Decreased H19, GAS5, and linc0597 Expression and Association Analysis of Related Gene Polymorphisms in Rheumatoid Arthritis
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Controls
2.2. Quantitative Real-Time Reverse Transcription Polymerase Chain Reaction
2.3. Genotyping
2.4. Statistical Analysis
3. Results
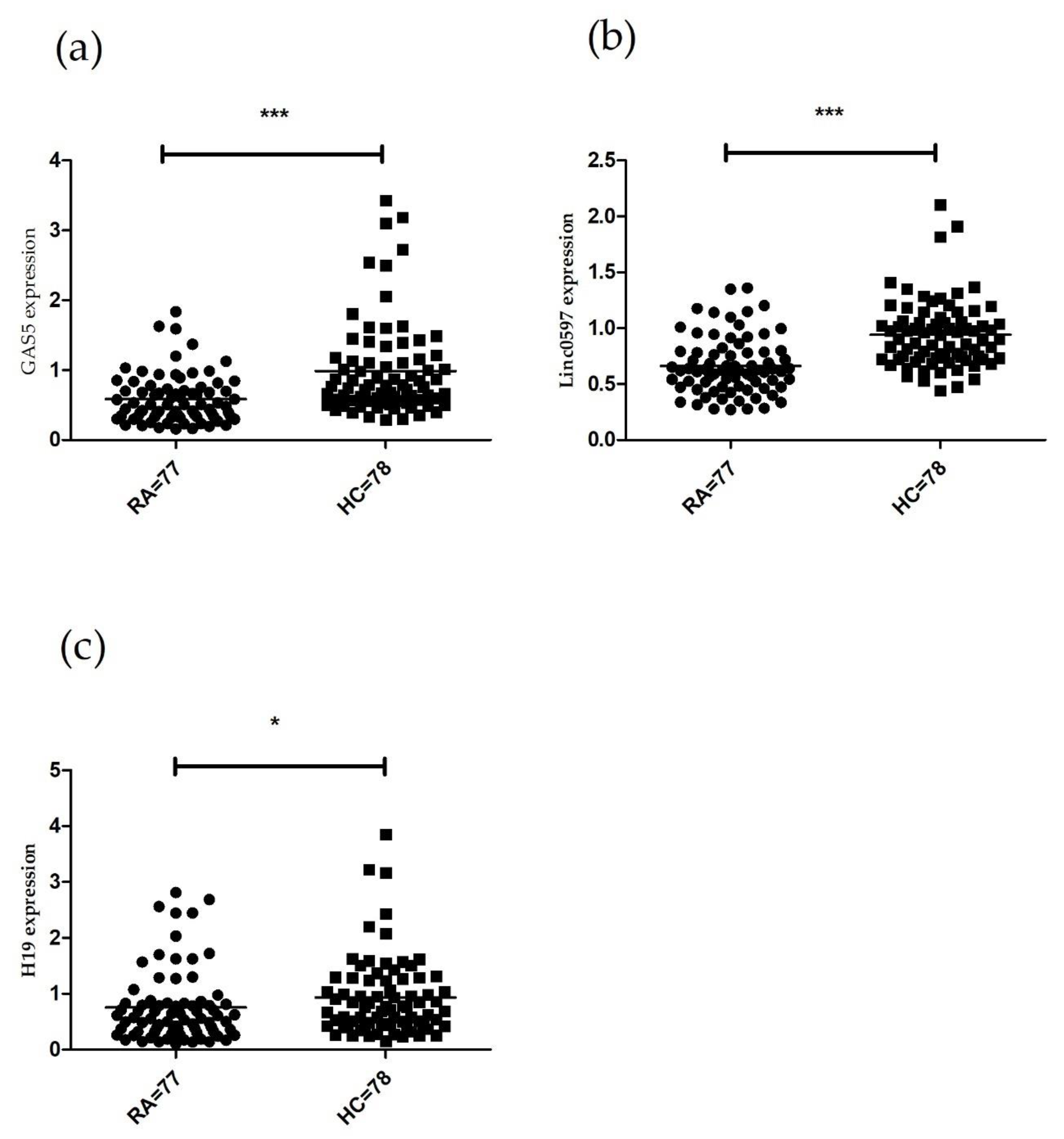
3.1. LncRNAs Expression in PBMCs of Patients with RA.
3.2. Association between lncRNAs Gene Single Nucleotide Polymorphisms and RA Susceptibility
3.3. Association of lncRNAs Expression Levels with Their Gene Single Nucleotide Polymorphisms in RA Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Scott, D.L.; Wolfe, F.; Huizinga, T.W. Rheumatoid arthritis. Lancet 2010, 376, 1094–1098. [Google Scholar] [CrossRef]
- Zhang, T.P.; Lv, T.T.; Xu, S.Z.; Pan, H.F.; Ye, D.Q. Association of interleukin-10 gene single nucleotide polymorphisms with rheumatoid arthritis in a Chinese population. Postgrad. Med. J. 2018, 94, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Prado, M.G.; Iversen, M.D.; Yu, Z.; Miller, K.R.; Triedman, N.A.; Kalia, S.S.; Lu, B.; Green, R.C.; Karlson, E.W.; Sparks, J.A. Effectiveness of a Web-Based Personalized Rheumatoid Arthritis Risk Tool With or Without a Health Educator for Knowledge of Rheumatoid Arthritis Risk Factors. Arthritis Care Res. (Hoboken) 2018, 70, 1421–1430. [Google Scholar] [CrossRef] [PubMed]
- Kallberg, H.; Padyukov, L.; Plenge, R.M.; Ronnelid, J.; Gregersen, P.K.; van der Helm-van Mil, A.H.; Toes, R.E.; Huizinga, T.W.; Klareskog, L.; Alfredsson, L.; et al. Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am. J. Hum. Genet. 2007, 80, 867–875. [Google Scholar] [CrossRef]
- Lindstrom, T.M.; Robinson, W.H. Rheumatoid arthritis: A role for immunosenescence? J. Am. Geriatr. Soc. 2010, 58, 1565–1575. [Google Scholar] [CrossRef]
- Furst, D.E.; Emery, P. Rheumatoid arthritis pathophysiology: Update on emerging cytokine and cytokine-associated cell targets. Rheumatology (Oxf.) 2014, 53, 1560–1569. [Google Scholar] [CrossRef]
- Mousavi, M.J.; Jamshidi, A.; Chopra, A.; Aslani, S.; Akhlaghi, M.; Mahmoudi, M. Implications of the noncoding RNAs in rheumatoid arthritis pathogenesis. J. Cell. Physiol. 2018, 234, 335–347. [Google Scholar] [CrossRef]
- Consortium, E.P. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007, 447, 799–816. [Google Scholar] [CrossRef]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Luo, Q.; Xu, C.; Li, X.; Zeng, L.; Ye, J.; Guo, Y.; Huang, Z.; Li, J. Comprehensive analysis of long non-coding RNA and mRNA expression profiles in rheumatoid arthritis. Exp. Ther. Med. 2017, 14, 5965–5973. [Google Scholar] [CrossRef]
- Mercer, T.R.; Mattick, J.S. Structure and function of long noncoding RNAs in epigenetic regulation. Nat. Struct. Mol. Biol. 2013, 20, 300–307. [Google Scholar] [CrossRef]
- Ceppi, M.; Pereira, P.M.; Dunand-Sauthier, I.; Barras, E.; Reith, W.; Santos, M.A.; Pierre, P. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2735–2740. [Google Scholar] [CrossRef]
- Filková, M.; Aradi, B.; Senolt, L.; Ospelt, C.; Vettori, S.; Mann, H.; Filer, A.; Raza, K.; Buckley, C.D.; Snow, M.; et al. Association of circulating miR-223 and miR-16 with disease activity in patients with early rheumatoid arthritis. Ann. Rheum. Dis. 2014, 73, 1898–1904. [Google Scholar] [CrossRef] [PubMed]
- Nakamachi, Y.; Kawano, S.; Takenokuchi, M.; Nishimura, K.; Sakai, Y.; Chin, T.; Saura, R.; Kurosaka, M.; Kumagai, S. MicroRNA-124a is a key regulator of proliferation and monocyte chemoattractant protein 1 secretion in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Rheum. 2009, 60, 1294–12304. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Hao, Y.; Dong, X.; Gong, Q.; Chen, J.; Zhang, J.; Tian, W. Molecular mechanisms and function prediction of long noncoding RNA. Sci. World J. 2012, 2012, 541786. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, L.; Ding, Y.; Lu, X.; Zhang, G.; Yang, J.; Zheng, H.; Wang, H.; Jiang, Y.; Xu, L. LncRNA Structural Characteristics in Epigenetic Regulation. Int. J. Mol. Sci. 2017, 18, 2659. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, Z.; Watabe, K.; Zhang, X.; Bai, C.; Xu, M.; Wu, F.; Mo, Y.Y. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ. 2013, 20, 1558–1568. [Google Scholar] [CrossRef] [PubMed]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Xing, D.; Liang, J.Q.; Li, Y.; Lu, J.; Jia, H.B.; Xu, L.Y.; Ma, X.L. Identification of long noncoding RNA associated with osteoarthritis in humans. Orthop. Surg. 2014, 6, 288–293. [Google Scholar] [CrossRef] [PubMed]
- Stuhlmüller, B.; Kunisch, E.; Franz, J.; Martinez-Gamboa, L.; Hernandez, M.M.; Pruss, A.; Ulbrich, N.; Erdmann, V.A.; Burmester, G.R.; Kinne, R.W. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am. J. Pathol. 2003, 163, 901–911. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, F.; Ma, J.; Zhang, X.; Wu, L.; Qu, B.; Xia, S.; Chen, S.; Tang, Y.; Shen, N. Association of large intergenic noncoding RNA expression with disease activity and organ damage in systemic lupus erythematosus. Arthritis Res. Ther. 2015, 17, 131. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.; King, R.M.; Philipson, L. Genes specifically expressed at growth arrest of mammalian cells. Cell 1988, 54, 787–793. [Google Scholar] [CrossRef]
- Mayama, T.; Marr, A.K.; Kino, T. Differential Expression of Glucocorticoid Receptor Noncoding RNA Repressor Gas5 in Autoimmune and Inflammatory Diseases. Horm. Metab. Res. 2016, 48, 550–557. [Google Scholar] [CrossRef] [PubMed]
- Moharamoghli, M.; Hassan-Zadeh, V.; Dolatshahi, E.; Alizadeh, Z.; Farazmand, A. The expression of GAS5, THRIL, and RMRP lncRNAs is increased in T cells of patients with rheumatoid arthritis. Clin. Rheumatol. 2019. [Google Scholar] [CrossRef]
- Arnett, F.C.; Edworthy, S.M.; Bloch, D.A.; McShane, D.J.; Fries, J.F.; Cooper, N.S.; Healey, L.A.; Kaplan, S.R.; Liang, M.H.; Luthra, H.S. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988, 31, 315–324. [Google Scholar] [CrossRef]
- Kuriya, B.; Schieir, O.; Lin, D.; Xiong, J.; Pope, J.; Boire, G.; Haraoui, B.; Thorne, J.C.; Tin, D.; Hitchon, C.; et al. Thresholds for the 28-joint disease activity score (DAS28) using C-reactive protein are lower compared to DAS28 using erythrocyte sedimentation rate in early rheumatoid arthritis. Clin. Exp. Rheumatol. 2017, 35, 799–803. [Google Scholar] [CrossRef]
- Sahu, A.; Singhal, U.; Chinnaiyan, A.M. Long noncoding RNAs in cancer: From function to translation. Trends Cancer 2015, 1, 93–109. [Google Scholar] [CrossRef]
- Steck, E.; Boeuf, S.; Gabler, J.; Werth, N.; Schnatzer, P.; Diederichs, S.; Richter, W. Regulation of H19 and its encoded microRNA-675 in osteoarthritis and under anabolic and catabolic in vitro conditions. J. Mol. Med. (Berl.) 2012, 90, 1185–1195. [Google Scholar] [CrossRef]
- Song, J.; Kim, D.; Han, J.; Kim, Y.; Lee, M.; Jin, E.J. PBMC and exosome-derived Hotair is a critical regulator and potent marker for rheumatoid arthritis. Clin. Exp. Med. 2015, 15, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal. 2010, 3, ra8. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.C.; Li, J.; Leng, R.X.; Li, X.P.; Li, X.M.; Wang, D.G.; Pan, H.F.; Ye, D.Q. Identification of long non-coding RNAs GAS5, linc0597 and lnc-DC in plasma as novel biomarkers for systemic lupus erythematosus. Oncotarget 2017, 8, 23650–23663. [Google Scholar] [CrossRef]
- Sigdel, K.R.; Cheng, A.; Wang, Y.; Duan, L.; Zhang, Y. The Emerging Functions of Long Noncoding RNA in Immune Cells: Autoimmune Diseases. J. Immunol. Res. 2015, 2015, 848790. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.W.; Kim, B.M.; Moon, H.W.; Lee, S.H.; Kim, H.R. Role of C-reactive protein in osteoclastogenesis in rheumatoid arthritis. Arthritis Res. Ther. 2015, 17, 41. [Google Scholar] [CrossRef]
- Holers, V.M.; Bana, N.K. Complement in the Initiation and Evolution of Rheumatoid Arthritis. Front. Immunol. 2018, 9, 1057. [Google Scholar] [CrossRef]
- Choy, E.H.; Isenberg, D.A.; Garrood, T.; Farrow, S.; Ioannou, Y.; Bird, H.; Cheung, N.; Williams, B.; Hazleman, B.; Price, R.; et al. Therapeutic benefit of blocking interleukin-6 activity with an anti-interleukin-6 receptor monoclonal antibody in rheumatoid arthritis: A randomized, double-blind, placebo-controlled, dose-escalation trial. Arthritis Rheum. 2002, 46, 3143–3150. [Google Scholar] [CrossRef]
- Nishimoto, N.; Yoshizaki, K.; Miyasaka, N.; Yamamoto, K.; Kawai, S.; Takeuchi, T.; Hashimoto, J.; Azuma, J.; Kishimoto, T. Treatment of rheumatoid arthritis with humanized anti-interleukin-6 receptor antibody: A multicenter, double-blind, placebo-controlled trial. Arthritis Rheum. 2004, 50, 1761–1769. [Google Scholar] [CrossRef]
- Li, Z.; Chao, T.C.; Chang, K.Y.; Lin, N.; Patil, V.S.; Shimizu, C.; Head, S.R.; Burns, J.C.; Rana, T.M. The long noncoding RNA THRIL regulates TNFalpha expression through its interaction with hnRNPL. Proc. Natl. Acad. Sci. USA 2014, 111, 1002–1007. [Google Scholar] [CrossRef]
- Li, Q.; Ma, G.; Sun, S.H.; Xu, Y.; Wang, B. Polymorphism in the promoter region of lncRNA GAS5 is functionally associated with the risk of gastric cancer. Clin. Res. Hepatol. Gastroenterol. 2018, 42, 478–482. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Y.; Zhao, Y.; Jin, Y.; An, L.; Wu, B.; Liu, Z.; Chen, X.; Zhou, H.; Wang, H.; et al. Genetic polymorphisms of long non-coding RNA GAS5 predict platinum-based concurrent chemoradiotherapy response in nasopharyngeal carcinoma patients. Oncotarget 2017, 8, 62286–62297. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Huang, K.; Wen, F.; Cui, G.; Guo, H.; Zhao, S. Genetic variation of lncRNA GAS5 contributes to the development of lung cancer. Oncotarget 2017, 8, 91025–91029. [Google Scholar] [CrossRef] [PubMed]
- Mahdi Eftekharian, M.; Noroozi, R.; Komaki, A.; Mazdeh, M.; Taheri, M.; Ghafouri-Fard, S. GAS5 genomic variants and risk of multiple sclerosis. Neurosci. Lett. 2019, 701, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Yu, Z.; Fang, X.; Liu, M.; Pu, Y.; Shao, Q.; Wang, D.; Zhao, X.; Huang, A.; Xiang, Z. LncRNA GAS5 inhibits microglial M2 polarization and exacerbates demyelination. EMBO Rep. 2017, 18, 1801–1816. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Xia, J.H.; Sipeky, C.; Dong, X.M.; Zhang, Q.; Yang, Y.; Zhang, P.; Cruz, S.P.; Zhang, K.; Zhu, J.; et al. Biology and Clinical Implications of the 19q13 Aggressive Prostate Cancer Susceptibility Locus. Cell 2018, 174, 576–589. [Google Scholar] [CrossRef]
- Hua, J.T.; Ahmed, M.; Guo, H.; Zhang, Y.; Chen, S.; Soares, F.; Lu, J.; Zhou, S.; Wang, M.; Li, H.; et al. Risk SNP-Mediated Promoter-Enhancer Switching Drives Prostate Cancer through lncRNA PCAT19. Cell 2018, 174, 564–575. [Google Scholar] [CrossRef]
- Huang, A.F.; Su, L.C.; Jia, H.; Liu, Y.; Xu, W.D. No association of single nucleotide polymorphisms within H19 and HOX transcript antisense RNA (HOTAIR) with genetic susceptibility to systemic lupus erythematosus, rheumatoid arthritis, and primary Sjogren’s syndrome in a Chinese Han population. Clin. Rheumatol. 2017, 36, 2447–2453. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Li, J.J.; Hua, D.J.; Huang, S.C.; Sun, Q.Q.; Huang, H.; Xin, X.F.; Cen, H. A study on associations of single-nucleotide polymorphisms within H19 and HOX transcript antisense RNA (HOTAIR) with genetic susceptibility to rheumatoid arthritis in a Chinese population. Inflamm. Res. 2017, 66, 515–521. [Google Scholar] [CrossRef]
- Lu, M.C.; Yu, H.C.; Yu, C.L.; Huang, H.B.; Koo, M.; Tung, C.H.; Lai, N.S. Increased expression of long noncoding RNAs LOC100652951 and LOC100506036 in T cells from patients with rheumatoid arthritis facilitates the inflammatory responses. Immunol. Res. 2016, 64, 576–583. [Google Scholar] [CrossRef]
- Ye, S.; Patodi, N.; Walker-Bone, K.; Reading, I.; Cooper, C.; Dennison, E. Variation in the matrix metalloproteinase-3, -7, -12 and -13 genes is associated with functional status in rheumatoid arthritis. Int. J. Immunogenet. 2007, 34, 81–85. [Google Scholar] [CrossRef]
- Spurlock, C.F., 3rd; Tossberg, J.T.; Matlock, B.K.; Olsen, N.J.; Aune, T.M. Methotrexate inhibits NF-kappaB activity via long intergenic (noncoding) RNA-p21 induction. Arthritis Rheumatol. (Hoboken NJ) 2014, 66, 2947–2957. [Google Scholar] [CrossRef] [PubMed]
- Messemaker, T.C.; Frank-Bertoncelj, M.; Marques, R.B.; Adriaans, A.; Bakker, A.M.; Daha, N.; Gay, S.; Huizinga, T.W.; Toes, R.E.; Mikkers, H.M.; et al. A novel long non-coding RNA in the rheumatoid arthritis risk locus TRAF1-C5 influences C5 mRNA levels. Genes Immun. 2016, 17, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Müller, N.; Doring, F.; Klapper, M.; Neumann, K.; Schulte, D.M.; Türk, K.; Schröder, J.O.; Zeuner, R.A.; Freitag-Wolf, S.; Schreiber, S.; et al. Interleukin-6 and tumour necrosis factor-alpha differentially regulate lincRNA transcripts in cells of the innate immune system in vivo in human subjects with rheumatoid arthritis. Cytokine 2014, 68, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Moran, V.A.; Perera, R.J.; Khalil, A.M. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012, 40, 6391–6400. [Google Scholar] [CrossRef] [PubMed]
- Khorkova, O.; Hsiao, J.; Wahlestedt, C. Basic biology and therapeutic implications of lncRNA. Adv. Drug. Deliv. Rev. 2015, 87, 15–24. [Google Scholar] [CrossRef] [PubMed]
| RNAs | Primers |
|---|---|
| H19 | F:5′TGCTGCACTTTACAACCACTG3′ |
| R:5′ATGGTGTCTTTGATGTTGGGC3′ | |
| GAS5 | F:5′TATGGTGCTGGGTGCGGAT3′ |
| R:5′CCAATGGCTTGAGTTAGGCTT3′ | |
| linc0597 | F:5′TTGGATTCATCCCGTTCACCTCCA3′ |
| R:5′CAGCATGACGATCAAGCGAGATTC3′ | |
| β-actin | F:5′CACGAAACTACCTTCAACTCC3′ |
| R:5′CATACTCCTGCTTGCTGATC3′ |
| Parameters | Number | GAS5 | linc0597 | H19 |
|---|---|---|---|---|
| RF | ||||
| Positive | 69 | 0.50 (0.34, 0.72) | 0.62 (0.47, 0.78) | 0.56 (0.35, 0.82) |
| Negative | 8 | 0.52 (0.33, 0.92) | 0.54 (0.34, 0.86) | 0.57 (0.16, 1.18) |
| Anti-CCP | ||||
| Positive | 65 | 0.51 (0.35, 0.74) | 0.61 (0.47, 0.79) | 0.61 (0.35, 0.83) |
| Negative | 12 | 0.34 (0.25, 0.77) | 0.63 (0.37, 0.73) | 0.36 (0.17, 0.74) |
| Low complement | ||||
| Positive | 14 | 0.35 (0.21, 0.46) | 0.50 (0.41, 0.69) | 0.52 (0.27, 1.23) |
| Negative | 52 | 0.53 (0.34, 0.77) | 0.62 (0.53, 0.80) | 0.60 (0.36, 0.82) |
| Parameters | GAS5 | linc0597 | H19 | |||
|---|---|---|---|---|---|---|
| rs | p Value | rs | p Value | rs | p Value | |
| ESR | −0.072 | 0.535 | −0.114 | 0.325 | −0.150 | 0.196 |
| CRP | −0.273 | 0.017 | −0.096 | 0.411 | 0.002 | 0.986 |
| TJC | −0.020 | 0.866 | −0.091 | 0.435 | −0.192 | 0.097 |
| SJC | −0.054 | 0.648 | 0.052 | 0.660 | −0.110 | 0.924 |
| DAS28-ESR | −0.112 | 0.337 | −0.012 | 0.920 | −0.166 | 0.153 |
| Group | Number | GAS5 | linc0597 | H19 |
|---|---|---|---|---|
| Prednisone (mg/day) | ||||
| ≤7.5 | 36 | 0.61 (0.37, 0.94) | 0.61 (0.46, 0.81) | 0.58 (0.30, 0.87) |
| >7.5 | 37 | 0.42 (0.34, 0.68) | 0.62 (0.48, 0.78) | 0.55 (0.31, 0.82) |
| DMARDS | ||||
| Yes | 44 | 0.51 (0.36, 0.81) | 0.62 (0.47, 0.78) | 0.61 (0.34, 0.84) |
| No | 29 | 0.53 (0.34, 0.69) | 0.59 (0.47, 0.80) | 0.43 (0.29, 0.84) |
| Botanical preparation | ||||
| Yes | 22 | 0.51 (0.36, 0.84) | 0.63 (0.46, 0.82) | 0.59 (0.46, 1.13) |
| No | 51 | 0.53 (0.34, 0.76) | 0.62 (0.48, 0.79) | 0.49 (0.27, 0.80) |
| SNPs | Analysis Model | RA n(%) | HC n(%) | χ2 | OR (95% CI) | padjust Value |
|---|---|---|---|---|---|---|
| rs6790 | Genotype | |||||
| GG | 362 (43.9) | 316 (40.8) | 1.103 | 0.848 (0.623–1.154) | 0.294 | |
| GA | 333 (40.4) | 365 (47.1) | 6.132 | 0.679 (0.499–0.922) | 0.013 | |
| AA | 130 (15.8) | 94 (12.1) | 1.000 | |||
| Allele | ||||||
| G | 1057 (63.9) | 997 (63.4) | 0.024 | 0.989 (0.856–1.142) | 0.877 | |
| A | 593 (36.1) | 553 (36.6) | 1.000 | |||
| Dominant model | ||||||
| AA + GA | 463 (56.1) | 459 (59.2) | 1.630 | 0.878 (0.719–1.072) | 0.202 | |
| GG | 362 (43.9) | 316 (40.8) | 1.000 | |||
| Recessive model | ||||||
| AA | 130 (15.8) | 94 (12.1) | 3.578 | 1.321 (0.990–1.762) | 0.059 | |
| GA + GG | 695 (84.2) | 681 (87.9) | 1.000 | |||
| rs2067051 | Genotypes | |||||
| TT | 101 (12.4) | 80 (10.7) | 1.059 | 1.189 (0.855–1.653) | 0.303 | |
| TC | 335 (41.1) | 306 (41.0) | 0.140 | 1.042 (0.842–1.289) | 0.708 | |
| CC | 380 (46.6) | 360 (48.3) | 1.000 | |||
| Allele | ||||||
| T | 537 (33.0) | 466 (31.6) | 0.998 | 1.080 (0.929–1.255) | 0.318 | |
| C | 1095 (67.0) | 1026 (68.4) | 1.000 | |||
| Dominant model | ||||||
| TT + TC | 436 (53.4) | 386 (51.7) | 0.464 | 1.072 (0.878–1.130) | 0.496 | |
| CC | 380 (46.6) | 360 (48.3) | 1.000 | |||
| Recessive model | ||||||
| TT | 101 (12.4) | 80 (10.7) | 0.923 | 1.167 (0.852–1.598) | 0.337 | |
| TC + CC | 715 (87.6) | 666 (89.3) | 1.000 | |||
| rs2070107 | Genotype | |||||
| GG | 576 (69.2) | 549 (72.0) | 0.022 | 0.955 (0.522–1.747) | 0.881 | |
| GC | 233 (28.0) | 193 (25.3) | 0.089 | 1.100 (0.590–2.050) | 0.765 | |
| CC | 23 (2.8) | 21 (2.8) | ||||
| Allele | ||||||
| G | 1385(83.2) | 1291(84.6) | 1.100 | 0.904 (0.748–1.092) | 0.294 | |
| C | 279 (16.8) | 235 (15.4) | 1.000 | |||
| Dominant model | ||||||
| GC + CC | 256 (30.8) | 214 (28.0) | 1.424 | 1.141 (0.919–1.418) | 0.233 | |
| GG | 576 (69.2) | 549 (72.0) | 1.000 | |||
| Recessive model | ||||||
| CC | 23 (2.8) | 21 (2.8) | 0.001 | 1.008(0.553–1.838) | 0.980 | |
| GC + GG | 809 (97.2) | 742 (97.2) | ||||
| rs2075745 | Genotype | |||||
| TT | 113 (14.1) | 106 (14.2) | 0.080 | 0.956 (0.703–1.302) | 0.777 | |
| TA | 340 (42.3) | 318 (42.6) | 0.026 | 0.982 (0.791–1.219) | 0.871 | |
| AA | 350 (43.6) | 323 (43.2) | 1.000 | |||
| Allele | ||||||
| T | 566 (35.2) | 530 (35.5) | 0.018 | 0.990 (0.854–1.147) | 0.892 | |
| A | 1040 (64.8) | 964 (64.5) | 1.000 | |||
| Dominant model | ||||||
| TT + TA | 453 (56.4) | 424 (56.8) | 0.056 | 0.976 (0.797–1.194) | 0.812 | |
| AA | 350 (43.6) | 323 (43.2) | 1.000 | |||
| Recessive model | ||||||
| TT | 113 (14.1) | 106 (14.2) | 0.058 | 0.965(0.722–1.289) | 0.809 | |
| TA + AA | 690 (85.9) | 641 (85.8) | 1.000 | |||
| rs2285991 | Genotype | |||||
| GG | 678 (86.1) | 612 (86.6) | 0.773 | 0.611 (0.203–1.834) | 0.379 | |
| GA | 100 (12.7) | 90 (12.7) | 0.719 | 0.613 (0.198–1.900) | 0.396 | |
| AA | 9 (1.1) | 5 (0.7) | 1.000 | |||
| Allele | ||||||
| G | 1456 (92.5) | 1314 (92.9) | 0.199 | 0.939 (0.712–1.238) | 0.656 | |
| A | 118 (7.5) | 100 (7.1) | 1.000 | |||
| Dominant model | ||||||
| GA + AA | 109 (13.9) | 95 (13.4) | 0.058 | 1.037 (0.770–1.397) | 0.809 | |
| GG | 678 (86.1) | 612 (86.6) | 1.000 | |||
| Recessive model | ||||||
| AA | 9 (1.1) | 5 (0.7) | 0.772 | 1.637 (0.545–4.914) | 0.380 | |
| GA + GG | 778 (98.9) | 702 (99.3) | 1.000 | |||
| rs2632516 | Genotype | |||||
| GG | 270 (32.6) | 229 (29.4) | 1.151 | 1.161 (0.884–1.527) | 0.283 | |
| GC | 379 (45.8)) | 373 (47.8) | 0.015 | 1.016 (0.789–1.309) | 0.901 | |
| CC | 179 (21.6) | 178 (22.8) | 1.000 | |||
| Allele | ||||||
| G | 919 (55.5) | 831 (53.3) | 1.604 | 1.094 (0.952–1.257) | 0.205 | |
| C | 737 (44.5) | 729 (46.7) | 1.000 | |||
| Dominant model | ||||||
| GG + GC | 649 (78.4) | 602 (77.2) | 0.329 | 1.072 (0.846–1.357) | 0.566 | |
| CC | 179 (21.6) | 178 (22.8) | 1.000 | |||
| Recessive model | ||||||
| GG | 270 (32.6) | 229 (29.4) | 1.632 | 1.149 (0.928–1.422) | 0.201 | |
| GC + CC | 558 (67.4) | 551 (70.6) | 1.000 | |||
| rs2877877 | Genotype | |||||
| GG | 50 (6.1) | 53 (7.3) | 1.270 | 0.790 (0.524–1.190) | 0.260 | |
| GA | 272 (33.1) | 248 (34.3) | 0.561 | 0.921 (0.741–1.143) | 0.454 | |
| AA | 500 (60.8) | 422 (58.4) | 1.000 | |||
| Allele | ||||||
| G | 372 (22.6) | 354 (24.5) | 1.470 | 0.902 (0.764–1.066) | 0.225 | |
| A | 1272 (77.4) | 1092 (75.5) | 1.000 | |||
| Dominant model | ||||||
| GG + GA | 322 (39.2) | 301 (41.6) | 1.070 | 0.898 (0.731–1.101) | 0.301 | |
| AA | 500 (60.8) | 422 (58.4) | 1.000 | |||
| Recessive model | ||||||
| GG | 50 (6.1) | 53 (7.3) | 1.006 | 0.814 (0.544–1.217) | 0.316 | |
| GA + AA | 772 (93.9) | 670 (92.7) | 1.000 | |||
| rs13414 | Genotype | |||||
| AA | 368 (51.0) | 316 (46.7) | 0.149 | 1.076 (0.741–1.563) | 0.700 | |
| AG | 283 (39.3) | 295 (43.6) | 0.403 | 0.885 (0.606–1.292) | 0.526 | |
| GG | 70 (9.7) | 65 (9.6) | 1.000 | |||
| Allele | ||||||
| A | 1019 (70.7) | 927 (68.6) | 1.456 | 1.104 (0.940–1.298) | 0.228 | |
| G | 423 (29.3) | 425 (31.4) | 1.000 | |||
| Dominant model | ||||||
| AG + GG | 353 (49.0) | 360 (53.3) | 2.551 | 0.841 (0.681–1.040) | 0.110 | |
| AA | 368 (51.0) | 316 (46.7) | 1.000 | |||
| Recessive model | ||||||
| GG | 70 (9.7) | 65 (9.6) | 0.008 | 1.016 (0.710–1.455) | 0.930 | |
| AG + AA | 651 (90.3) | 611 (90.4) | 1.000 | |||
| rs4372750 | Genotype | |||||
| AA | 168 (23.5) | 156 (23.0) | 0.184 | 1.070 (0.786–1.456) | 0.668 | |
| AC | 378 (52.8) | 352 (51.9) | 0.370 | 1.084 (0.836–1.405) | 0.543 | |
| CC | 170 (23.7) | 170 (25.1) | 1.000 | |||
| Allele | ||||||
| A | 714 (49.9) | 664 (49.0) | 0.222 | 1.036 (0.893–1.202) | 0.637 | |
| C | 718 (50.1) | 692 (51.0) | 1.000 | |||
| Dominant model | ||||||
| AA + AC | 546 (76.3) | 508 (74.9) | 0.318 | 1.074 (0.839–1.374) | 0.573 | |
| CC | 170 (23.7) | 170 (25.1) | 1.000 | |||
| Recessive model | ||||||
| AA | 168 (23.5) | 156 (23.0) | 0.006 | 1.007 (0.999–1.015) | 0.937 | |
| AC+CC | 548 (76.5) | 522 (77.0) | 1.000 | |||
| rs12601867 | Genotype | |||||
| CC | 165 (22.6) | 166 (24.0) | 0.953 | 0.860 (0.634–1.165) | 0.329 | |
| CG | 381 (52.1) | 361 (52.2) | 0.274 | 0.934 (0.723–1.206) | 0.601 | |
| GG | 185 (25.3) | 165 (23.8) | 1.000 | |||
| Allele | ||||||
| C | 711 (48.6) | 693 (50.1) | 0.590 | 0.944 (0.815–1.094) | 0.442 | |
| G | 751 (51.4) | 691 (49.9) | 1.000 | |||
| Dominant model | ||||||
| CG + GG | 566 (77.4) | 526 (76.0) | 0.681 | 1.111 (0.866–1.425) | 0.409 | |
| CC | 165 (22.6) | 166 (24.0) | 1.000 | |||
| Recessive model | ||||||
| GG | 185 (25.3) | 165 (23.8) | 0.571 | 1.098 (0.861–1.400) | 0.450 | |
| CG + CC | 546 (74.7) | 527 (76.2) | 1.000 | |||
| rs16847206 | Genotype | |||||
| AA | 338 (46.5) | 296 (43.7) | 1.358 | 1.247 (0.860–1.808) | 0.244 | |
| AT | 325 (44.7) | 308 (45.5) | 0.679 | 1.169 (0.806–1.694) | 0.410 | |
| TT | 64 (8.8) | 73 (10.8) | 1.000 | |||
| Allele | ||||||
| A | 1001 (68.8) | 900 (66.5) | 1.808 | 1.115 (0.952–1.306) | 0.179 | |
| T | 453 (31.2) | 454 (33.5) | 1.000 | |||
| Dominant model | ||||||
| AT + TT | 389 (53.5) | 381 (56.3) | 0.735 | 0.911 (0.737–1.127) | 0.391 | |
| AA | 338 (46.5) | 296 (43.7) | 1.000 | |||
| Recessive model | ||||||
| TT | 64 (8.8) | 73 (10.8) | 1.086 | 0.828 (0.581–1.180) | 0.297 | |
| AT + AA | 663 (91.2) | 604 (89.2) | ||||
| rs6692753 | Genotype | |||||
| GG | 339 (46.6) | 287 (42.0) | 1.624 | 1.272 (0.878–1.843) | 0.203 | |
| GT | 323 (44.4) | 323 (47.3) | 0.241 | 1.097 (0.758–1.587) | 0.623 | |
| TT | 65 (8.9) | 73 (10.7) | 1.000 | |||
| Allele | ||||||
| G | 1001 (68.8) | 897 (65.7) | 3.231 | 1.155 (0.987–1.352) | 0.072 | |
| T | 453 (31.2) | 469 (34.3) | 1.000 | |||
| Dominant model | ||||||
| GT + TT | 388 (53.4) | 396 (58.0) | 2.315 | 0.848 (0.686–1.049) | 0.128 | |
| GG | 339 (46.6) | 287 (42.0) | 1.000 | |||
| Recessive model | ||||||
| TT | 65 (8.9) | 73 (10.7) | 0.842 | 0.848 (0.596–1.206) | 0.359 | |
| GT + GG | 662 (91.1) | 610 (89.3) | 1.000 | |||
| rs2680700 | Genotype | |||||
| GG | 412 (57.3) | 376 (55.1) | 1.773 | 0.755 (0.499–1.142) | 0.183 | |
| GT | 243 (33.8) | 263 (38.6) | 4.477 | 0.630 (0.411–0.967) | 0.034 | |
| TT | 64 (8.9) | 43 (6.3) | 1.000 | |||
| Allele | ||||||
| G | 1067 (74.2) | 1015 (74.4) | 0.017 | 0.989 (0.835–1.172) | 0.897 | |
| T | 371 (25.8) | 349 (25.6) | 1.000 | |||
| Dominant model | ||||||
| GT + TT | 307 (42.7) | 306 (44.9) | 0.857 | 0.904 (0.731–1.119) | 0.355 | |
| GG | 412 (57.3) | 376 (55.1) | 1.000 | |||
| Recessive model | ||||||
| TT | 64 (8.9) | 43 (6.3) | 2.896 | 1.421 (0.948–2.131) | 0.089 | |
| GT+GG | 655 (91.1) | 639 (93.7) | 1.000 | |||
| rs8071916 | Genotype | |||||
| AA | 175 (24.1) | 162 (23.4) | 0.016 | 1.020 (0.751–1.384) | 0.900 | |
| AG | 378 (52.0) | 366 (53.0) | 0.015 | 0.984 (0.758–1.277) | 0.904 | |
| GG | 174 (23.9) | 163 (23.6) | 1.000 | |||
| Allele | ||||||
| A | 728 (50.1) | 690 (49.9) | 0.006 | 1.006 (0.868–1.165) | 0.940 | |
| G | 726 (49.9) | 692 (50.1) | 1.000 | |||
| Dominant model | ||||||
| AG + GG | 552 (75.9) | 529 (76.6) | 0.059 | 0.970 (0.758–1.241) | 0.808 | |
| AA | 175 (24.1) | 162 (23.4) | 1.000 | |||
| Recessive model | ||||||
| GG | 174 (23.9) | 163 (23.6) | 0.002 | 1.005 (0.785–1.287) | 0.969 | |
| AG + AA | 553 (76.1) | 528 (76.4) | 1.000 | |||
| SNPs | GAS5 | linc0597 | H19 | |||
|---|---|---|---|---|---|---|
| rs | p Value | rs | p Value | rs | p Value | |
| rs2067051 | −0.030 | 0.823 | −0.076 | 0.569 | −0.011 | 0.935 |
| rs2075745 | −0.025 | 0.850 | −0.055 | 0.681 | −0.107 | 0.420 |
| rs2877877 | 0.085 | 0.523 | 0.088 | 0.509 | −0.031 | 0.815 |
| rs2070107 | −0.077 | 0.560 | −0.142 | 0.285 | −0.051 | 0.703 |
| rs2632516 | 0.096 | 0.470 | −0.094 | 0.478 | 0.031 | 0.816 |
| rs6790 | −0.010 | 0.941 | 0.029 | 0.828 | 0.068 | 0.611 |
| rs2285991 | 0.103 | 0.438 | −0.079 | 0.551 | 0.055 | 0.677 |
| rs13414 | −0.149 | 0.360 | −0.041 | 0.803 | 0.014 | 0.930 |
| rs4372750 | 0.344 | 0.030 | −0.010 | 0.950 | 0.202 | 0.211 |
| rs12601867 | 0.216 | 0.180 | 0.010 | 0.950 | 0.244 | 0.129 |
| rs16847206 | −0.135 | 0.414 | −0.090 | 0.588 | −0.136 | 0.408 |
| rs6692753 | −0.143 | 0.378 | −0.100 | 0.538 | −0.145 | 0.373 |
| rs2680700 | −0.149 | 0.358 | −0.135 | 0.406 | −0.013 | 0.939 |
| rs8071916 | −0.279 | 0.081 | −0.027 | 0.870 | −0.250 | 0.120 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Zhang, T.-P.; Zhao, Y.-L.; Li, B.-Z.; Leng, R.-X.; Pan, H.-F.; Ye, D.-Q. Decreased H19, GAS5, and linc0597 Expression and Association Analysis of Related Gene Polymorphisms in Rheumatoid Arthritis. Biomolecules 2020, 10, 55. https://doi.org/10.3390/biom10010055
Wu J, Zhang T-P, Zhao Y-L, Li B-Z, Leng R-X, Pan H-F, Ye D-Q. Decreased H19, GAS5, and linc0597 Expression and Association Analysis of Related Gene Polymorphisms in Rheumatoid Arthritis. Biomolecules. 2020; 10(1):55. https://doi.org/10.3390/biom10010055
Chicago/Turabian StyleWu, Jun, Tian-Ping Zhang, Yu-Lan Zhao, Bao-Zhu Li, Rui-Xue Leng, Hai-Feng Pan, and Dong-Qing Ye. 2020. "Decreased H19, GAS5, and linc0597 Expression and Association Analysis of Related Gene Polymorphisms in Rheumatoid Arthritis" Biomolecules 10, no. 1: 55. https://doi.org/10.3390/biom10010055
APA StyleWu, J., Zhang, T.-P., Zhao, Y.-L., Li, B.-Z., Leng, R.-X., Pan, H.-F., & Ye, D.-Q. (2020). Decreased H19, GAS5, and linc0597 Expression and Association Analysis of Related Gene Polymorphisms in Rheumatoid Arthritis. Biomolecules, 10(1), 55. https://doi.org/10.3390/biom10010055





