HLA-G Polymorphisms Are Associated with Non-Segmental Vitiligo among Brazilians
Abstract
:1. Introduction
2. Material and Methods
2.1. Population Sample
2.2. Extended HLA-G Analysis
2.3. Bioinformatics Analysis
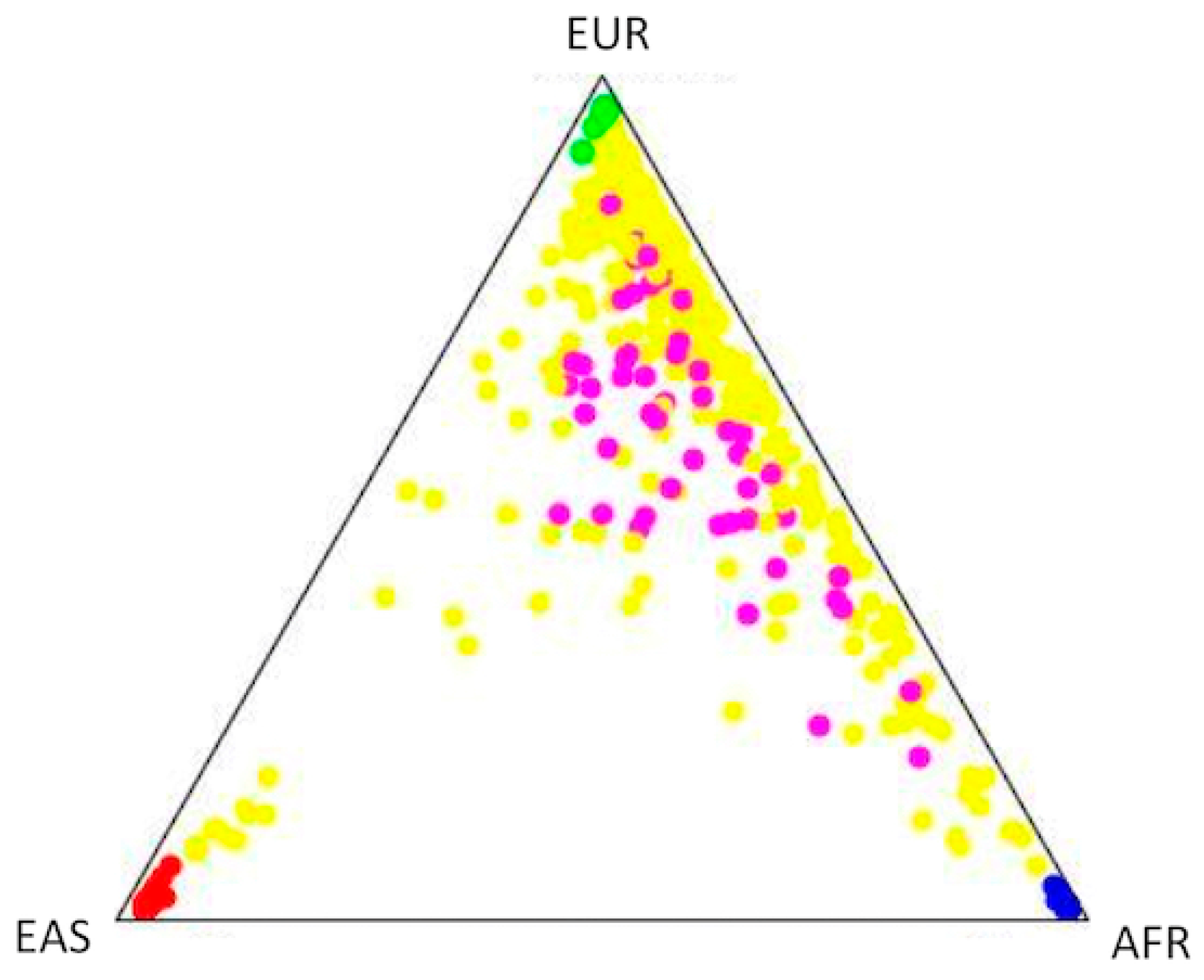
2.4. Population Structure and Association Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Alikhan, A.; Felsten, L.M.; Daly, M.; Petronic-Rosic, V. Vitiligo: A comprehensive overview Part I. Introduction, epidemiology, quality of life, diagnosis, differential diagnosis, associations, histopathology, etiology, and work-up. J. Am. Acad. Dermatol. 2011, 65, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Harris, J.E. Vitiligo, An Issue of Dermatologic Clinics, 1st ed.; Elsevier: Philadelphia, PA, USA, 2017; Volume 1. [Google Scholar]
- Oyarbide-Valencia, K.; van den Boorn, J.G.; Denman, C.J.; Li, M.; Carlson, J.M.; Hernandez, C.; Nishimura, M.I.; Das, P.K.; Luiten, R.M.; Le Poole, I.C. Therapeutic implications of autoimmune vitiligo T cells. Autoimmun. Rev. 2006, 5, 486–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ongenae, K.; Van Geel, N.; Naeyaert, J.M. Evidence for an autoimmune pathogenesis of vitiligo. Pigment Cell Res. 2003, 16, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Alkhateeb, A.; Fain, P.R.; Thody, A.; Bennett, D.C.; Spritz, R.A. Epidemiology of vitiligo and associated autoimmune diseases in Caucasian probands and their families. Pigment Cell Res. 2003, 16, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, Y.; Nie, H.; Zhang, X.; Shao, Q.; Hou, X.; Xu, W.; Hong, W.; Xu, A. The changes of gene expression profiling between segmental vitiligo, generalized vitiligo and healthy individual. J. Dermatol. Sci. 2016, 84, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Zamani, M.; Spaepen, M.; Sghar, S.S.; Huang, C.; Westerhof, W.; Nieuweboer-Krobotova, L.; Cassiman, J.J. Linkage and association of HLA class II genes with vitiligo in a Dutch population. Br. J. Dermatol. 2001, 145, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Misri, R.; Khopkar, U.; Shankarkumar, U.; Ghosh, K. Comparative case control study of clinical features and human leukocyte antigen susceptibility between familial and nonfamilial vitiligo. Indian J. Dermatol. Venereol. Leprol. 2009, 75, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Ramire, L.D.; Marcos, E.V.; Godoy, D.A.; de Souza-Santana, F.C. Association of class I and II HLA alleles and haplotypes with susceptibility to vitiligo: A study of patients with vitiligo from southeast Brazil. Int. J. Dermatol. 2016, 55, e347–e355. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Hayashi, M.; Fain, P.R.; Suzuki, T.; Fukai, K.; Oiso, N.; Tanemura, A.; Holcomb, C.L.; Rastrou, M.; Erlich, H.A.; et al. Major association of vitiligo with HLA-A*02:01 In Japanese. Pigment Cell Melanoma Res. 2015, 28, 360–362. [Google Scholar] [CrossRef] [PubMed]
- Cui, T.; Yi, X.; Guo, S.; Zhou, F.; Liu, L.; Li, C.; Li, K.; Gao, T. Identification of Novel HLA-A*0201-Restricted CTL Epitopes in Chinese Vitiligo Patients. Sci. Rep. 2016, 6, 36360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carlini, F.; Ferreira, V.; Buhler, S.; Tous, A.; Eliaou, J.F.; Rene, C.; Chiaroni, J.; Picard, C.; Di Cristofaro, J. Association of HLA-A and Non-Classical HLA Class I Alleles. PLoS ONE 2016, 11, e0163570. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Jin, Y.; Yorgov, D.; Santorico, S.A.; Hagman, J.; Ferrara, T.M.; Jones, K.L.; Cavalli, G.; Dinarello, C.A.; Spritz, R.A. Autoimmune vitiligo is associated with gain-of-function by a transcriptional regulator that elevates expression of HLA-A*02:01 in vivo. Proc. Natl. Acad. Sci. USA 2016, 113, 1357–1362. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Sharma, P.; Kar, H.K.; Sharma, V.K.; Tembhre, M.K.; Gupta, S.; Laddha, N.C.; Dwivedi, M.; Begum, R.; Indian Genome Variation, C.; et al. HLA alleles and amino-acid signatures of the peptide-binding pockets of HLA molecules in vitiligo. J. Investig. Dermatol. 2012, 132, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Hayashi, M.; Jin, Y.; Yorgov, D.; Santorico, S.A.; Holcomb, C.; Rastrou, M.; Erlich, H.; Tengesdal, I.W.; Dagna, L.; et al. MHC class II super-enhancer increases surface expression of HLA-DR and HLA-DQ and affects cytokine production in autoimmune vitiligo. Proc. Natl. Acad. Sci. USA 2016, 113, 1363–1368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, A.; Hiby, S.E.; Gardner, L.; Joseph, S.; Bowen, J.M.; Verma, S.; Burrows, T.D.; Loke, Y.W. Recognition of trophoblast HLA class I molecules by decidual NK cell receptors—A review. Placenta 2000, 21 (Suppl. A), S81–S85. [Google Scholar] [CrossRef]
- Donadi, E.A.; Castelli, E.C.; Arnaiz-Villena, A.; Roger, M.; Rey, D.; Moreau, P. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell. Mol. Life Sci. 2011, 68, 369–395. [Google Scholar] [CrossRef] [PubMed]
- Jalel, A.; Ridha, A.; Laurent, D.; Philippe, M.; Hamdaoui, M.H. Impact of HLA-G in the outcome of vitiligo in Tunisian patients. Indian J. Dermatol. 2010, 55, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Wu, J.; Zhang, X.; Wen, L.; Sun, J.; Cheng, Y.; Tang, X.; Liang, B.; Chen, G.; Zhou, F.; et al. Fine-mapping analysis of the MHC region for vitiligo based on a new Han-MHC reference panel. Gene 2018, 648, 76–81. [Google Scholar] [CrossRef]
- Hassab El Naby, H.M.; Alnaggar, M.R.; Abdelhamid, M.F.; Alsaid, K.; Al Shawadfy, E.M.; Elsaie, M.L. Study of human leukocyte antigen-cw in Egyptian patients with vitiligo. J. Drugs Dermatol. JDD 2015, 14, 359–364. [Google Scholar]
- Bouayad, A.; Benzekri, L.; Hamada, S.; Brick, C.; Hassam, B.; Essakalli, M. Association of HLA alleles and haplotypes with vitiligo in Moroccan patients: A case-control study. Arch. Dermatol. Res. 2013, 305, 925–932. [Google Scholar] [CrossRef]
- Kim, S.K.; Hong, M.S.; Shin, M.K.; Uhm, Y.K.; Chung, J.H.; Lee, M.H. Promoter polymorphisms of the HLA-G gene, but not the HLA-E and HLA-F genes, is associated with non-segmental vitiligo patients in the Korean population. Arch. Dermatol. Res. 2011, 303, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Jeong, K.H.; Kim, S.K.; Kang, B.K.; Chung, J.H.; Shin, M.K.; Lee, M.H. Association between an HLA-G 14 bp insertion/deletion polymorphism and non-segmental vitiligo in the Korean population. Arch. Dermatol. Res. 2014, 306, 577–582. [Google Scholar] [CrossRef]
- Tan, Z.; Randall, G.; Fan, J.; Camoretti-Mercado, B.; Brockman-Schneider, R.; Pan, L.; Solway, J.; Gern, J.E.; Lemanske, R.F.; Nicolae, D.; et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 2007, 81, 829–834. [Google Scholar] [CrossRef]
- Yie, S.M.; Li, L.H.; Xiao, R.; Librach, C.L. A single base-pair mutation in the 3’-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol. Hum. Reprod. 2008, 14, 649–653. [Google Scholar] [CrossRef] [PubMed]
- Castelli, E.C.; Mendes-Junior, C.T.; Deghaide, N.H.; de Albuquerque, R.S.; Muniz, Y.C.; Simoes, R.T.; Carosella, E.D.; Moreau, P.; Donadi, E.A. The genetic structure of 3’untranslated region of the HLA-G gene: Polymorphisms and haplotypes. Genes Immun. 2010, 11, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Castelli, E.C.; Mendes-Junior, C.T.; Veiga-Castelli, L.C.; Roger, M.; Moreau, P.; Donadi, E.A. A Comprehensive Study of Polymorphic Sites along the HLA-G Gene: Implication for Gene Regulation and Evolution. Mol. Biol. Evol. 2011, 28, 3069–3086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martelli-Palomino, G.; Pancotto, J.A.; Muniz, Y.C.; Mendes-Junior, C.T.; Castelli, E.C.; Massaro, J.D.; Krawice-Radanne, I.; Poras, I.; Rebmann, V.; Carosella, E.D.; et al. Polymorphic Sites at the 3’ Untranslated Region of the HLA-G Gene Are Associated with Differential hla-g Soluble Levels in the Brazilian and French Population. PLoS ONE 2013, 8, e71742. [Google Scholar] [CrossRef]
- Yaghi, L.; Poras, I.; Simoes, R.T.; Donadi, E.A.; Tost, J.; Daunay, A.; de Almeida, B.S.; Carosella, E.D.; Moreau, P. Hypoxia inducible factor-1 mediates the expression of the immune checkpoint HLA-G in glioma cells through hypoxia response element located in exon 2. Oncotarget 2016, 7, 63690–63707. [Google Scholar] [CrossRef]
- Castelli, E.C.; Gerasimou, P.; Paz, M.A.; Ramalho, J.; Porto, I.O.P.; Lima, T.H.A.; Souza, A.S.; Veiga-Castelli, L.C.; Collares, C.V.A.; Donadi, E.A.; et al. HLA-G variability and haplotypes detected by massively parallel sequencing procedures in the geographicaly distinct population samples of Brazil and Cyprus. Mol. Immunol. 2017, 83, 115–126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poras, I.; Yaghi, L.; Martelli-Palomino, G.; Mendes-Junior, C.T.; Muniz, Y.C.; Cagnin, N.F.; Sgorla de Almeida, B.; Castelli, E.C.; Carosella, E.D.; Donadi, E.A.; et al. Haplotypes of the HLA-G 3’ Untranslated Region Respond to Endogenous Factors of HLA-G+ and HLA-G- Cell Lines Differentially. PLoS ONE 2017, 12, e0169032. [Google Scholar] [CrossRef]
- Phillips, C.; Salas, A.; Sanchez, J.J.; Fondevila, M.; Gomez-Tato, A.; Alvarez-Dios, J.; Calaza, M.; de Cal, M.C.; Ballard, D.; Lareu, M.V.; et al. Inferring ancestral origin using a single multiplex assay of ancestry-informative marker SNPs. Forensic Sci. Int. Genet. 2007, 1, 273–280. [Google Scholar] [CrossRef]
- Oliveira, M.L.G.; Veiga-Castelli, L.C.; Marcorin, L.; Debortoli, G.; Pereira, A.L.E.; Fracasso, N.C.A.; Silva, G.D.V.; Souza, A.S.; Massaro, J.D.; Simoes, A.L.; et al. Extended HLA-G genetic diversity and ancestry composition in a Brazilian admixed population sample: Implications for HLA-G transcriptional control and for case-control association studies. Hum. Immunol. 2018, 79, 790–799. [Google Scholar] [CrossRef] [PubMed]
- Castelli, E.C.; Paz, M.A.; Souza, A.S.; Ramalho, J.; Mendes-Junior, C.T. Hla-mapper: An application to optimize the mapping of HLA sequences produced by massively parallel sequencing procedures. Hum. Immunol. 2018, 79, 678–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef] [PubMed]
- Santos, K.E.; Lima, T.H.; Felicio, L.P.; Massaro, J.D.; Palomino, G.M.; Silva, A.C.; Oliveira, S.F.; Sabbagh, A.; Garcia, A.; Moreau, P.; et al. Insights on the HLA-G evolutionary history provided by a nearby Alu insertion. Mol. Biol. Evol. 2013, 30, 2423–2434. [Google Scholar] [CrossRef] [PubMed]
- Sonon, P.; Sadissou, I.; Tokplonou, L.; M’Po K, K.G.; Glitho, S.S.C.; Agniwo, P.; Ibikounle, M.; Massaro, J.D.; Massougbodji, A.; Moreau, P.; et al. HLA-G, -E and -F regulatory and coding region variability and haplotypes in the Beninese Toffin population sample. Mol. Immunol. 2018, 104, 108–127. [Google Scholar] [CrossRef]
- Ibrahim, E.C.; Aractingi, S.; Allory, Y.; Borrini, F.; Dupuy, A.; Duvillard, P.; Carosella, E.D.; Avril, M.F.; Paul, P. Analysis of HLA antigen expression in benign and malignant melanocytic lesions reveals that upregulation of HLA-G expression correlates with malignant transformation, high inflammatory infiltration and HLA-A1 genotype. Int. J. Cancer 2004, 108, 243–250. [Google Scholar] [CrossRef]
- Cardili, R.N.; Alves, T.G.; Freitas, J.C.; Soares, C.P.; Mendes-Junior, C.T.; Soares, E.G.; Donadi, E.A.; Silva-Souza, C. Expression of human leucocyte antigen-G primarily targets affected skin of patients with psoriasis. Br. J. Dermatol. 2010, 163, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Yari, F.; Zavaran Hosseini, A.; Nemat Gorgani, M.; Khorramizadeh, M.R.; Mansouri, P.; Kazemnejad, A. Expression of HLA-G in the skin of patients with pemphigus vulgaris. Iran. J. Allergy Asthma Immunol. 2008, 7, 7–12. [Google Scholar] [PubMed]
- Verloes, A.; Spits, C.; Vercammen, M.; Geens, M.; LeMaoult, J.; Sermon, K.; Coucke, W.; Van de Velde, H. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem. Cell Res. 2017, 19, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Schwich, E.; Rebmann, V.; Michita, R.T.; Rohn, H.; Voncken, J.W.; Horn, P.A.; Kimmig, R.; Kasimir-Bauer, S.; Buderath, P. HLA-G 3’ untranslated region variants +3187G/G, +3196G/G and +3035T define diametrical clinical status and disease outcome in epithelial ovarian cancer. Sci. Rep. 2019, 9, 5407. [Google Scholar] [CrossRef] [PubMed]
- Porto, I.O.; Mendes-Junior, C.T.; Felicio, L.P.; Georg, R.C.; Moreau, P.; Donadi, E.A.; Chies, J.A.; Castelli, E.C. MicroRNAs targeting the immunomodulatory HLA-G gene: A new survey searching for microRNAs with potential to regulate HLA-G. Mol. Immunol. 2015, 65, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Ober, C.; Aldrich, C.L.; Chervoneva, I.; Billstrand, C.; Rahimov, F.; Gray, H.L.; Hyslop, T. Variation in the HLA-G promoter region influences miscarriage rates. Am. J. Hum. Genet. 2003, 72, 1425–1435. [Google Scholar] [CrossRef] [PubMed]
- Ober, C.; Billstrand, C.; Kuldanek, S.; Tan, Z. The miscarriage-associated HLA-G -725G allele influences transcription rates in JEG-3 cells. Hum. Reprod. 2006, 21, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Grundberg, E.; Small, K.S.; Hedman, A.K.; Nica, A.C.; Buil, A.; Keildson, S.; Bell, J.T.; Yang, T.P.; Meduri, E.; Barrett, A.; et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nat. Genet. 2012, 44, 1084–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajagopalan, S.; Long, E.O. KIR2DL4 (CD158d): An activation receptor for HLA-G. Front. Immunol. 2012, 3, 258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birol, A.; Kisa, U.; Kurtipek, G.S.; Kara, F.; Kocak, M.; Erkek, E.; Caglayan, O. Increased tumor necrosis factor alpha (TNF-alpha) and interleukin 1 alpha (IL1-alpha) levels in the lesional skin of patients with nonsegmental vitiligo. Int. J. Dermatol. 2006, 45, 992–993. [Google Scholar] [CrossRef]
- Tu, C.X.; Gu, J.S.; Lin, X.R. Increased interleukin-6 and granulocyte-macrophage colony stimulating factor levels in the sera of patients with non-segmental vitiligo. J. Dermatol. Sci. 2003, 31, 73–78. [Google Scholar] [CrossRef]
- Jin, Y.; Andersen, G.; Yorgov, D.; Ferrara, T.M.; Ben, S.; Brownson, K.M.; Holland, P.J.; Birlea, S.A.; Siebert, J.; Hartmann, A.; et al. Genome-wide association studies of autoimmune vitiligo identify 23 new risk loci and highlight key pathways and regulatory variants. Nat. Genet. 2016, 48, 1418–1424. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Birlea, S.A.; Fain, P.R.; Ferrara, T.M.; Ben, S.; Riccardi, S.L.; Cole, J.B.; Gowan, K.; Holland, P.J.; Bennett, D.C.; et al. Genome-wide association analyses identify 13 new susceptibility loci for generalized vitiligo. Nat. Genet. 2012, 44, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Birlea, S.A.; Fain, P.R.; Gowan, K.; Riccardi, S.L.; Holland, P.J.; Mailloux, C.M.; Sufit, A.J.; Hutton, S.M.; Amadi-Myers, A.; et al. Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N. Engl. J. Med. 2010, 362, 1686–1697. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Ferrara, T.; Gowan, K.; Holcomb, C.; Rastrou, M.; Erlich, H.A.; Fain, P.R.; Spritz, R.A. Next-generation DNA re-sequencing identifies common variants of TYR and HLA-A that modulate the risk of generalized vitiligo via antigen presentation. J. Investig. Dermatol. 2012, 132, 1730–1733. [Google Scholar] [CrossRef] [PubMed]
| Variant | HLA-G Position | Associated Allele | Model | OR | L95 | U95 | P | Known HLA-G Alleles or Haplotypes |
|---|---|---|---|---|---|---|---|---|
| Before ancestry adjustment | ||||||||
| rs9380142 | 3187 | G | Dominant | 2.096 | 1.145 | 3.838 | 0.0164 | Distal-010101a, Proximal-010101a, G*01:01:01:01, UTR-1 * |
| After ancestry adjustment | ||||||||
| rs9380142 | 3187 | G | Additive | 1.590 | 1.020 | 2.479 | 0.0404 | Distal-010101a, Proximal-010101a, G*01:01:01:01, UTR-1 * |
| rs9380142 | 3187 | G | Dominant | 2.270 | 1.222 | 4.217 | 0.0095 | Distal-010101a, Proximal-010101a, G*01:01:01:01, UTR-1 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veiga-Castelli, L.; de Oliveira, M.L.; Pereira, A.; Debortoli, G.; Marcorin, L.; Fracasso, N.; Silva, G.; Souza, A.; Massaro, J.; Simões, A.L.; et al. HLA-G Polymorphisms Are Associated with Non-Segmental Vitiligo among Brazilians. Biomolecules 2019, 9, 463. https://doi.org/10.3390/biom9090463
Veiga-Castelli L, de Oliveira ML, Pereira A, Debortoli G, Marcorin L, Fracasso N, Silva G, Souza A, Massaro J, Simões AL, et al. HLA-G Polymorphisms Are Associated with Non-Segmental Vitiligo among Brazilians. Biomolecules. 2019; 9(9):463. https://doi.org/10.3390/biom9090463
Chicago/Turabian StyleVeiga-Castelli, Luciana, Maria Luiza de Oliveira, Alison Pereira, Guilherme Debortoli, Letícia Marcorin, Nádia Fracasso, Guilherme Silva, Andreia Souza, Juliana Massaro, Aguinaldo Luiz Simões, and et al. 2019. "HLA-G Polymorphisms Are Associated with Non-Segmental Vitiligo among Brazilians" Biomolecules 9, no. 9: 463. https://doi.org/10.3390/biom9090463
APA StyleVeiga-Castelli, L., de Oliveira, M. L., Pereira, A., Debortoli, G., Marcorin, L., Fracasso, N., Silva, G., Souza, A., Massaro, J., Simões, A. L., Sabbagh, A., Cardili, R., Donadi, E., Castelli, E., & Mendes-Junior, C. (2019). HLA-G Polymorphisms Are Associated with Non-Segmental Vitiligo among Brazilians. Biomolecules, 9(9), 463. https://doi.org/10.3390/biom9090463





