Interaction of a Novel Zn2Cys6 Transcription Factor DcGliZ with Promoters in the Gliotoxin Biosynthetic Gene Cluster of the Deep-Sea-Derived Fungus Dichotomomyces cejpii
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome Sequencing and Assembly
2.2. Gli Cluster Prediction and Annotation
2.3. Conserved Domain Prediction and Phylogenetic Analysis of DcGliZ
2.4. Heterologous Expression and Purification of Full-Length DcGliZ and DcGliZ Core Proteins from E. coli
2.5. Interaction Between Transcriptional Factor DcGliZ and the Promoters of the Gli Cluster
2.6. Surface Plasmon Resonance Assay
3. Results
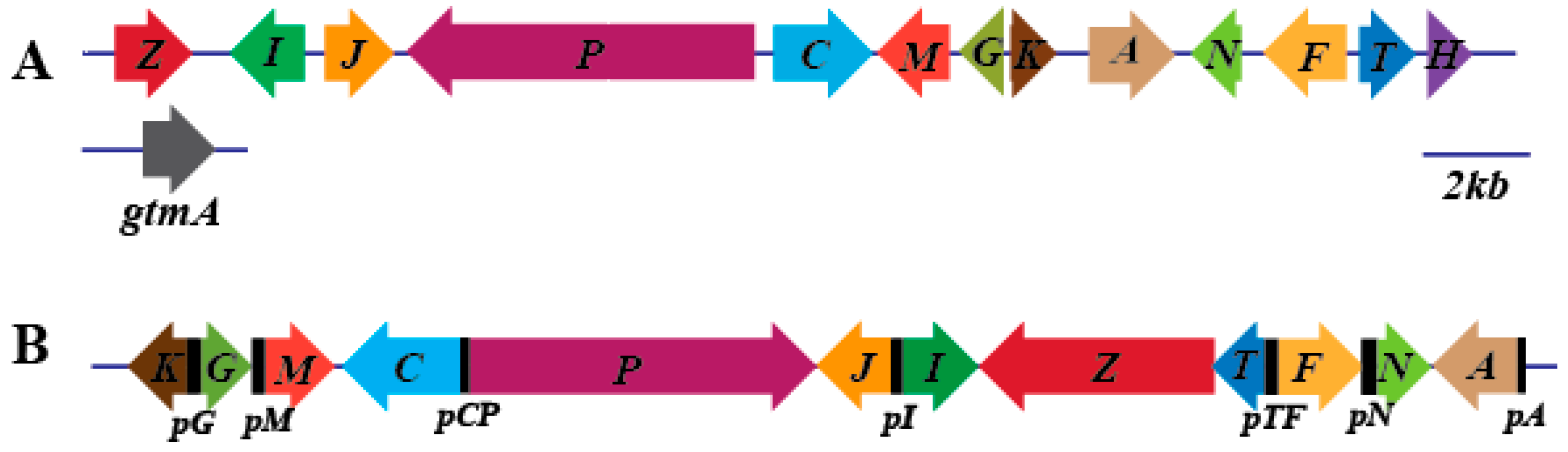
3.1. Prediction and Analysis of the Gliotoxin Biosynthesis Cluster of D. cejpii
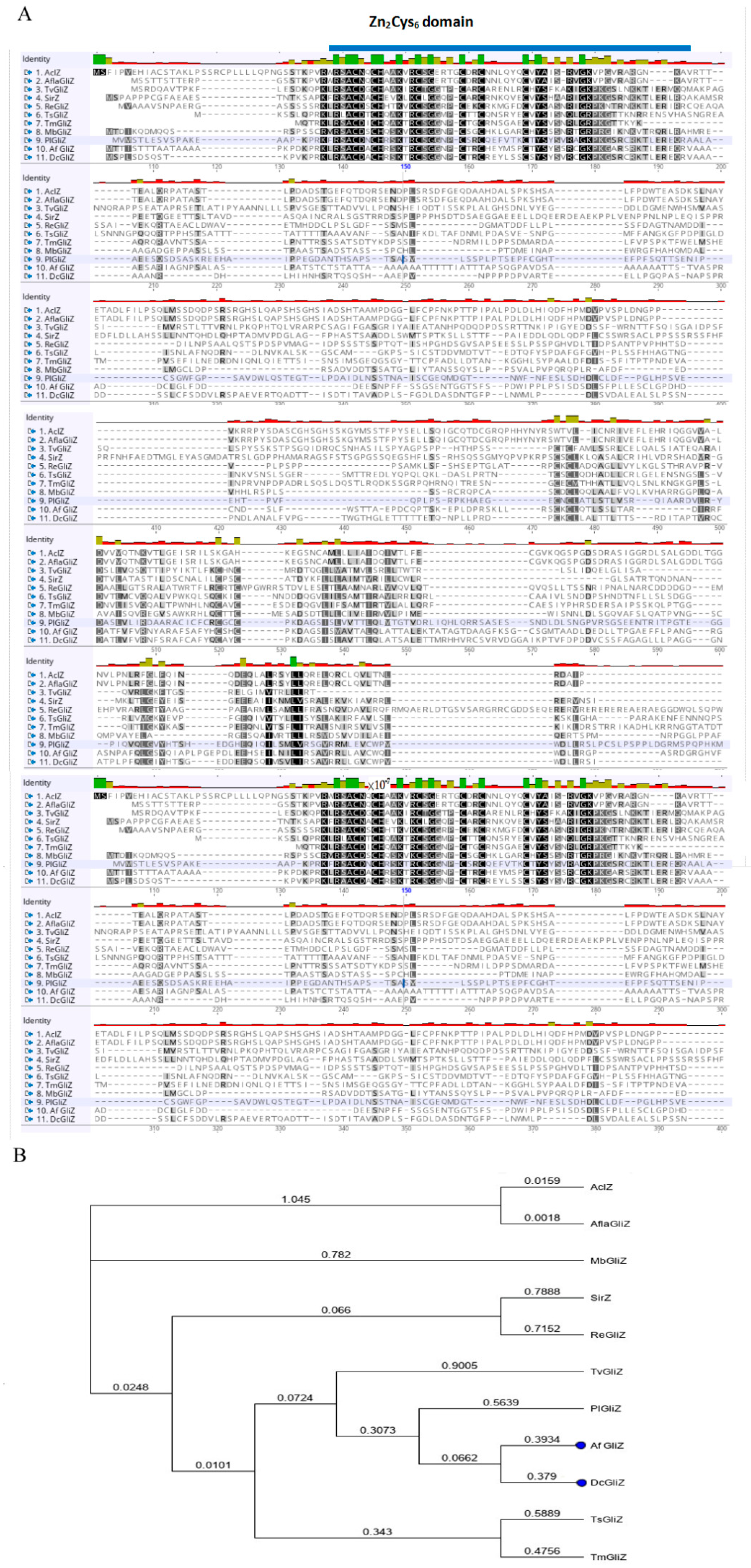
3.2. Conserved Domain of Zn2Cys6 Transcriptional Factor DcGliZ
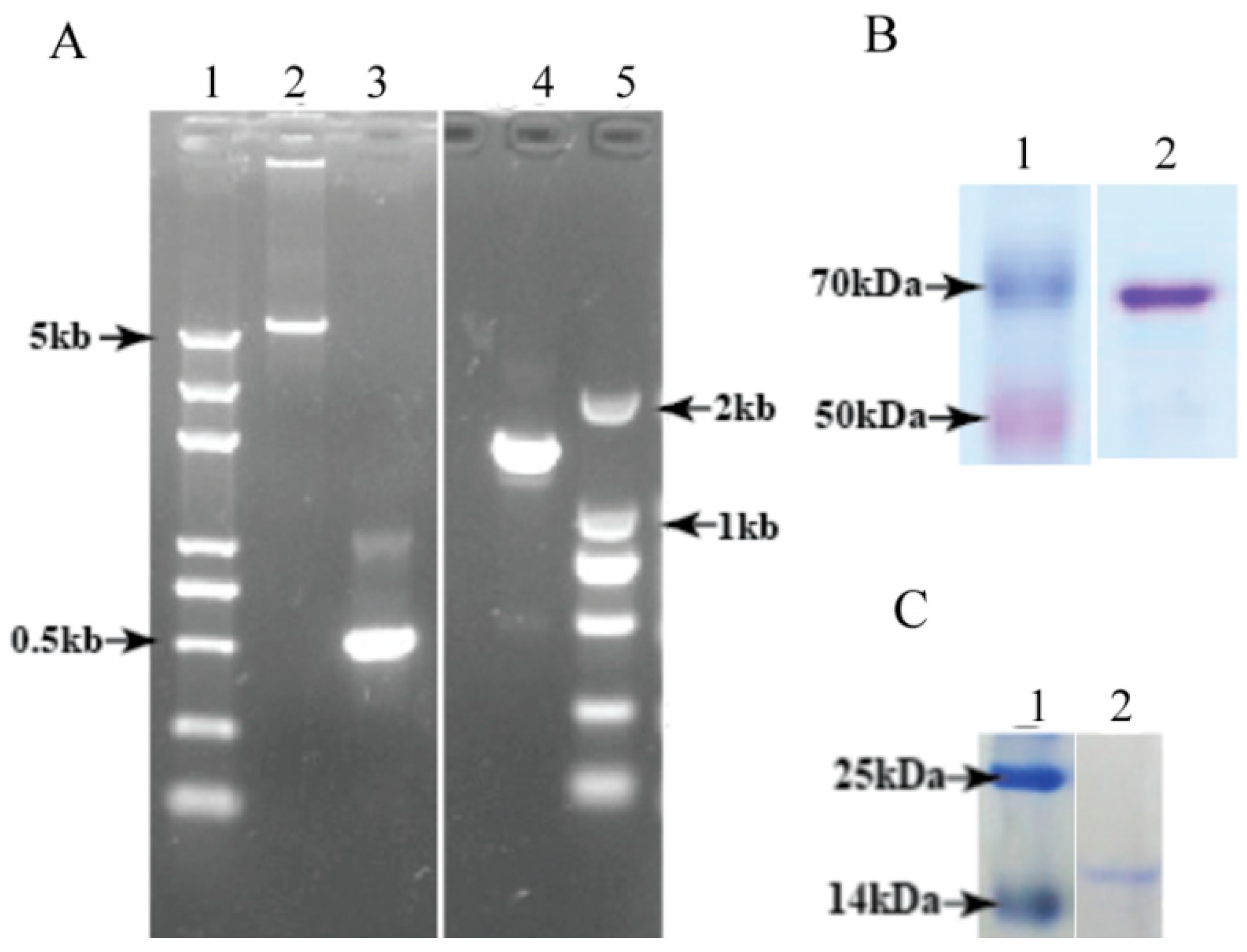
3.3. Heterologous Expression and Purification of Full-Length DcGliZ and DcGliZ Core Proteins from E. coli
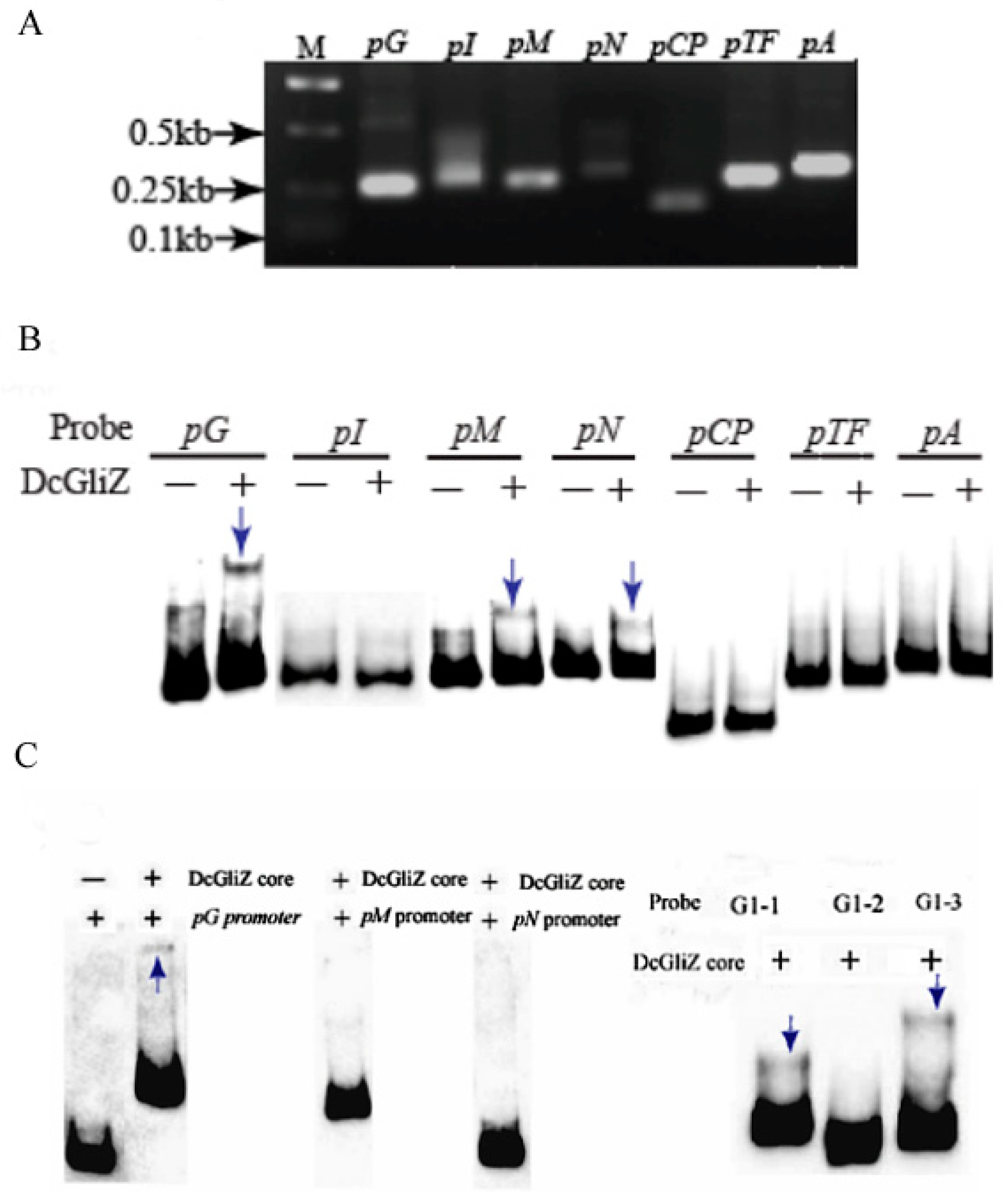
3.4. Electrophoretic Mobility Shift Assay between Full-Length DcGliZ or DcGliZ Core Protein and Functional Gene Promoters of the Gli Cluster
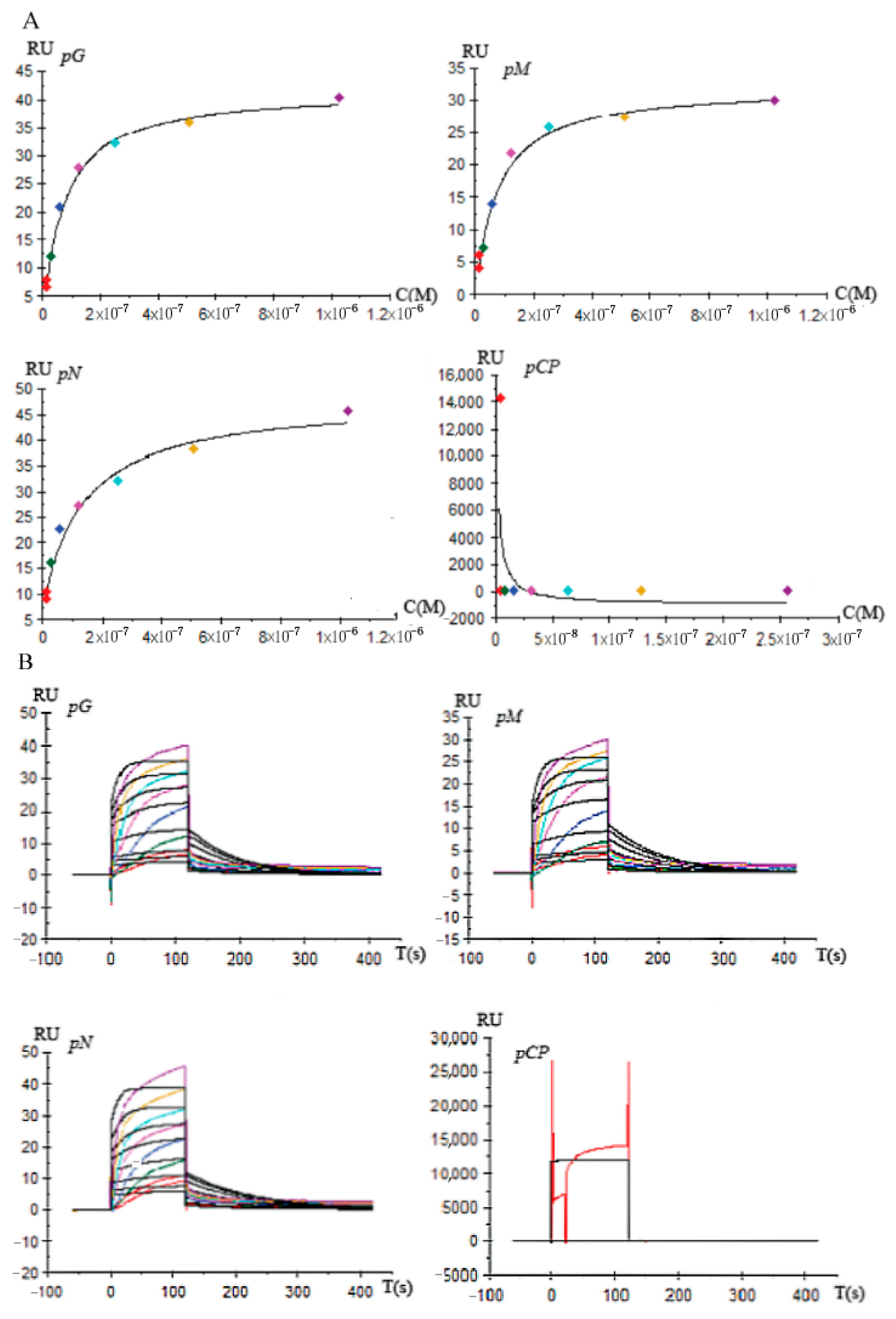
3.5. Affinity and Kinetics Analysis between DcGliZ and Gli Cluster Gene Promoters
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Imhoff, J.F. Natural Products from Marine Fungi—Still an Underrepresented Resource. Mar. Drugs 2016, 14, 19. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Wei, X.; Tu, Z.; Zhou, X.; Wang, J.; He, W.; Tian, X.; Liao, L.; Yang, B.; Chen, B.; et al. Three new indolyl diketopiperazine metabolites from the antarctic soil-derived fungus Penicillium sp. SCSIO 05705. RSC Adv. 2015, 5, 68736–68742. [Google Scholar]
- Tian, Z.; Chu, Y.; Wang, H.; Zhong, L.; Deng, M.; Li, W. Biological activity and interaction mechanism of the diketopiperazine derivatives as tubulin polymerization inhibitors. RSC Adv. 2018, 8, 1055–1064. [Google Scholar] [CrossRef]
- Gerwick, W.H.; Moore, B.S. Lessons from the Past and Charting the Future of Marine Natural Products Drug Discovery and Chemical Biology. Chem. Boil. 2012, 19, 1631. [Google Scholar] [CrossRef]
- Mayer, A.M.; Glaser, K.B.; Cuevas, C.; Jacobs, R.S.; Kem, W.; Little, R.D.; McIntosh, J.M.; Newman, D.J.; Potts, B.C.; Shuster, D.E. The odyssey of marine pharmaceuticals: A current pipeline perspective. Trends Pharmacol. Sci. 2010, 31, 255–265. [Google Scholar] [CrossRef]
- Zhong, W.M.; Wang, J.F.; Shi, X.F.; Wei, X.Y.; Chen, Y.C.; Zeng, Q.; Xiang, Y.; Chen, X.Y.; Tian, X.P.; Xiao, Z.H.; et al. Eurotiumins A-E, five new alkaloids from the marine-derived fungus Eurotium sp SCSIO F452. Mar. Drugs 2018, 16, 136. [Google Scholar] [CrossRef]
- Scharf, D.H.; Heinekamp, T.; Remme, N.; Hortschansky, P.; Brakhage, A.A.; Hertweck, C. Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 2012, 93, 467–472. [Google Scholar] [CrossRef]
- Gardiner, D.M.; Howlett, B.J. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster ofAspergillus fumigatus. FEMS Microbiol. Lett. 2005, 248, 241–248. [Google Scholar] [CrossRef]
- Dolan, S.K.; O’Keeffe, G.; Jones, G.W.; Doyle, S. Resistance is not futile: Gliotoxin biosynthesis, functionality and utility. Trends Microbiol. 2015, 23, 419–428. [Google Scholar] [CrossRef]
- Balibar, C.J.; Walsh, C.T. GliP, a Multimodular Nonribosomal Peptide Synthetase inAspergillus fumigatus, Makes the Diketopiperazine Scaffold of Gliotoxin†. Biochemistry 2006, 45, 15029–15038. [Google Scholar] [CrossRef]
- Chang, S.-L.; Chiang, Y.-M.; Yeh, H.-H.; Wu, T.-K.; Wang, C.C.C. Reconstitution of the early steps of gliotoxin biosynthesis in Aspergillus nidulans reveals the role of the monooxygenase GliC. Bioorganic Med. Chem. Lett. 2013, 23, 2155–2157. [Google Scholar] [CrossRef]
- Davis, C.; Carberry, S.; Schrettl, M.; Singh, I.; Stephens, J.C.; Barry, S.M.; Kavanagh, K.; Challis, G.L.; Brougham, D.; Doyle, S. The Role of Glutathione S-Transferase GliG in Gliotoxin Biosynthesis in Aspergillus fumigatus. Chem. Boil. 2011, 18, 542–552. [Google Scholar] [CrossRef]
- Scharf, D.H.; Remme, N.; Habel, A.; Chankhamjon, P.; Scherlach, K.; Heinekamp, T.; Hortschansky, P.; Brakhage, A.A.; Hertweck, C. A Dedicated GlutathioneS-Transferase Mediates Carbon–Sulfur Bond Formation in Gliotoxin Biosynthesis. J. Am. Chem. Soc. 2011, 133, 12322–12325. [Google Scholar] [CrossRef]
- Gallagher, L.; Owens, R.A.; Dolan, S.K.; O’Keeffe, G.; Schrettl, M.; Kavanagh, K.; Jones, G.W.; Doyle, S. The Aspergillus fumigatus Protein GliK Protects against Oxidative Stress and Is Essential for Gliotoxin Biosynthesis. Eukaryot. Cell 2012, 11, 1226–1238. [Google Scholar] [CrossRef]
- Scharf, D.H.; Chankhamjon, P.; Scherlach, K.; Heinekamp, T.; Willing, K.; Brakhage, A.A.; Hertweck, C. Epidithiodiketopiperazine Biosynthesis: A Four-Enzyme Cascade Converts Glutathione Conjugates into Transannular Disulfide Bridges. Angew. Chem. 2013, 125, 11298–11301. [Google Scholar] [CrossRef]
- Scharf, D.H.; Chankhamjon, P.; Scherlach, K.; Heinekamp, T.; Roth, M.; Brakhage, A.A.; Hertweck, C. Epidithiol Formation by an Unprecedented Twin Carbon-Sulfur Lyase in the Gliotoxin Pathway. Angew. Chem. 2012, 124, 10211–10215. [Google Scholar] [CrossRef]
- Scharf, D.H.; Habel, A.; Heinekamp, T.; Brakhage, A.A.; Hertweck, C. Opposed Effects of Enzymatic Gliotoxin N- and S-Methylations. J. Am. Chem. Soc. 2014, 136, 11674–11679. [Google Scholar] [CrossRef]
- Scharf, D.H.; Remme, N.; Heinekamp, T.; Hortschansky, P.; Brakhage, A.A.; Hertweck, C. Transannular Disulfide Formation in Gliotoxin Biosynthesis and Its Role in Self-Resistance of the Human PathogenAspergillus fumigatus. J. Am. Chem. Soc. 2010, 132, 10136–10141. [Google Scholar] [CrossRef]
- Wang, D.-N.; Toyotome, T.; Muraosa, Y.; Watanabe, A.; Wuren, T.; Bunsupa, S.; Aoyagi, K.; Yamazaki, M.; Takino, M.; Kamei, K. GliA in Aspergillus fumigatus is required for its tolerance to gliotoxin and affects the amount of extracellular and intracellular gliotoxin. Med Mycol. 2014, 52, 506–518. [Google Scholar] [CrossRef]
- Dolan, S.K.; Owens, R.A.; O’Keeffe, G.; Hammel, S.; Fitzpatrick, D.A.; Jones, G.W.; Doyle, S. Regulation of Nonribosomal Peptide Synthesis: Bis-Thiomethylation Attenuates Gliotoxin Biosynthesis in Aspergillus fumigatus. Chem. Boil. 2014, 21, 999–1012. [Google Scholar] [CrossRef]
- Bok, J.W.; Chung, D.; Balajee, S.A.; Marr, K.A.; Andes, D.; Nielsen, K.F.; Frisvad, J.C.; Kirby, K.A.; Keller, N.P. GliZ, a Transcriptional Regulator of Gliotoxin Biosynthesis, Contributes to Aspergillus fumigatus Virulence. Infect. Immun. 2006, 74, 6761–6768. [Google Scholar] [CrossRef]
- Schoberle, T.J.; Nguyen-Coleman, C.K.; Herold, J.; Yang, A.; Weirauch, M.; Hughes, T.R.; McMurray, J.S.; May, G.S. A Novel C2H2 Transcription Factor that Regulates gliA Expression Interdependently with GliZ in Aspergillus fumigatus. PLoS Genet. 2014, 10, e1004336. [Google Scholar] [CrossRef]
- Fox, E.M.; Gardiner, D.M.; Keller, N.P.; Howlett, B.J. A Zn(II)(2)Cys(6) DNA binding protein regulates the sirodesmin PL biosynthetic gene cluster in Leptosphaeria maculans. Fungal Genet. Biol. 2008, 45, 671–682. [Google Scholar] [CrossRef]
- Fan, Z.; Sun, Z.-H.; Liu, Z.; Chen, Y.-C.; Liu, H.-X.; Li, H.-H.; Zhang, W.-M. Dichotocejpins A–C: New Diketopiperazines from a Deep-Sea-Derived Fungus Dichotomomyces cejpii FS110. Mar. Drugs 2016, 14, 164. [Google Scholar] [CrossRef]
- Sun, Z.-H.; Gu, J.; Ye, W.; Wen, L.-X.; Lin, Q.-B.; Li, S.-N.; Chen, Y.-C.; Li, H.-H.; Zhang, W.-M. Geospallins A–C: New Thiodiketopiperazines with Inhibitory Activity against Angiotensin-Converting Enzyme from a Deep-Sea-Derived Fungus Geosmithia pallida FS140. Mar. Drugs 2018, 16, 464. [Google Scholar] [CrossRef]
- Liu, Z.; Fan, Z.; Sun, Z.; Liu, H.; Zhang, W. Dechdigliotoxins A–C, Three Novel Disulfide-Bridged Gliotoxin Dimers from Deep-Sea Sediment Derived Fungus Dichotomomyces cejpii. Mar. Drugs 2019, 17, 596. [Google Scholar] [CrossRef]
- Ye, W.; Zhang, W.; Liu, T.; Huang, Z.; Zhu, M.; Chen, Y.; Li, H.; Li, S. De Novo Transcriptome Sequencing of the Deep-Sea-Derived Fungus Dichotomomyces cejpii and Analysis of Gliotoxin Biosynthesis Genes. Int. J. Mol. Sci. 2018, 19, 1910. [Google Scholar] [CrossRef]
- Luo, R.B.; Liu, B.H.; Xie, Y.L.; Li, Z.Y.; Huang, W.H.; Yuan, J.Y.; He, G.Z.; Chen, Y.X.; Pan, Q.; Liu, Y.J.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18. [Google Scholar] [CrossRef]
- Lomsadze, A.; Burns, P.D.; Borodovsky, M. Integration of mapped RNA-Seq reads into automatic training of eukaryotic gene finding algorithm. Nucleic Acids Res. 2014, 42, e119. [Google Scholar] [CrossRef]
- Holt, C.; Yandell, M. MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinform. 2011, 12, 491. [Google Scholar] [CrossRef]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef]
- Chankhamjon, P.; Scherlach, K.; Urbansky, B.; Lackner, G.; Kalb, D.; Dahse, H.-M.; Hoffmeister, D.; Hertweck, C.; Boettger-Schmidt, D. Biosynthesis of the Halogenated Mycotoxin Aspirochlorine in Koji Mold Involves a Cryptic Amino Acid Conversion. Angew. Chem. 2014, 126, 13627–13631. [Google Scholar] [CrossRef]
- Patron, N.J.; Waller, R.F.; Cozijnsen, A.J.; Straney, D.C.; Gardiner, D.M.; Nierman, W.C.; Howlett, B.J. Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes. BMC Evol. Boil. 2007, 7, 174. [Google Scholar] [CrossRef]
- Vargas, W.A.; Mukherjee, P.K.; Laughlin, D.; Wiest, A.; Moran-Diez, M.E.; Kenerley, C.M. Role of gliotoxin in the symbiotic and pathogenic interactions of Trichoderma virens. Microbiology 2014, 160, 2319–2330. [Google Scholar] [CrossRef]
- Quan, J.; Tian, J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 2011, 6, 242–251. [Google Scholar] [CrossRef]
- Behr, S.; Heermann, R.; Jung, K. Insights into the DNA-binding mechanism of a LytTR-type transcription regulator. Biosci. Rep. 2016, 36, e00326. [Google Scholar] [CrossRef]
- Wang, S.; Dong, Y.; Liang, X. Development of a SPR aptasensor containing oriented aptamer for direct capture and detection of tetracycline in multiple honey samples. Biosens. Bioelectron. 2018, 109, 1–7. [Google Scholar] [CrossRef]
- Nissink, J.W.M.; Bista, M.; Breed, J.; Carter, N.; Embrey, K.; Read, J.; Winter-Holt, J.J. MTH1 Substrate Recognition—An Example of Specific Promiscuity. PLoS ONE 2016, 11, 0151154. [Google Scholar] [CrossRef]
- Chen, J.; Wang, C.; Lan, W.; Huang, C.; Lin, M.; Wang, Z.; Liang, W.; Iwamoto, A.; Yang, X.; Liu, H. Gliotoxin Inhibits Proliferation and Induces Apoptosis in Colorectal Cancer Cells. Mar. Drugs 2015, 13, 6259–6273. [Google Scholar] [CrossRef]
- Van-Tinh, N.; Lee, J.S.; Qian, Z.J.; Li, Y.X.; Kim, K.N.; Heo, S.J.; Jeon, Y.J.; Park, W.S.; Choi, I.W.; Je, J.Y.; et al. Gliotoxin isolated from marine fungus Aspergillus sp. induces apoptosis of human cervical cancer and chondrosarcoma cells. Mar. Drugs 2014, 12, 69–87. [Google Scholar]
- Hubmann, R.; Hilgarth, M.; Schnabl, S.; Ponath, E.; Reiter, M.; Demirtas, D.; Sieghart, W.; Valent, P.; Zielinski, C.; Jaeger, U.; et al. Gliotoxin is a potent NOTCH2 transactivation inhibitor and efficiently induces apoptosis in chronic lymphocytic leukaemia (CLL) cells. Br. J. Haematol. 2013, 160, 618–629. [Google Scholar] [CrossRef] [PubMed]
- Geissler, A.; Haun, F.; O Frank, D.; Wieland, K.; Simon, M.M.; Idzko, M.; Davis, R.J.; Maurer, U.; Borner, C. Apoptosis induced by the fungal pathogen gliotoxin requires a triple phosphorylation of Bim by JNK. Cell Death Differ. 2013, 20, 1317–1329. [Google Scholar] [CrossRef]
- Arias, M.; Santiago, L.; Vidal-García, M.; Redrado, S.; Lanuza, P.; Comas, L.; Domingo, M.P.; Rezusta, A.; Gálvez, E.M. Preparations for Invasion: Modulation of Host Lung Immunity During Pulmonary Aspergillosis by Gliotoxin and Other Fungal Secondary Metabolites. Front. Immunol. 2018, 9, 2549. [Google Scholar] [CrossRef]
- Todd, R.B.; Andrianopoulos, A. Evolution of a Fungal Regulatory Gene Family: The Zn(II)2Cys6 Binuclear Cluster DNA Binding Motif. Fungal Genet. Boil. 1997, 21, 388–405. [Google Scholar] [CrossRef]
- Liang, S.D.; Marmorstein, R.; Harrison, S.C.; Ptashine, M. DNA sequence preferences of GAL4 and PPR1: How a subset of Zn2 Cys6 binuclear cluster proteins recognizes DNA. Mol. Cell. Boil. 1996, 16, 3773–3780. [Google Scholar] [CrossRef][Green Version]
- Ehrlich, K.; Montalbano, B.; Cary, J. Binding of the C6-zinc cluster protein, AFLR, to the promoters of aflatoxin pathway biosynthesis genes in Aspergillus parasiticus. Gene 1999, 230, 249–257. [Google Scholar] [CrossRef]
- Huang, Z.L.; Zhang, W.M.; Ye, W.; Li, S.N.; Li, H.H.; Zhu, M.Z. Cloning and identification of gene promoter for gliotoxin giosynthesis from deep-sea-derived fungus Dichotomomyces cejpii. Biotechnol. Bull. 2018, 309, 150–156. [Google Scholar]
- Homola, J. Surface Plasmon Resonance Sensors for Detection of Chemical and Biological Species. Chem. Rev. 2008, 108, 462–493. [Google Scholar] [CrossRef]
- Meza-Menchaca, T.; Ramos-Ligonio, A.; López-Monteon, A.; Limón, A.V.; Kaluzhskiy, L.A.; Shkel, T.V.; Strushkevich, N.V.; Jiménez-García, L.F.; Moreno, L.T.A.; Gallegos-García, V.; et al. Insights into Ergosterol Peroxide’s Trypanocidal Activity. Biomolecules 2019, 9, 484. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.-L.; Ye, W.; Zhu, M.-Z.; Kong, Y.-L.; Li, S.-N.; Liu, S.; Zhang, W.-M. Interaction of a Novel Zn2Cys6 Transcription Factor DcGliZ with Promoters in the Gliotoxin Biosynthetic Gene Cluster of the Deep-Sea-Derived Fungus Dichotomomyces cejpii. Biomolecules 2020, 10, 56. https://doi.org/10.3390/biom10010056
Huang Z-L, Ye W, Zhu M-Z, Kong Y-L, Li S-N, Liu S, Zhang W-M. Interaction of a Novel Zn2Cys6 Transcription Factor DcGliZ with Promoters in the Gliotoxin Biosynthetic Gene Cluster of the Deep-Sea-Derived Fungus Dichotomomyces cejpii. Biomolecules. 2020; 10(1):56. https://doi.org/10.3390/biom10010056
Chicago/Turabian StyleHuang, Zi-Lei, Wei Ye, Mu-Zi Zhu, Ya-Li Kong, Sai-Ni Li, Shan Liu, and Wei-Min Zhang. 2020. "Interaction of a Novel Zn2Cys6 Transcription Factor DcGliZ with Promoters in the Gliotoxin Biosynthetic Gene Cluster of the Deep-Sea-Derived Fungus Dichotomomyces cejpii" Biomolecules 10, no. 1: 56. https://doi.org/10.3390/biom10010056
APA StyleHuang, Z.-L., Ye, W., Zhu, M.-Z., Kong, Y.-L., Li, S.-N., Liu, S., & Zhang, W.-M. (2020). Interaction of a Novel Zn2Cys6 Transcription Factor DcGliZ with Promoters in the Gliotoxin Biosynthetic Gene Cluster of the Deep-Sea-Derived Fungus Dichotomomyces cejpii. Biomolecules, 10(1), 56. https://doi.org/10.3390/biom10010056




