J.; Eckersdorff, M.; Gleason, M.; Bronson, R.; Lee, C.; Alt, F.W. Histone H2AX: A dosage-dependent suppressor of oncogenic translocations and tumors. Cell 2003, 114, 359–370. [Google Scholar] [CrossRef]
- Lou, Z.; Minter-Dykhouse, K.; Franco, S.; Gostissa, M.; Rivera, M.A.; Celeste, A.; Manis, J.P.; van Deursen, J.; Nussenzweig, A.; Paull, T.T.; et al. MDC1 maintains genomic stability by participating in the amplification of ATM-dependent DNA damage signals. Mol. Cell 2006, 21, 187–200. [Google Scholar] [CrossRef]
- Manis, J.P.; Morales, J.C.; Xia, Z.; Kutok, J.L.; Alt, F.W.; Carpenter, P.B. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 2004, 5, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Zha, S.; Guo, C.; Boboila, C.; Oksenych, V.; Cheng, H.L.; Zhang, Y.; Wesemann, D.R.; Yuen, G.; Patel, H.; Goff, P.H.; et al. ATM damage response and XLF repair factor are functionally redundant in joining DNA breaks. Nature 2011, 469, 250–254. [Google Scholar] [CrossRef]
- Oksenych, V.; Alt, F.W.; Kumar, V.; Schwer, B.; Wesemann, D.R.; Hansen, E.; Patel, H.; Su, A.; Guo, C. Functional redundancy between repair factor XLF and damage response mediator 53BP1 in V(D)J recombination and DNA repair. Proc. Natl. Acad. Sci. USA 2012, 109, 2455–2460. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, W.; Dubois, R.L.; Yamamoto, K.; Wolner, Z.; Zha, S. Overlapping functions between XLF repair protein and 53BP1 DNA damage response factor in end joining and lymphocyte development. Proc. Natl. Acad. Sci. USA 2012, 109, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Peng, R.; Bjorkman, A.; Filipe de Miranda, N.; Rosner, C.; Kotnis, A.; Berglund, M.; Liu, C.; Rosenquist, R.; Enblad, G.; et al. Cernunnos influences human immunoglobulin class switch recombination and may be associated with B cell lymphomagenesis. J. Exp. Med. 2012, 209, 291–305. [Google Scholar] [CrossRef] [PubMed]
- Valencia, M.; Bentele, M.; Vaze, M.B.; Herrmann, G.; Kraus, E.; Lee, S.E.; Schar, P.; Haber, J.E. NEJ1 controls non-homologous end joining in Saccharomyces cerevisiae. Nature 2001, 414, 666–669. [Google Scholar] [CrossRef] [PubMed]
- Salguero, I.; Belotserkovskaya, R.; Coates, J.; Sczaniecka-Clift, M.; Demir, M.; Jhujh, S.; Wilson, M.D.; Jackson, S.P. MDC1 PST-repeat region promotes histone H2AX-independent chromatin association and DNA damage tolerance. Nat. Commun. 2019, 10, 5191. [Google Scholar] [CrossRef] [PubMed]
- Leimbacher, P.A.; Jones, S.E.; Shorrocks, A.K.; de Marco Zompit, M.; Day, M.; Blaauwendraad, J.; Bundschuh, D.; Bonham, S.; Fischer, R.; Fink, D.; et al. MDC1 Interacts with TOPBP1 to Maintain Chromosomal Stability during Mitosis. Mol. Cell 2019, 74, 571–583.e8. [Google Scholar] [CrossRef]
- Bredemeyer, A.L.; Sharma, G.G.; Huang, C.Y.; Helmink, B.A.; Walker, L.M.; Khor, K.C.; Nuskey, B.; Sullivan, K.E.; Pandita, T.K.; Bassing, C.H.; et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 2006, 442, 466–470. [Google Scholar] [CrossRef]
- Lenden Hasse, H.; Lescale, C.; Bianchi, J.J.; Yu, W.; Bedora-Faure, M.; Deriano, L. Generation and CRISPR/Cas9 editing of transformed progenitor B cells as a pseudo-physiological system to study DNA repair gene function in V(D)J recombination. J. Immunol. Methods 2017, 451, 71–77. [Google Scholar] [CrossRef]
- Dewan, A.; Xing, M.; Lundbaek, M.B.; Gago-Fuentes, R.; Beck, C.; Aas, P.A.; Liabakk, N.B.; Saeterstad, S.; Chau, K.T.P.; Kavli, B.M.; et al. Robust DNA repair in PAXX-deficient mammalian cells. FEBS Open Bio 2018, 8, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef]
- Strasser, A.; Harris, A.W.; Cory, S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell 1991, 67, 889–899. [Google Scholar] [CrossRef]
- Xing, M.; Oksenych, V. Genetic interaction between DNA repair factors PAXX, XLF, XRCC4 and DNA-PKcs in human cells. FEBS Open Bio 2019. [Google Scholar] [CrossRef] [PubMed]
- Helmink, B.A.; Tubbs, A.T.; Dorsett, Y.; Bednarski, J.J.; Walker, L.M.; Feng, Z.; Sharma, G.G.; McKinnon, P.J.; Zhang, J.; Bassing, C.H.; et al. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature 2011, 469, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Alt, F.W.; Oksenych, V. Reprint of “Functional overlaps between XLF and the ATM-dependent DNA double strand break response”. DNA Repair 2014, 17, 52–63. [Google Scholar] [CrossRef]
- Gapud, E.J.; Dorsett, Y.; Yin, B.; Callen, E.; Bredemeyer, A.; Mahowald, G.K.; Omi, K.Q.; Walker, L.M.; Bednarski, J.J.; McKinnon, P.J.; et al. Ataxia telangiectasia mutated (Atm) and DNA-PKcs kinases have overlapping activities during chromosomal signal joint formation. Proc. Natl. Acad. Sci. USA 2011, 108, 2022–2027. [Google Scholar] [CrossRef] [PubMed]
- Dev, H.; Chiang, T.W.; Lescale, C.; de Krijger, I.; Martin, A.G.; Pilger, D.; Coates, J.; Sczaniecka-Clift, M.; Wei, W.; Ostermaier, M.; et al. Shieldin complex promotes DNA end-joining and counters homologous recombination in BRCA1-null cells. Nat. Cell Biol. 2018, 20, 954–965. [Google Scholar] [CrossRef]
- Innes, C.L.; Hesse, J.E.; Morales, A.J.; Helmink, B.A.; Schurman, S.H.; Sleckman, B.P.; Paules, R.S. DNA damage responses in murine Pre-B cells with genetic deficiencies in damage response genes. Cell Cycle 2020, 19, 67–83. [Google Scholar] [CrossRef]
- Deriano, L.; Chaumeil, J.; Coussens, M.; Multani, A.; Chou, Y.; Alekseyenko, A.V.; Chang, S.; Skok, J.A.; Roth, D.B. The RAG2 C terminus suppresses genomic instability and lymphomagenesis. Nature 2011, 471, 119–123. [Google Scholar] [CrossRef]
| Genotypes | Live Born | Expected (1:2:1) | Expected * (1:2:0) |
|---|---|---|---|
| MDC1+/+XLF−/− | 34 | 26 | 35 |
| MDC1+/−XLF−/− | 70 | 52 | 69 |
| MDC1−/−XLF−/− | 0 | 26 | 0 |
| Total | 104 | 104 | 104 |
| Genotypes | V(D)J Recombination | Mice |
|---|---|---|
| Single Knockouts | ||
| DNA-PKcs−/− [23] | No | Alive |
| PAXX−/− [9,10,11,27] | Normal | Alive |
| Mri−/− [12,13] | Normal | Alive |
| XLF−/− [24,25] | Normal | Alive |
| ATM−/− [37] | Normal | Alive |
| H2AX−/− [38,39] | Reduced | Alive |
| MDC1−/− [40] | Normal | Alive |
| 53BP1−/− [41] | Normal | Alive |
| RAG2Δ/Δ [60] | Reduced | Alive |
| Double Knockouts | ||
| XLF−/− DNA-PKcs−/− [34,35] | No | Embryonic lethality |
| XLF−/− PAXX−/− [26,28,29,33] | No | Embryonic lethality |
| XLF−/− Mri−/− [12] | No | Embryonic lethality |
| XLF−/− ATM−/− [42] | Very low | Alive, small |
| XLF−/− H2AX−/− [42] | Reduced | Embryonic lethality |
| XLF−/− MDC1−/− [*] | Reduced | Embryonic lethality |
| XLF−/−53BP1−/− [43,44] | Very low | Alive, small |
| XLF−/− RAG2Δ/Δ [36] | Very low | Alive |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beck, C.; Castañeda-Zegarra, S.; Huse, C.; Xing, M.; Oksenych, V. Mediator of DNA Damage Checkpoint Protein 1 Facilitates V(D)J Recombination in Cells Lacking DNA Repair Factor XLF. Biomolecules 2020, 10, 60. https://doi.org/10.3390/biom10010060
Beck C, Castañeda-Zegarra S, Huse C, Xing M, Oksenych V. Mediator of DNA Damage Checkpoint Protein 1 Facilitates V(D)J Recombination in Cells Lacking DNA Repair Factor XLF. Biomolecules. 2020; 10(1):60. https://doi.org/10.3390/biom10010060
Chicago/Turabian StyleBeck, Carole, Sergio Castañeda-Zegarra, Camilla Huse, Mengtan Xing, and Valentyn Oksenych. 2020. "Mediator of DNA Damage Checkpoint Protein 1 Facilitates V(D)J Recombination in Cells Lacking DNA Repair Factor XLF" Biomolecules 10, no. 1: 60. https://doi.org/10.3390/biom10010060
APA StyleBeck, C., Castañeda-Zegarra, S., Huse, C., Xing, M., & Oksenych, V. (2020). Mediator of DNA Damage Checkpoint Protein 1 Facilitates V(D)J Recombination in Cells Lacking DNA Repair Factor XLF. Biomolecules, 10(1), 60. https://doi.org/10.3390/biom10010060






