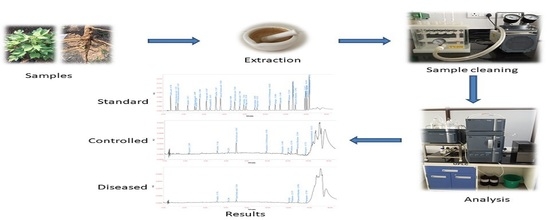
Simultaneous Estimation of Twenty Eight Phenolic Compounds by a Novel and Expeditious Method Developed on Quaternary Ultra-Performance Liquid Chromatography System with a Photodiode Array Detector
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Standards
2.3. Samples
2.4. Instrumentation
2.5. Analytical Method Validation
2.5.1. Selectivity and System Suitability
2.5.2. Linearity
2.5.3. Precision and Accuracy
2.5.4. Limit of Quantification (LOQ) and Limit of Detection (LOD)
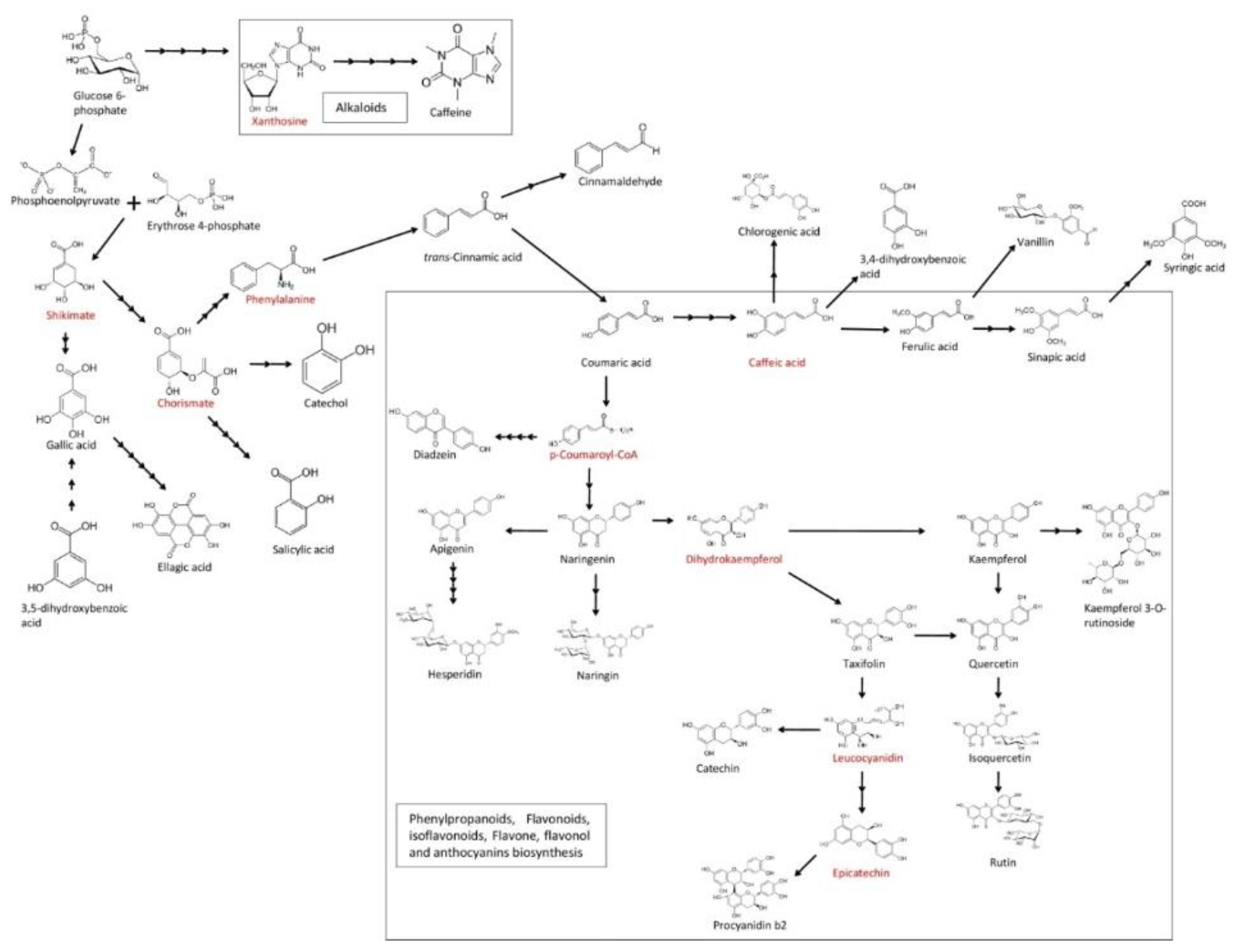
3. Results and Discussion
3.1. UPLC Method Development
3.2. Analytical Method Validation
3.2.1. Selectivity and System Suitability
3.2.2. Linearity, LOQ and LOD
3.2.3. Accuracy and Precision
3.2.4. Application of Method
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, J.J.; Woodward, A.W.; Chen, Z.J. Gene expression changes and early events in cotton fibre development. Ann. Bot. 2007, 100, 1391–1401. [Google Scholar] [CrossRef] [PubMed]
- Wendel, J.F.; Cronn, R.C. Polyploidy and the evolutionary history of cotton. Adv. Agron. 2003, 78, 139–186. [Google Scholar]
- Razaq, M.; Aslam, M.; Shad, S.A.; Aslam, M.N.; Saeed, N.A. Evaluation of some new promising cotton strains against bollworm complex. J. Res. Sci. 2004, 15, 313–318. [Google Scholar]
- Dua, I.S.; Kumar, V.; Bhavneet, D.E.A. Genetically modified cotton and its Biosafety concerns: A review. In Current Concepts in Botany (Book), 1st ed.; Mukerji, K.G., Manoharachary, C., Eds.; I.K. International Publishing House Pvt. Ltd.: New Delhi, India, 2006; pp. 447–459. [Google Scholar]
- Lawo, N.C.; Wackers, F.L.; Romeis, J. Indian Bt cotton varieties do not affect the performance of cotton aphids. PLoS ONE 2009, 4, e4804. [Google Scholar] [CrossRef] [PubMed]
- Satpute, U.S.; Patil, V.N.; Katole, S.R.; Men, U.B.; Bhagwat, V.R.; Thakare, A.Y. Avoidable field losses due to sucking pests and bollworms in cotton. J. Appl. Zool. Res. 1990, 1, 67–72. [Google Scholar]
- Kempema, L.A.; Cui, X.; Holzer, F.M.; Walling, L.L. Arabidopsis transcriptome changes in response to phloem-feeding Silverleaf whitefly nymphs. Similarities and distinctions in responses to aphids. Plant Physiol. 2007, 143, 849–865. [Google Scholar] [CrossRef] [PubMed]
- Shalash, M.; Makahleh, A.; Salhimi, S.M.; Saad, B. Vortex-assisted liquid-liquid–liquid microextraction followed by high performance liquid chromatography for the simultaneous determination of fourteen phenolic acids in honey, iced tea and canned coffee drinks. Talanta 2017, 174, 428–435. [Google Scholar] [CrossRef]
- Association of Official Analytical Chemists, Official Methods of the Association of the Agricultural Chemists, eighteen Ed., Gaithersburg, 2007
- Peled-Zehavi, H.; Oliva, M.; Xie, Q.; Tzin, V.; Oren-Shamir, M.; Aharoni, A.; Galili, G. Metabolic engineering of the phenylpropanoid and its primary, precursor pathway to enhance the flavor of fruits and the aroma of flowers. Bioengineering (Basel) 2015, 2, 204–212. [Google Scholar] [CrossRef]
- Cocuron, J.C.; Casas, M.I.; Yang, F.; Grotewold, E.; Alonso, A.P. Beyond the wall: High-throughput quantification of plant soluble and cell-wall bound phenolics by liquid chromatography tandem mass spectrometry. J. Chromatogr. A 2019, 1589, 93–104. [Google Scholar] [CrossRef]
- Lege, K.E.; Cothren, J.T.; Smith, C.W. Phenolic acid and condensed tannin concentrations of six cotton genotypes. Environ. Exp. Bot. 1995, 35, 241–249. [Google Scholar] [CrossRef]
- Liu, Q.; Cai, W.; Shao, X. Determination of seven polyphenols in water by high performance liquid chromatography combined with preconcentration. Talanta 2008, 77, 679–683. [Google Scholar] [CrossRef]
- Naczk, M.; Shahidi, F. Review: Extraction and analysis of phenolics in food. J. Chromatogr. A 2004, 1054, 95–111. [Google Scholar] [CrossRef]
- Khoddami, A.; Wilkes, M.A.; Roberts, T.H. Techniques for analysis of plant phenolic compounds. Molecules 2013, 18, 2328–2375. [Google Scholar] [CrossRef]
- Mattila, P.; Kumpulainen, J. Determination of free and total phenolic acids in plant-derived foods by HPLC with diode-array detection. J. Agric. Food Chem. 2002, 50, 3660–3667. [Google Scholar] [CrossRef]
- Santiago, R.; Lopez-Malvar, A.; Souto, C.; Barros-Rios, J. Methods for determining cell wall-bound phenolics in maize stem tissues. J. Agric. Food Chem. 2018, 66, 1279–1284. [Google Scholar] [CrossRef]
- Garzon, G.A.; Narvaez-Cuenca, C.E.; Vincken, J.P.; Gruppen, H. Polyphenolic composition and antioxidant activity of Acai (Euterpe oleracea Mart.) from Colombia. Food Chem. 2017, 217, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Ambrosi, M.; Abad-Garcia, B.; Viloria-Bernal, M.; Garmon-Lobato, S.; Berrueta, L.A.; Gallo, B. A new ultra high performance liquid chromatography with diode array detection coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry analytical strategy for fast analysis and improved characterization of phenolic compounds in apple products. J. Chromatogr. A 2013, 1316, 78–91. [Google Scholar] [PubMed]
- Simirgiotis, M.J.; Quispe, C.; Borquez, J.; Areche, C.; Sepulveda, B. Fast detection of phenolic compounds in extracts of easter pears (Pyrus communis) from the Atacama desert by ultrahigh-performance liquid chromatography and mass spectrometry (UHPLC-Q/Orbitrap/MS/MS). Molecules 2016, 21, 92. [Google Scholar] [CrossRef] [PubMed]
- Vagiri, M.; Ekholm, A.; Andersson, S.C.; Johansson, E.; Rumpunen, K. An optimized method for analysis of phenolic compounds in buds, leaves, and fruits of black currant (Ribes nigrum L.). J. Agric. Food Chem. 2012, 60, 10501–10510. [Google Scholar] [CrossRef]
- Jaini, R.; Wang, P.; Dudareva, N.; Chapple, C.; Morgan, J.A. Targeted metabolomics of the phenylpropanoid pathway in Arabidopsis thaliana using reversed phase liquid chromatography coupled with tandem mass spectrometry. Phytochem. Anal. 2017, 28, 267–276. [Google Scholar] [CrossRef]
- Casas, M.I.; Duarte, S.; Doseff, A.I.; Grotewold, E. Flavone-rich maize: an opportunity to improve the nutritional value of an important commodity crop. Front. Plant. Sci. 2014, 5, 440. [Google Scholar] [CrossRef] [PubMed]
- Orcic, D.; Franciskovic, M.; Bekvalac, K.; Svircev, E.; Beara, I.; Lesjak, M.; Mimica-Dukic, N. Quantitative determination of plant phenolics in Urticadioica extracts by high-performance liquid chromatography coupled with tandem mass spectrometric detection. Food Chem. 2014, 143, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Vieira, G.S.; Marques, A.S.F.; Machado, M.T.C.; Silva, V.M.; Hubinger, M.D. Determination of anthocyanins and non-anthocyanin polyphenols by ultra performance liquid chromatography/electrospray ionization mass spectrometry (UPLC/ESI–MS) in Jussara (Euterpe edulis) extracts. J. Food Sci. Technol. 2017, 54, 2135–2144. [Google Scholar] [CrossRef] [PubMed]
- Adom, K.K.; Liu, R.H. Antioxidant activity of grains. J. Agric. Food Chem. 2002, 50, 6182–6187. [Google Scholar] [CrossRef] [PubMed]
- Irakli, M.N.; Samanidou, V.F.; Biliaderis, C.G.; Papadoyannis, I.N. Simultaneous determination of phenolic acids and flavonoids in rice using solid-phase extraction and RP-HPLC with photodiode array detection. J. Sep. Sci. 2012, 35, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Amarowicz, R.; Weidner, S. Content of phenolic acids in rye caryopses determined using DAD-HPLC method. Czech. J. Food Sci. 2001, 19, 201–205. [Google Scholar] [CrossRef]
- Fracassetti, D.; Lawrence, N.; Tredoux, A.G.J.; Tirelli, A.; Nieuwoudt, H.H.; Du Toit, W.J. Quantification of glutathione, catechin and caffeic acid in grape juice and wine by a novel ultra-performance liquid chromatography method. Food Chem. 2011, 128, 1136–1142. [Google Scholar] [CrossRef]
- Dias, F.S.; Lovillo, M.P.; Barroso, C.G.; David, J.M. Optimization and validation of a method for the direct determination of catechin and epicatechin in red wines by HPLC/fluorescence. Microchem. J. 2010, 96, 17–20. [Google Scholar] [CrossRef]
- Escarpa, A.; González, M.C. Optimization strategy and validation of one chromatographic method as approach to determine the phenolic compounds from different sources. J. Chromatogr. A 2000, 897, 161–170. [Google Scholar] [CrossRef]
- SemanticScholar. Available online: https://pdfs.semanticscholar.org/5d48/0d7b3e906131b2cb5eba3c2cc87192cb78df.pdf?_ga=2.122522187.1122837214.1576659057-437361496.1575990593 (accessed on 16 November 2019).
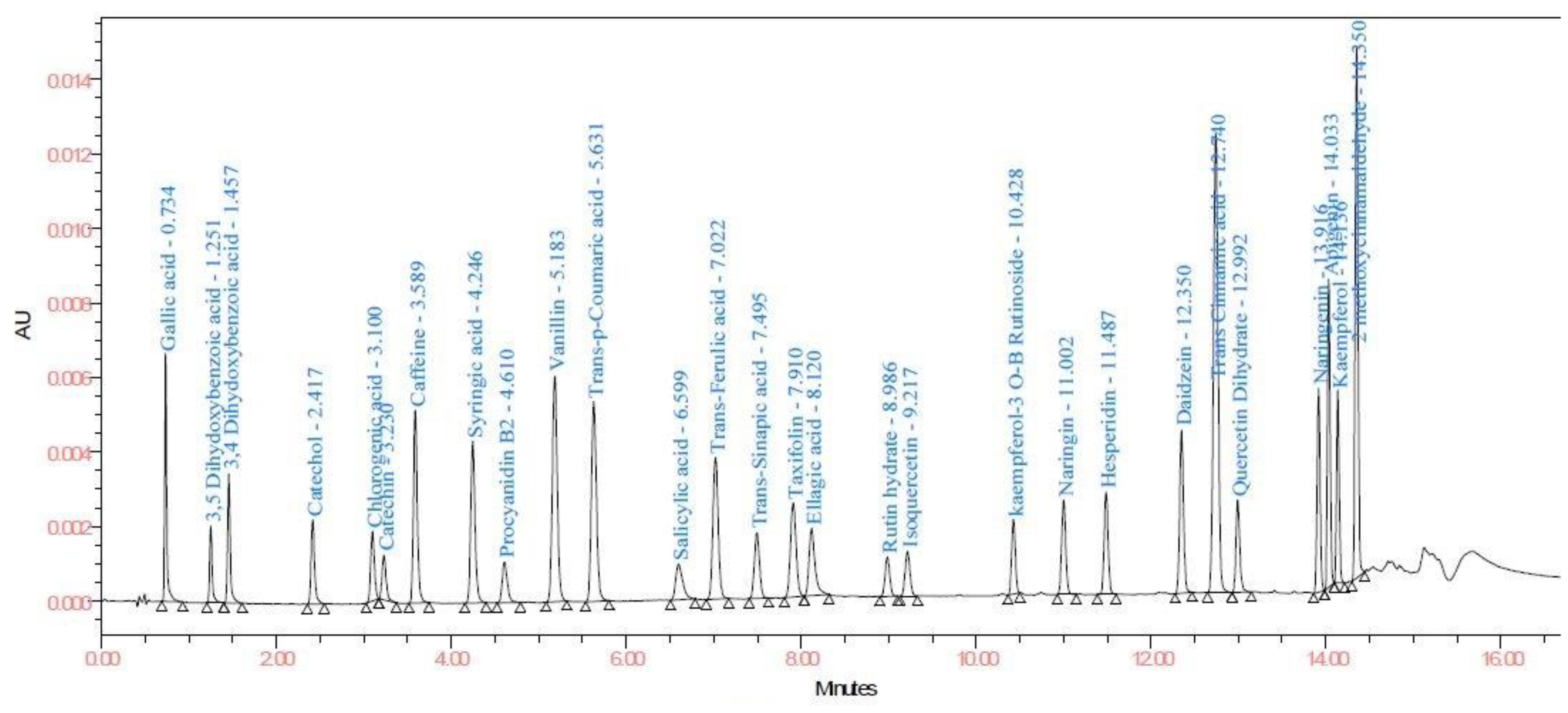
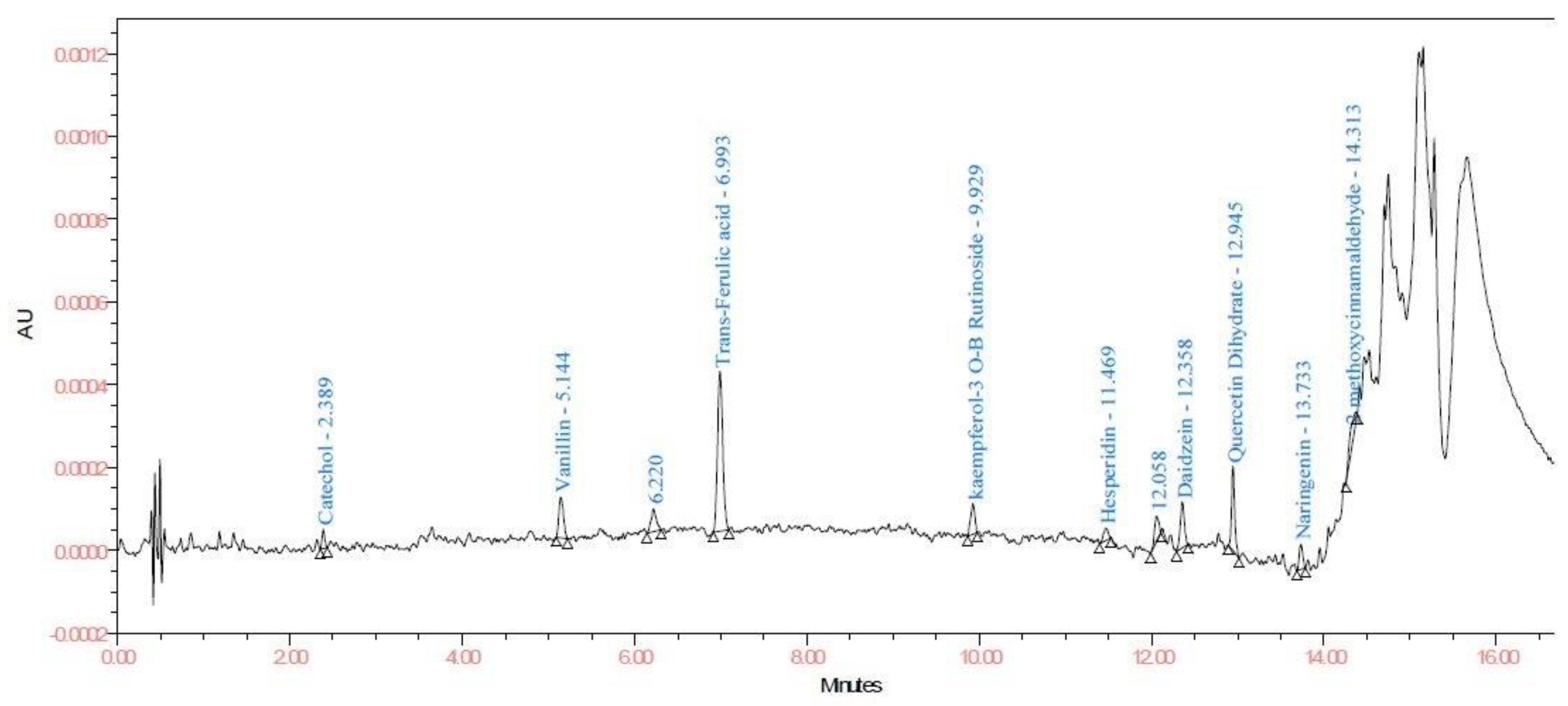
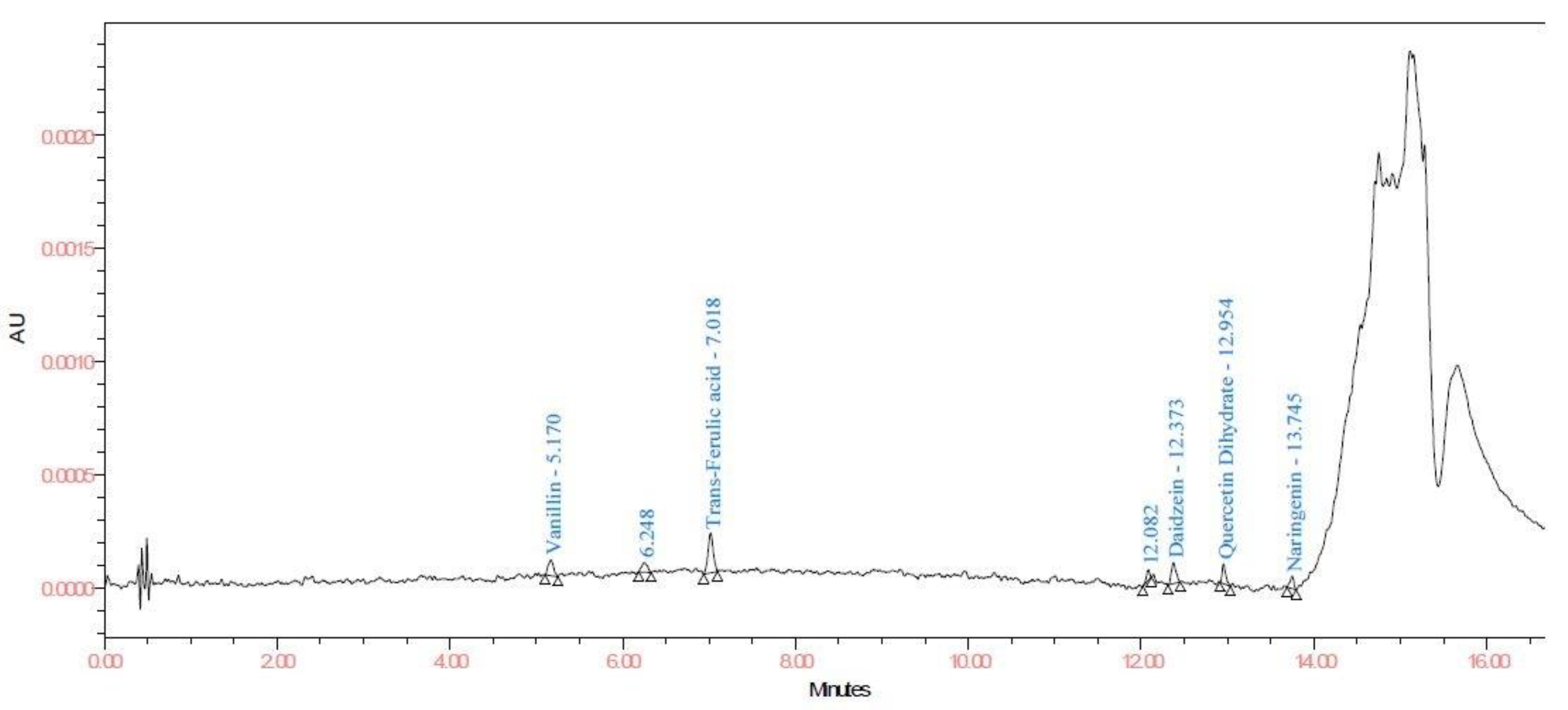
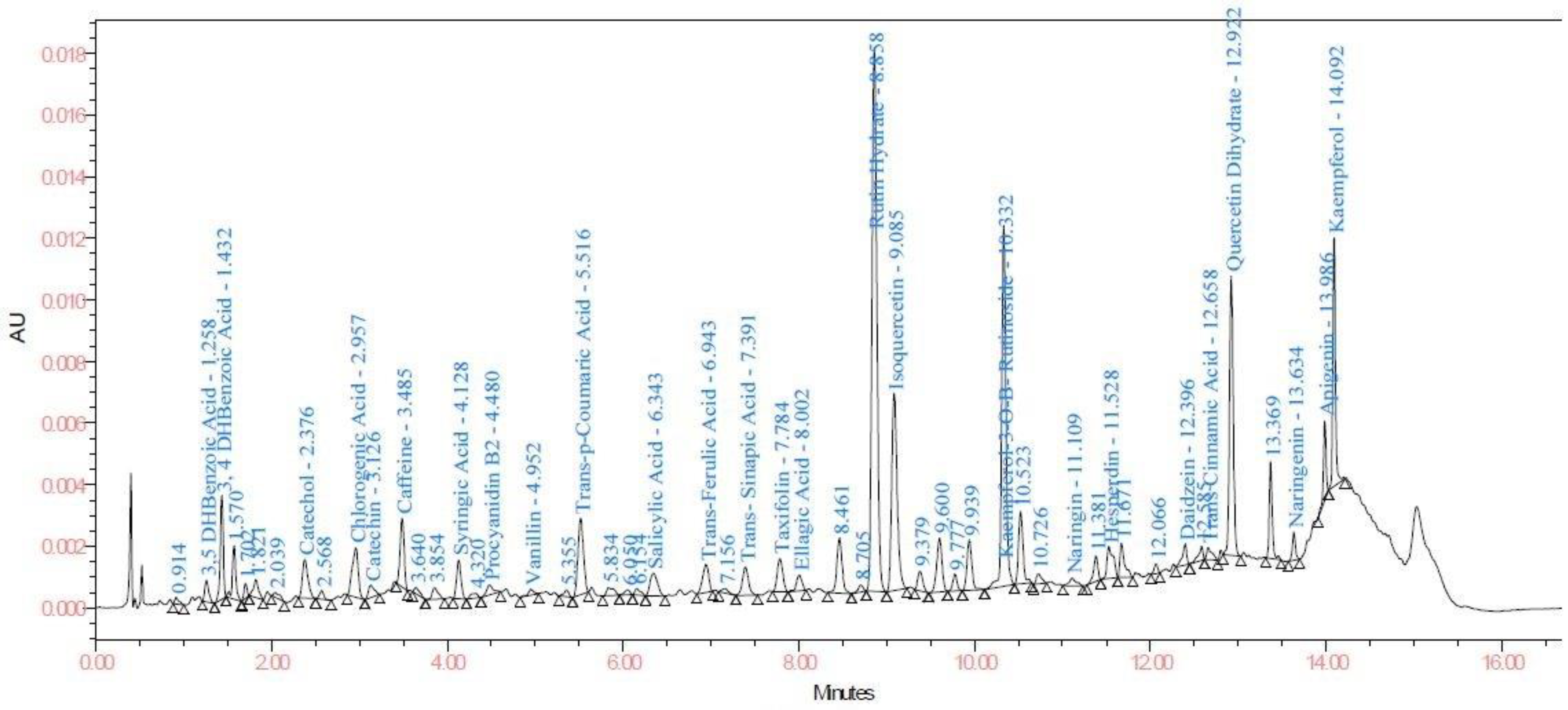
| Sr. No. | Compound | LOD and LOQ (μg/mL) | Linearity and Recovery (μg/mL) | λmax |
|---|---|---|---|---|
| 1 | Gallic acid | 8.0 | 40.0 | 214.7 |
| 2 | 3,5DHBA | 40.0 | 120.0 | 209.7 |
| 3 | 3,4DHBA | 15.0 | 40.0 | 209.7 |
| 4 | Catechol | 20.0 | 40.0 | 209.7 |
| 5 | Chlorogenic acid | 20.0 | 40.0 | 324.9 |
| 6 | Catechin | 45.0 | 80.0 | 209.7 |
| 7 | Caffeine | 8.0 | 40.0 | 209.7 |
| 8 | Syringic acid | 10.0 | 40.0 | 216.8 |
| 9 | ProcyanidinB2 | 30.0 | 40.0 | 209.7 |
| 10 | Vanillin | 20.0 | 40.0 | 229.7 |
| 11 | Trans-p-coumaric acid | 8.0 | 40.0 | 309.3 |
| 12 | Salicylic acid | 40.0 | 80.0 | 209.7 |
| 13 | Trans-ferulic acid | 15.0 | 40.0 | 321.3 |
| 14 | Trans-sinapicacid | 40.0 | 40.0 | 322,5 |
| 15 | Taxifolin | 8.0 | 40.0 | 209.7 |
| 16 | Ellagic acid | 40.0 | 40.0 | 253.4 |
| 17 | Rutinhydrate | 40.0 | 40.0 | 209.7 |
| 18 | Isoquercetin | 30.0 | 40.0 | 209.7 |
| 19 | K-3-O-βR | 30.0 | 40.0 | 209.7 |
| 20 | Naringin | 15.0 | 40.0 | 213.2 |
| 21 | Hesperidin | 15.0 | 40.0 | 209.7 |
| 22 | Daidzein | 15.0 | 40.0 | 248.7 |
| 23 | Trans-cinnamic acid | 8.0 | 40.0 | 277.1 |
| 24 | Quercetin dihydrate | 40.0 | 40.0 | 209.7 |
| 25 | Naringenin | 15.0 | 40.0 | 212.0 |
| 26 | Apigenin | 15.0 | 40.0 | 209.7 |
| 27 | Kaempferol | 20.0 | 40.0 | 209.7 |
| 28 | 2-MC | 4.0 | 40.0 | 286.7 |
| Sr. No. | Compound | Peak Purity of Standard | Peak purity of Spiked Sample C | Peak Purity of Spiked Sample D | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RT | Area | Purity Angle | Purity Threshold | RT | Area | Purity Angle | Purity Threshold | RT | Area | Purity Angle | Purity Threshold | ||
| 1 | Gallic acid | 0.739 | 11215 | 0.719 | 2.660 | 0.739 | 10690 | 0.855 | 2.754 | 0.738 | 10905 | 0.758 | 2.798 |
| 2 | 3,5 DHBA | 1.256 | 4009 | 0.230 | 1.966 | 1.258 | 4047 | 0.189 | 1.983 | 1.258 | 4119 | 0.249 | 2.058 |
| 3 | 3,4 DHBA | 1.460 | 7211 | 0.436 | 2.288 | 1.463 | 7402 | 0.499 | 2.557 | 1.462 | 7332 | 0.454 | 2.525 |
| 4 | Catechol | 2.421 | 6245 | 2.004 | 4.969 | 2.412 | 6520 | 3.355 | 6.878 | 2.413 | 6597 | 4.331 | 6.885 |
| 5 | Chlorogenic acid | 3.100 | 5562 | 0.866 | 2.316 | 3.072 | 5758 | 1.054 | 2.586 | 3.079 | 5862 | 0.772 | 2.501 |
| 6 | Catechin | 3.232 | 3938 | 8.175 | 7.158 | 3.204 | 4306 | 9.411 | 9.754 | 3.211 | 4083 | 6.609 | 8.747 |
| 7 | Caffeine | 3.589 | 15622 | 0.648 | 2.712 | 3.552 | 16310 | 2.094 | 3.415 | 3.558 | 16204 | 1.370 | 3.516 |
| 8 | Syringic acid | 4.246 | 14732 | 0.635 | 2.705 | 4.208 | 15648 | 0.836 | 3.268 | 4.215 | 15669 | 0.761 | 3.246 |
| 9 | Procyanidin B2 | 4.609 | 4456 | 2.879 | 6.805 | 4.567 | 4818 | 4.279 | 10.237 | 4.574 | 4650 | 4.204 | 10.033 |
| 10 | Vanillin | 5.186 | 23682 | 0.315 | 2.103 | 5.151 | 25149 | 0.370 | 2.301 | 5.158 | 25133 | 0.361 | 2.270 |
| 11 | Trans-p-coumaric acid | 5.634 | 21509 | 0.263 | 1.957 | 5.601 | 22689 | 0.285 | 2.049 | 5.609 | 22487 | 0.234 | 2.026 |
| 12 | Salicylic acid | 6.605 | 5213 | 0.905 | 3.261 | 6.575 | 5581 | 0.950 | 3.454 | 6.582 | 5444 | 0.901 | 3.465 |
| 13 | Trans-ferulic acid | 7.028 | 16178 | 0.258 | 1.991 | 6.996 | 18347 | 0.213 | 1.984 | 7.005 | 17770 | 0.257 | 2.063 |
| 14 | Trans-sinapicacid | 7.501 | 7192 | 0.276 | 2.013 | 7.468 | 7315 | 0.291 | 2.127 | 7.477 | 7289 | 0.278 | 2.125 |
| 15 | Taxifolin | 7.918 | 11366 | 0.727 | 2.826 | 7.884 | 11727 | 0.880 | 3.072 | 7.894 | 11904 | 0.776 | 3.183 |
| 16 | Ellagic acid | 8.130 | 8676 | 2.871 | 2.903 | 8.095 | 8598 | 3.021 | 3.438 | 8.106 | 8621 | 3.082 | 3.410 |
| 17 | Rutin hydrate | 8.997 | 3836 | 0.587 | 2.683 | 8.951 | 4099 | 0.666 | 2.972 | 8.959 | 3865 | 0.533 | 2.832 |
| 18 | Isoquercetin | 9.228 | 4574 | 0.647 | 2.679 | 9.182 | 4668 | 0.550 | 2.693 | 9.189 | 4696 | 0.550 | 2.797 |
| 19 | K-3-O-βR | 10.437 | 6280 | 0.597 | 2.337 | 10.399 | 6003 | 0.373 | 2.285 | 10.402 | 6071 | 0.361 | 2.317 |
| 20 | Naringin | 11.009 | 8451 | 1.786 | 3.553 | 10.978 | 8729 | 1.897 | 4.113 | 10.978 | 8811 | 1.975 | 4.261 |
| 21 | Hesperidin | 11.495 | 8761 | 1.077 | 3.313 | 11.466 | 9162 | 1.266 | 3.886 | 11.465 | 9140 | 1.284 | 3.776 |
| 22 | Daidzein | 12.358 | 13358 | 0.395 | 2.126 | 12.332 | 14063 | 0.499 | 2.215 | 12.333 | 13816 | 0.404 | 2.205 |
| 23 | Trans-cinnamic acid | 12.749 | 45275 | 0.255 | 1.950 | 12.719 | 47365 | 0.384 | 2.075 | 12.720 | 47448 | 0.295 | 2.104 |
| 24 | Quercetin dihydrate | 12.999 | 7423 | 0.359 | 2.056 | 12.977 | 8034 | 1.629 | 2.095 | 12.977 | 8022 | 0.957 | 2.215 |
| 25 | Naringenin | 13.925 | 13065 | 0.316 | 2.167 | 13.905 | 13839 | 0.434 | 2.314 | 13.905 | 13835 | 0.336 | 2.357 |
| 26 | Apigenin | 14.041 | 17240 | 0.130 | 1.699 | 14.024 | 18187 | 0.195 | 1.726 | 14.024 | 18054 | 0.155 | 1.741 |
| 27 | Kaempferol | 14.143 | 10491 | 0.223 | 1.756 | 14.128 | 10972 | 0.280 | 1.784 | 14.128 | 10976 | 0.231 | 1.798 |
| 28 | 2-MC | 14.358 | 32624 | 0.549 | 1.762 | 14.340 | 33852 | 0.404 | 1.807 | 14.340 | 33499 | 0.255 | 1.787 |
| Sr. No. | Compound | System Suitability | Correlation Coefficient | Linearity Range (μg/mL) | LOD (μg/mL) | LOQ (μg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RT | RSD | Area | RSD | USP Resolution | USP Tailing | USP Plate Count | ||||||
| 1 | Gallic acid | 0.739 | 0.2 | 11215 | 0.91 | 1.43 | 5384 | 0.998 | 2–6 | 0.080 | 0.264 | |
| 2 | 3,5 DHBA | 1.256 | 0.3 | 4009 | 1.44 | 11.25 | 1.46 | 10198 | 0.995 | 6–18 | 0.400 | 1.320 |
| 3 | 3,4 DHBA | 1.460 | 0.2 | 7211 | 1.23 | 3.91 | 1.40 | 12274 | 0.996 | 2–6 | 0.150 | 0.495 |
| 4 | Catechol | 2.421 | 0.0 | 6245 | 1.04 | 15.42 | 1.27 | 19593 | 0.995 | 2–6 | 0.200 | 0.660 |
| 5 | Chlorogenic acid | 3.100 | 0.1 | 5562 | 0.37 | 8.97 | 1.08 | 24397 | 0.996 | 2–6 | 0.200 | 0.660 |
| 6 | Catechin | 3.232 | 0.1 | 3938 | 0.53 | 1.57 | 1.46 | 23646 | 0.997 | 4–12 | 0.450 | 1.485 |
| 7 | Caffeine | 3.589 | 0.2 | 15622 | 0.55 | 4.31 | 1.21 | 34002 | 0.997 | 2– 6 | 0.080 | 0.264 |
| 8 | Syringic acid | 4.246 | 0.2 | 14732 | 0.29 | 7.74 | 1.19 | 37086 | 0.998 | 2–6 | 0.100 | 0.330 |
| 9 | Procyanidin B2 | 4.609 | 0.2 | 4456 | 2.26 | 3.63 | 1.26 | 29671 | 0.998 | 2- 6 | 0.300 | 0.990 |
| 10 | Vanillin | 5.186 | 0.1 | 23682 | 0.31 | 5.39 | 1.11 | 41111 | 0.997 | 2–6 | 0.200 | 0.660 |
| 11 | Trans-p-coumaric acid | 5.634 | 0.1 | 21509 | 0.31 | 4.25 | 1.15 | 46333 | 0.997 | 2–6 | 0.080 | 0.264 |
| 12 | Salicylic acid | 6.605 | 0.1 | 5213 | 1.67 | 7.62 | 1.41 | 32991 | 0.994 | 4–12 | 0.400 | 1.320 |
| 13 | Trans-ferulic Acid | 7.028 | 0.1 | 16178 | 0.46 | 3.23 | 1.10 | 63206 | 0.997 | 2–6 | 0.150 | 0.495 |
| 14 | Trans-sinapic acid | 7.501 | 0.1 | 7192 | 0.18 | 4.25 | 1.08 | 76938 | 0.996 | 2–6 | 0.400 | 1.320 |
| 15 | Taxifolin | 7.918 | 0.1 | 11366 | 0.44 | 3.59 | 1.08 | 69063 | 0.996 | 2- 6 | 0.080 | 0.264 |
| 16 | Ellagic acid | 8.130 | 0.1 | 8676 | 2.07 | 1.72 | 1.63 | 74926 | 0.995 | 2–6 | 0.400 | 1.320 |
| 17 | Rutin hydrate | 8.997 | 0.1 | 3836 | 1.22 | 7.80 | 1.09 | 133785 | 0.997 | 2–6 | 0.400 | 1.320 |
| 18 | Isoquercetin | 9.228 | 0.1 | 4574 | 0.61 | 2.26 | 1.11 | 127943 | 0.996 | 2–6 | 0.300 | 0.990 |
| 19 | K-3-O-βR | 10.437 | 0.0 | 6280 | 3.71 | 12.98 | 1.15 | 269816 | 0.997 | 2- 6 | 0.300 | 0.990 |
| 20 | Naringin | 11.009 | 0.0 | 8451 | 0.62 | 6.76 | 1.05 | 258318 | 0.998 | 2- 6 | 0.150 | 0.495 |
| 21 | Hesperidin | 11.495 | 0.0 | 8761 | 0.17 | 5.59 | 1.09 | 297168 | 0.998 | 2- 6 | 0.150 | 0.495 |
| 22 | Daidzein | 12.358 | 0.0 | 13358 | 0.43 | 10.33 | 1.10 | 378827 | 0.997 | 2–6 | 0.150 | 0.495 |
| 23 | Trans-cinnamic acid | 12.749 | 0.0 | 45275 | 0.13 | 4.33 | 1.05 | 274170 | 0.997 | 2–6 | 0.080 | 0.264 |
| 24 | Quercetin dehydrate | 12.999 | 0.0 | 7423 | 0.28 | 2.79 | 1.24 | 430037 | 0.996 | 2–6 | 0.400 | 1.320 |
| 25 | Naringenin | 13.925 | 0.0 | 13065 | 0.24 | 12.73 | 1.07 | 764867 | 0.997 | 2–6 | 0.150 | 0.495 |
| 26 | Apigenin | 14.041 | 0.0 | 17240 | 0.27 | 1.90 | 1.13 | 990979 | 0.994 | 2–6 | 0.150 | 0.495 |
| 27 | Kaempferol | 14.143 | 0.0 | 10491 | 0.21 | 1.81 | 1.18 | 1050030 | 0.996 | 2–6 | 0.200 | 0.660 |
| 28 | 2-MC | 14.358 | 0.0 | 32624 | 0.47 | 3.66 | 1.05 | 899325 | 0.997 | 2–6 | 0.040 | 0.132 |
| Sr. No. | Compound | Spiked Amount (μg/mL) | Recovery % (Sample C) | Recovery % (Sample D) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 75% | 100% | 125% | 75% | 100% | 125% | 75% | 100% | 125% | ||
| 1 | Gallic acid | 3 | 4 | 5 | 102.06 | 96.28 | 95.53 | 100.83 | 100.54 | 93.47 |
| 2 | 3,5 DHBA | 9 | 12 | 15 | 101.69 | 98.00 | 96.58 | 100.13 | 99.22 | 92.66 |
| 3 | 3,4 DHBA | 3 | 4 | 5 | 105.81 | 101.53 | 99.91 | 103.01 | 102.39 | 96.35 |
| 4 | Catechol | 3 | 4 | 5 | 90.29 | 88.74 | 91.24 | 104.56 | 103.04 | 96.83 |
| 5 | Chlorogenic acid | 3 | 4 | 5 | 105.18 | 101.84 | 100.05 | 104.61 | 103.13 | 96.08 |
| 6 | Catechin | 6 | 8 | 10 | 106.97 | 103.34 | 101.20 | 103.43 | 103.61 | 96.21 |
| 7 | Caffeine | 3 | 4 | 5 | 94.11 | 93.09 | 93.62 | 111.86 | 103.63 | 96.72 |
| 8 | Syringic acid | 3 | 4 | 5 | 105.63 | 101.81 | 100.67 | 103.43 | 103.10 | 96.18 |
| 9 | Procyanidin B2 | 3 | 4 | 5 | 103.99 | 101.05 | 99.12 | 101.57 | 102.74 | 93.68 |
| 10 | Vanillin | 3 | 4 | 5 | 92.92 | 92.19 | 92.63 | 90.54 | 93.03 | 88.13 |
| 11 | Trans-p-coumaric acid | 3 | 4 | 5 | 93.46 | 93.29 | 93.86 | 104.07 | 103.29 | 96.98 |
| 12 | Salicylic acid | 6 | 8 | 10 | 108.88 | 101.88 | 102.82 | 101.01 | 102.64 | 98.79 |
| 13 | Trans-ferulic acid | 3 | 4 | 5 | 93.38 | 92.61 | 93.79 | 90.88 | 93.90 | 88.86 |
| 14 | Trans-sinapic acid | 3 | 4 | 5 | 105.12 | 101.19 | 100.08 | 103.27 | 103.19 | 95.87 |
| 15 | Taxifolin | 3 | 4 | 5 | 105.45 | 101.96 | 100.57 | 103.73 | 102.86 | 96.12 |
| 16 | Ellagic acid | 3 | 4 | 5 | 104.55 | 100.46 | 100.02 | 102.01 | 101.39 | 94.23 |
| 17 | Rutin hydrate | 3 | 4 | 5 | 106.91 | 103.03 | 102.01 | 100.60 | 102.81 | 96.45 |
| 18 | Isoquercetin | 3 | 4 | 5 | 105.70 | 101.14 | 100.28 | 101.86 | 101.86 | 95.25 |
| 19 | K-3-O-βR | 3 | 4 | 5 | 84.85 | 85.41 | 86.52 | 86.59 | 86.81 | 84.95 |
| 20 | Naringin | 3 | 4 | 5 | 105.43 | 101.18 | 100.35 | 102.77 | 102.83 | 95.58 |
| 21 | Hesperidin | 3 | 4 | 5 | 94.34 | 92.91 | 93.21 | 104.43 | 103.58 | 96.13 |
| 22 | Daidzein | 3 | 4 | 5 | 92.03 | 91.27 | 92.08 | 89.38 | 92.19 | 87.70 |
| 23 | Trans-cinnamic acid | 3 | 4 | 5 | 105.93 | 102.18 | 100.70 | 103.32 | 103.03 | 95.92 |
| 24 | Quercetin dihydrate | 3 | 4 | 5 | 88.30 | 88.62 | 90.79 | 84.89 | 89.65 | 84.86 |
| 25 | Naringenin | 3 | 4 | 5 | 91.58 | 91.32 | 92.10 | 88.73 | 92.23 | 87.64 |
| 26 | Apigenin | 3 | 4 | 5 | 108.33 | 104.63 | 103.17 | 104.91 | 104.71 | 98.04 |
| 27 | Kaempferol | 3 | 4 | 5 | 105.96 | 102.63 | 101.07 | 103.24 | 103.18 | 96.52 |
| 28 | 2-MC | 3 | 4 | 5 | 90.95 | 91.14 | 92.41 | 88.17 | 91.69 | 87.12 |
| Sr. No. | Compound | Precision | Intermediate Method Precision | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Spiked Sample C | Spiked Sample D | Spiked Sample C | Spiked Sample D | ||||||
| RT (%RSD) | AREA (%RSD) | RT (%RSD) | AREA (%RSD) | RT (%RSD) | AREA (%RSD) | RT (%RSD) | AREA (%RSD) | ||
| 1 | Gallic acid | 0.4 | 0.58 | 0.1 | 3.19 | 0.3 | 2.05 | 0.3 | 4.0 |
| 2 | 3,5 DHBA | 0.3 | 0.46 | 0.0 | 1.12 | 0.3 | 0.75 | 0.4 | 2.38 |
| 3 | 3,4 DHBA | 0.3 | 0.74 | 0.0 | 1.25 | 0.3 | 0.82 | 0.4 | 1.09 |
| 4 | Catechol | 0.2 | 0.68 | 0.1 | 0.83 | 0.2 | 1.05 | 0.3 | 1.15 |
| 5 | Chlorogenic acid | 0.2 | 0.75 | 0.2 | 0.95 | 0.4 | 1.33 | 0.5 | 1.84 |
| 6 | Catechin | 0.2 | 0.73 | 0.2 | 1.62 | 0.4 | 1.01 | 0.5 | 2.09 |
| 7 | Caffeine | 0.2 | 0.12 | 0.2 | 1.20 | 0.4 | 0.56 | 0.5 | 1.26 |
| 8 | Syringic acid | 0.2 | 0.51 | 0.1 | 0.62 | 0.4 | 0.49 | 0.4 | 1.04 |
| 9 | Procyanidin B2 | 0.2 | 2.96 | 0.2 | 1.99 | 0.4 | 2.42 | 0.4 | 2.07 |
| 10 | Vanillin | 0.1 | 0.14 | 0.1 | 1.34 | 0.3 | 0.5 | 0.3 | 1.27 |
| 11 | Trans-p-coumaric acid | 0.1 | 0.69 | 0.1 | 0.81 | 0.3 | 0.58 | 0.3 | 1.34 |
| 12 | Salicylic acid | 0.1 | 1.89 | 0.1 | 4.09 | 0.2 | 1.58 | 0.3 | 3.06 |
| 13 | Trans-ferulic acid | 0.1 | 0.57 | 0.1 | 0.97 | 0.2 | 0.40 | 0.2 | 1.22 |
| 14 | Trans-sinapic acid | 0.1 | 0.89 | 0.1 | 0.75 | 0.2 | 1.35 | 0.3 | 1.97 |
| 15 | Taxifolin | 0.1 | 0.38 | 0.1 | 1.25 | 0.2 | 0.33 | 0.3 | 1.34 |
| 16 | Ellagic acid | 0.1 | 1.35 | 0.1 | 2.21 | 0.2 | 1.31 | 0.3 | 1.94 |
| 17 | Rutin hydrate | 0.1 | 2.38 | 0.1 | 2.34 | 0.2 | 1.63 | 0.3 | 2.33 |
| 18 | Isoquercetin | 0.1 | 0.91 | 0.0 | 1.95 | 0.2 | 1.10 | 0.3 | 1.91 |
| 19 | K-3-O-βR | 0.1 | 1.14 | 0.0 | 1.11 | 0.2 | 0.93 | 0.2 | 1.19 |
| 20 | Naringin | 0.1 | 0.35 | 0.0 | 0.80 | 0.1 | 0.74 | 0.1 | 1.96 |
| 21 | Hesperidin | 0.1 | 0.54 | 0.0 | 1.20 | 0.1 | 0.49 | 0.1 | 1.25 |
| 22 | Daidzein | 0.0 | 0.26 | 0.0 | 1.44 | 0.1 | 1.10 | 0.1 | 1.62 |
| 23 | Trans-cinnamic acid | 0.0 | 0.17 | 0.0 | 1.04 | 0.1 | 0.32 | 0.1 | 1.19 |
| 24 | Quercetin dehydrate | 0.0 | 0.89 | 0.0 | 1.47 | 0.1 | 0.96 | 0.1 | 1.94 |
| 25 | Naringenin | 0.0 | 0.23 | 0.0 | 1.29 | 0.1 | 0.67 | 0.0 | 1.43 |
| 26 | Apigenin | 0.0 | 0.15 | 0.0 | 1.16 | 0.0 | 0.46 | 0.0 | 1.33 |
| 27 | Kaempferol | 0.0 | 0.21 | 0.0 | 0.92 | 0.0 | 0.36 | 0.0 | 1.04 |
| 28 | 2-MC | 0.0 | 0.81 | 0.0 | 1.76 | 0.0 | 0.93 | 0.0 | 1.46 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandhania, S.; Pal, A.; Saharan, V. Simultaneous Estimation of Twenty Eight Phenolic Compounds by a Novel and Expeditious Method Developed on Quaternary Ultra-Performance Liquid Chromatography System with a Photodiode Array Detector. Biomolecules 2020, 10, 6. https://doi.org/10.3390/biom10010006
Mandhania S, Pal A, Saharan V. Simultaneous Estimation of Twenty Eight Phenolic Compounds by a Novel and Expeditious Method Developed on Quaternary Ultra-Performance Liquid Chromatography System with a Photodiode Array Detector. Biomolecules. 2020; 10(1):6. https://doi.org/10.3390/biom10010006
Chicago/Turabian StyleMandhania, Shiwani, Ajay Pal, and Vinod Saharan. 2020. "Simultaneous Estimation of Twenty Eight Phenolic Compounds by a Novel and Expeditious Method Developed on Quaternary Ultra-Performance Liquid Chromatography System with a Photodiode Array Detector" Biomolecules 10, no. 1: 6. https://doi.org/10.3390/biom10010006
APA StyleMandhania, S., Pal, A., & Saharan, V. (2020). Simultaneous Estimation of Twenty Eight Phenolic Compounds by a Novel and Expeditious Method Developed on Quaternary Ultra-Performance Liquid Chromatography System with a Photodiode Array Detector. Biomolecules, 10(1), 6. https://doi.org/10.3390/biom10010006