Development of Human Monoclonal Antibody for Claudin-3 Overexpressing Carcinoma Targeting
Abstract
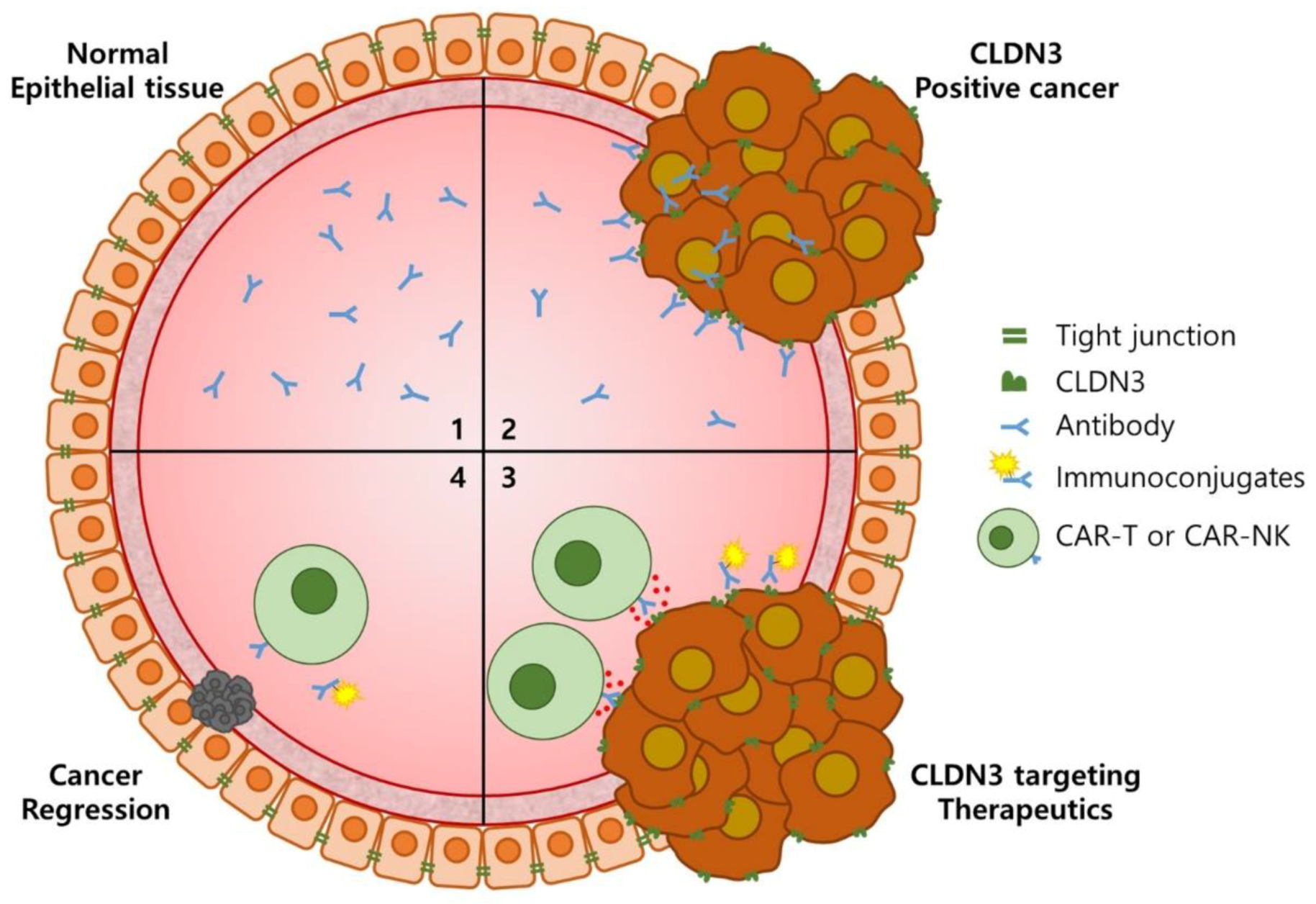
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Establishment of Stable CLDN Transfectants
2.3. Establishment of NK-92MI-CD16a Cell Line
2.4. Production of Human Monoclonal Antibody (mAb)
2.5. Flow Cytometry Analysis
2.6. Western Blotting and Immunoprecipitation
2.7. Immunofluorescence
2.8. Cell-Based Affinity Kinetics
2.9. In Vitro Antibody-Dependent Cellular Cytotoxicity (ADCC)
2.10. Biodistribution in a Nude Mouse Xenograft Tumor Model
2.11. Statistical Analysis
3. Results
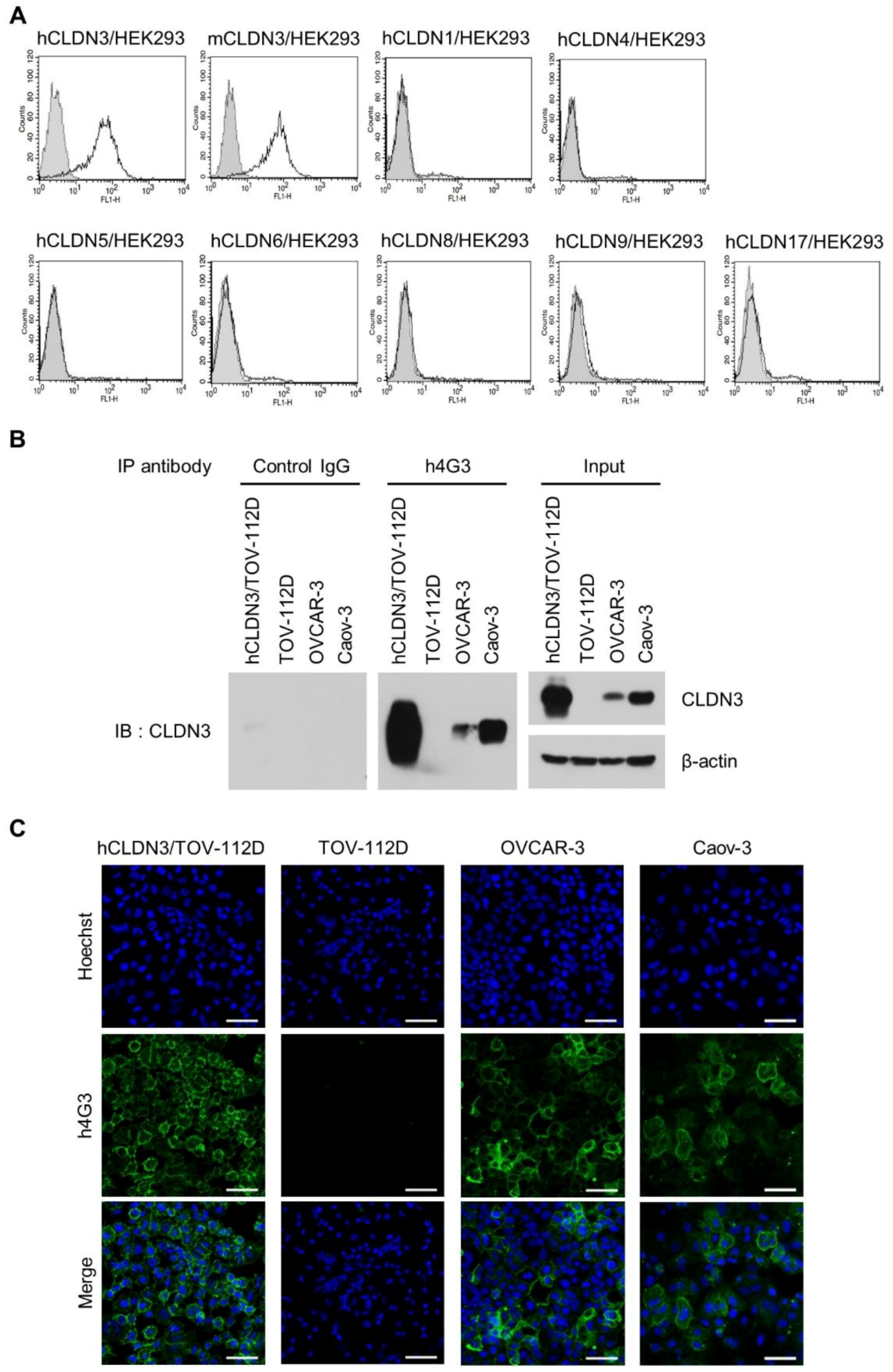
3.1. Generation of a Human mAb Against Human CLDN3
3.2. h4G3 Recognizes the ECL2 Domain of CLDN3
3.3. Binding Kinetics of h4G3 to CLDN3 on the Cell Membrane
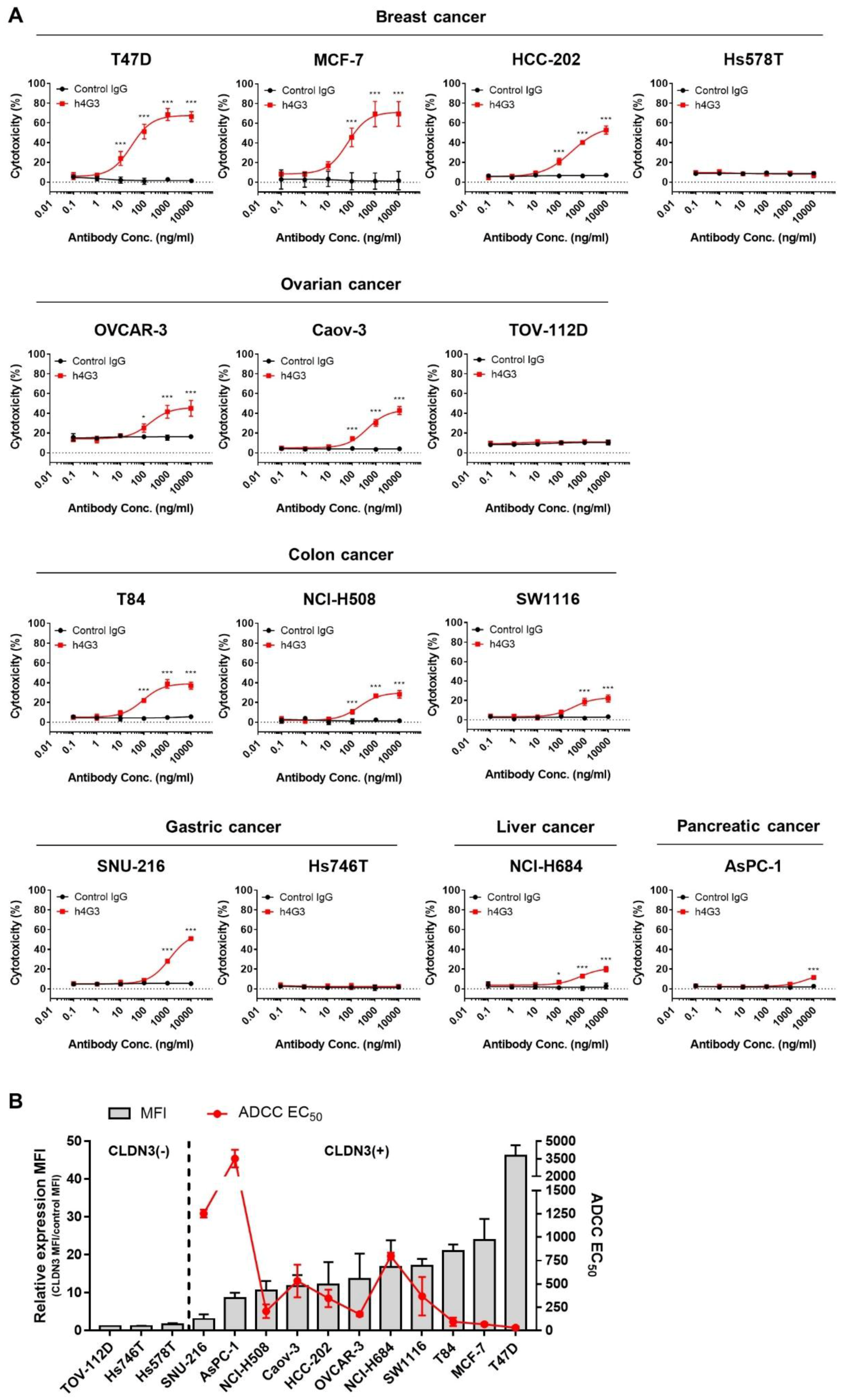
3.4. Evaluation of the Anti-Tumor Activity of h4G3
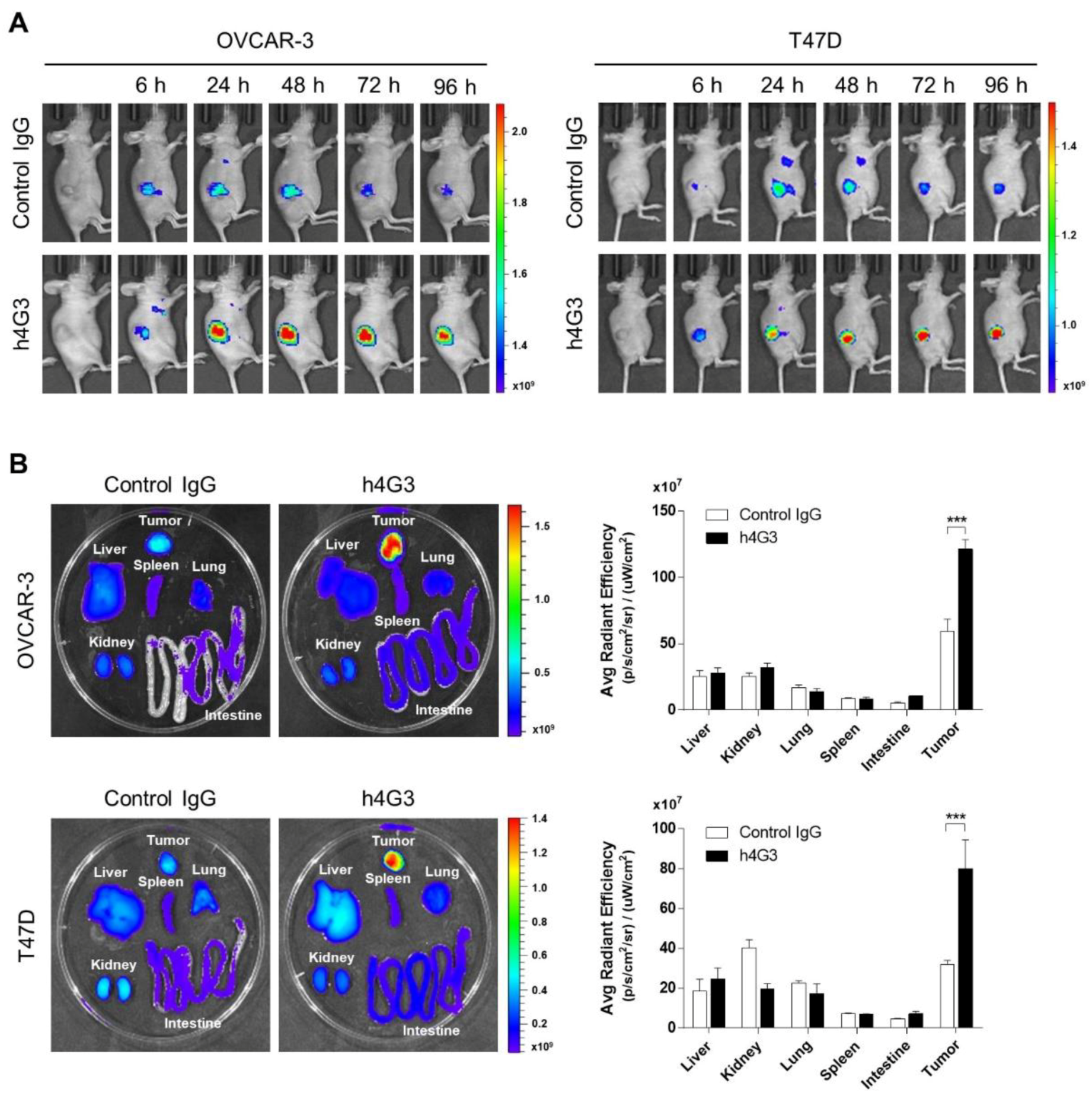
3.5. Tumor Targeting and Accumulation of h4G3 in Xenograft Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shin, K.; Fogg, V.C.; Margolis, B. Tight junctions and cell polarity. Annu. Rev. Cell Dev. Biol. 2006, 22, 207–235. [Google Scholar] [CrossRef] [PubMed]
- Wodarz, A.; Näthke, I. Cell polarity in development and cancer. Nat. Cell Biol. 2007, 9, 1016. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Jiang, W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Beamish, H.; De Boer, L.; Giles, N.; Stevens, F.; Oakes, V.; Gabrielli, B. Cyclin a/cdk2 regulates adenomatous polyposis coli-dependent mitotic spindle anchoring. J. Biol. Chem. 2009, 284, 29015–29023. [Google Scholar]
- Todd, M.C.; Petty, H.M.; King, J.M.; Piana Marshall, B.N.; Sheller, R.A.; Cuevas, M.E. Overexpression and delocalization of claudin-3 protein in mcf-7 and mda-mb-415 breast cancer cell lines. Oncol. Lett. 2015, 10, 156–162. [Google Scholar] [CrossRef]
- Kominsky, S.L. Claudins: Emerging targets for cancer therapy. Expert Rev. Mol. Med. 2006, 8, 1–11. [Google Scholar] [CrossRef]
- Saeki, R.; Kondoh, M.; Uchida, H.; Yagi, K. Potency of claudin-targeting as antitumor therapy. Mol. Cell. Pharmacol. 2010, 2, 47–51. [Google Scholar]
- Maeda, T.; Murata, M.; Chiba, H.; Takasawa, A.; Tanaka, S.; Kojima, T.; Masumori, N.; Tsukamoto, T.; Sawada, N. Claudin-4-targeted therapy using clostridium perfringens enterotoxin for prostate cancer. Prostate 2012, 72, 351–360. [Google Scholar] [CrossRef]
- Van Itallie, C.M.; Anderson, J.M. The molecular physiology of tight junction pores. Physiology 2004, 19, 331–338. [Google Scholar] [CrossRef]
- Krause, G.; Winkler, L.; Mueller, S.L.; Haseloff, R.F.; Piontek, J.; Blasig, I.E. Structure and function of claudins. Biochim. Biophys. Acta 2008, 1778, 631–645. [Google Scholar] [CrossRef]
- Gunzel, D.; Yu, A.S. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [PubMed]
- Agarwal, R.; D’Souza, T.; Morin, P.J. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Res. 2005, 65, 7378–7385. [Google Scholar]
- De Souza, W.F.; Fortunato-Miranda, N.; Robbs, B.K.; de Araujo, W.M.; de-Freitas-Junior, J.C.; Bastos, L.G.; Viola, J.P.; Morgado-Diaz, J.A. Claudin-3 overexpression increases the malignant potential of colorectal cancer cells: Roles of erk1/2 and pi3k-akt as modulators of egfr signaling. PLoS ONE 2013, 8, e74994. [Google Scholar] [CrossRef] [PubMed]
- Kwon, M.J. Emerging roles of claudins in human cancer. Int. J. Mol. Sci. 2013, 14, 18148–18180. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Lin, X.; Alvarez, E.; Manorek, G.; Howell, S.B. Tight junction proteins claudin-3 and claudin-4 control tumor growth and metastases. Neoplasia 2012, 14, 974. [Google Scholar] [CrossRef]
- Lin, X.; Shang, X.; Manorek, G.; Howell, S.B. Regulation of the epithelial-mesenchymal transition by claudin-3 and claudin-4. PLoS ONE 2013, 8, e67496. [Google Scholar] [CrossRef]
- Morin, P.J. Claudin proteins in human cancer: Promising new targets for diagnosis and therapy. Cancer Res. 2005, 65, 9603–9606. [Google Scholar] [CrossRef]
- Singh, A.B.; Sharma, A.; Dhawan, P. Claudin family of proteins and cancer: An overview. J. Oncol. 2010, 2010, 541957. [Google Scholar] [CrossRef]
- Tsukita, S.; Yamazaki, Y.; Katsuno, T.; Tamura, A. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene 2008, 27, 6930. [Google Scholar] [CrossRef]
- Ding, L.; Lu, Z.; Lu, Q.; Chen, Y.-H. The claudin family of proteins in human malignancy: A clinical perspective. Cancer Manag. Res. 2013, 5, 367. [Google Scholar]
- Choi, Y.L.; Kim, J.; Kwon, M.J.; Choi, J.S.; Kim, T.J.; Bae, D.S.; Koh, S.S.; In, Y.H.; Park, Y.W.; Kim, S.H.; et al. Expression profile of tight junction protein claudin 3 and claudin 4 in ovarian serous adenocarcinoma with prognostic correlation. Histol. Histopathol. 2007, 22, 1185–1195. [Google Scholar] [PubMed]
- Kleinberg, L.; Holth, A.; Trope, C.G.; Reich, R.; Davidson, B. Claudin upregulation in ovarian carcinoma effusions is associated with poor survival. Hum. Pathol. 2008, 39, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Konecny, G.E.; Agarwal, R.; Keeney, G.A.; Winterhoff, B.; Jones, M.B.; Mariani, A.; Riehle, D.; Neuper, C.; Dowdy, S.C.; Wang, H.J.; et al. Claudin-3 and claudin-4 expression in serous papillary, clear-cell, and endometrioid endometrial cancer. Gynecol. Oncol. 2008, 109, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Lechpammer, M.; Resnick, M.B.; Sabo, E.; Yakirevich, E.; Greaves, W.O.; Sciandra, K.T.; Tavares, R.; Noble, L.C.; DeLellis, R.A.; Wang, L.J. The diagnostic and prognostic utility of claudin expression in renal cell neoplasms. Mod. Pathol. 2008, 21, 1320. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Katahira, J.; Horiguchi, Y.; Sonoda, N.; Furuse, M.; Tsukita, S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 2000, 476, 258–261. [Google Scholar] [CrossRef]
- Mitchell, L.A.; Koval, M. Specificity of interaction between clostridium perfringens enterotoxin and claudin-family tight junction proteins. Toxins 2010, 2, 1595–1611. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, S.L.; Vali, M.; Korz, D.; Gabig, T.G.; Weitzman, S.A.; Argani, P.; Sukumar, S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am. J. Pathol. 2004, 164, 1627–1633. [Google Scholar] [CrossRef]
- Santin, A.D.; Cané, S.; Bellone, S.; Palmieri, M.; Siegel, E.R.; Thomas, M.; Roman, J.J.; Burnett, A.; Cannon, M.J.; Pecorelli, S. Treatment of chemotherapy-resistant human ovarian cancer xenografts in cb-17/scid mice by intraperitoneal administration of clostridium perfringens enterotoxin. Cancer Res. 2005, 65, 4334–4342. [Google Scholar] [CrossRef]
- Gao, Z.; Xu, X.; McClane, B.; Zeng, Q.; Litkouhi, B.; Welch, W.R.; Berkowitz, R.S.; Mok, S.C.; Garner, E.I. C-terminus of clostridium perfringens enterotoxin downregulates cldn4 and sensitizes ovarian cancer cells to taxol and carboplatin. Clin. Cancer Res. 2011, 17, 1065–1074. [Google Scholar] [CrossRef]
- Saeki, R.; Kondoh, M.; Kakutani, H.; Tsunoda, S.; Mochizuki, Y.; Hamakubo, T.; Tsutsumi, Y.; Horiguchi, Y.; Yagi, K. A novel tumor-targeted therapy using a claudin-4-targeting molecule. Mol. Pharmacol. 2009, 76, 918–926. [Google Scholar] [CrossRef]
- Yuan, X.; Lin, X.; Manorek, G.; Kanatani, I.; Cheung, L.H.; Rosenblum, M.G.; Howell, S.B. Recombinant cpe fused to tumor necrosis factor targets human ovarian cancer cells expressing the claudin-3 and claudin-4 receptors. Mol. Cancer Ther. 2009, 8, 1906–1915. [Google Scholar] [CrossRef] [PubMed]
- Nagase, S.; Doyama, R.; Yagi, K.; Kondoh, M. Recent advances in claudin-targeting technology. Biol. Pharm. Bull. 2013, 36, 708–714. [Google Scholar] [CrossRef] [PubMed]
- Cocco, E.; Casagrande, F.; Bellone, S.; Richter, C.E.; Bellone, M.; Todeschini, P.; Holmberg, J.C.; Fu, H.H.; Montagna, M.K.; Mor, G.; et al. Clostridium perfringens enterotoxin carboxy-terminal fragment is a novel tumor-homing peptide for human ovarian cancer. BMC Cancer 2010, 10, 349. [Google Scholar] [CrossRef] [PubMed]
- Cocco, E.; Shapiro, E.M.; Gasparrini, S.; Lopez, S.; Schwab, C.L.; Bellone, S.; Bortolomai, I.; Sumi, N.J.; Bonazzoli, E.; Nicoletti, R.; et al. Clostridium perfringens enterotoxin c-terminal domain labeled to fluorescent dyes for in vivo visualization of micrometastatic chemotherapy-resistant ovarian cancer. Int. J. Cancer 2015, 137, 2618–2629. [Google Scholar] [CrossRef]
- Suzuki, H.; Kondoh, M.; Takahashi, A.; Yagi, K. Proof of concept for claudin-targeted drug development. Ann. N. Y. Acad. Sci. 2012, 1258, 65–70. [Google Scholar] [CrossRef]
- Gao, Z.; McClane, B.A. Use of clostridium perfringens enterotoxin and the enterotoxin receptor-binding domain (c-cpe) for cancer treatment: Opportunities and challenges. J. Toxicol. 2012, 2012. [Google Scholar] [CrossRef]
- Suzuki, H.; Kondoh, M.; Li, X.; Takahashi, A.; Matsuhisa, K.; Matsushita, K.; Kakamu, Y.; Yamane, S.; Kodaka, M.; Isoda, K. A toxicological evaluation of a claudin modulator, the c-terminal fragment of clostridium perfringens enterotoxin, in mice. Pharmazie 2011, 66, 543–546. [Google Scholar]
- Li, X.; Saeki, R.; Watari, A.; Yagi, K.; Kondoh, M. Tissue distribution and safety evaluation of a claudin-targeting molecule, the c-terminal fragment of clostridium perfringens enterotoxin. Eur. J. Pharm. Sci. 2014, 52, 132–137. [Google Scholar] [CrossRef]
- Liu, J.K. The history of monoclonal antibody development-progress, remaining challenges and future innovations. Ann. Med. Surg. 2014, 3, 113–116. [Google Scholar] [CrossRef]
- Rudnick, S.I.; Adams, G.P. Affinity and avidity in antibody-based tumor targeting. Cancer Biother. Radiopharm. 2009, 24, 155–161. [Google Scholar] [CrossRef]
- Cruz, E.; Kayser, V. Monoclonal antibody therapy of solid tumors: Clinical limitations and novel strategies to enhance treatment efficacy. Biologics 2019, 13, 33–51. [Google Scholar] [CrossRef]
- Weiner, L.M.; Surana, R.; Wang, S. Monoclonal antibodies: Versatile platforms for cancer immunotherapy. Nat. Rev. Immunol. 2010, 10, 317–327. [Google Scholar] [CrossRef]
- Scott, A.M.; Wolchok, J.D.; Old, L.J. Antibody therapy of cancer. Nat. Rev. Cancer 2012, 12, 278–287. [Google Scholar] [CrossRef]
- Coats, S.; Williams, M.; Kebble, B.; Dixit, R.; Tseng, L.; Yao, N.S.; Tice, D.A.; Soria, J.C. Antibody-drug conjugates: Future directions in clinical and translational strategies to improve the therapeutic index. Clin. Cancer Res. 2019, 25, 5441–5448. [Google Scholar] [CrossRef]
- Larson, S.M.; Carrasquillo, J.A.; Cheung, N.K.; Press, O.W. Radioimmunotherapy of human tumours. Nat. Rev. Cancer 2015, 15, 347–360. [Google Scholar] [CrossRef]
- Akbari, B.; Farajnia, S.; Ahdi Khosroshahi, S.; Safari, F.; Yousefi, M.; Dariushnejad, H.; Rahbarnia, L. Immunotoxins in cancer therapy: Review and update. Int. Rev. Immunol. 2017, 36, 207–219. [Google Scholar] [CrossRef]
- Chitgupi, U.; Qin, Y.; Lovell, J.F. Targeted nanomaterials for phototherapy. Nanotheranostics 2017, 1, 38–58. [Google Scholar] [CrossRef]
- Fass, L. Imaging and cancer: A review. Mol. Oncol. 2008, 2, 115–152. [Google Scholar] [CrossRef]
- Warram, J.M.; de Boer, E.; Sorace, A.G.; Chung, T.K.; Kim, H.; Pleijhuis, R.G.; van Dam, G.M.; Rosenthal, E.L. Antibody-based imaging strategies for cancer. Cancer Metastasis Rev. 2014, 33, 809–822. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Yagi, K.; Kondoh, M. Current progress in a second-generation claudin binder, anti-claudin antibody, for clinical applications. Drug Discov. Today 2016, 21, 1711–1718. [Google Scholar] [CrossRef]
- Bai, X.; Kim, J.; Kang, S.; Kim, W.; Shim, H. A novel human scfv library with non-combinatorial synthetic cdr diversity. PLoS ONE 2015, 10, e0141045. [Google Scholar] [CrossRef]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef]
- Bondza, S.; Foy, E.; Brooks, J.; Andersson, K.; Robinson, J.; Richalet, P.; Buijs, J. Real-time characterization of antibody binding to receptors on living immune cells. Front. Immunol. 2017, 8, 455. [Google Scholar] [CrossRef]
- Strome, S.E.; Sausville, E.A.; Mann, D. A mechanistic perspective of monoclonal antibodies in cancer therapy beyond target-related effects. Oncologist 2007, 12, 1084–1095. [Google Scholar] [CrossRef]
- Zhang, J.; Zheng, H.; Diao, Y. Natural killer cells and current applications of chimeric antigen receptor-modified nk-92 cells in tumor immunotherapy. Int. J. Mol. Sci. 2019, 20, 317. [Google Scholar] [CrossRef]
- Ling, J.; Liao, H.; Clark, R.; Wong, M.S.; Lo, D.D. Structural constraints for the binding of short peptides to claudin-4 revealed by surface plasmon resonance. J. Biol. Chem. 2008, 283, 30585–30595. [Google Scholar] [CrossRef]
- Ando, H.; Suzuki, M.; Kato-Nakano, M.; Kawamoto, S.; Misaka, H.; Kimoto, N.; Furuya, A.; Nakamura, K. Generation of specific monoclonal antibodies against the extracellular loops of human claudin-3 by immunizing mice with target-expressing cells. Biosci. Biotechnol. Biochem. 2015, 79, 1272–1279. [Google Scholar] [CrossRef]
- Kato-Nakano, M.; Suzuki, M.; Kawamoto, S.; Furuya, A.; Ohta, S.; Nakamura, K.; Ando, H. Characterization and evaluation of the antitumour activity of a dual-targeting monoclonal antibody against claudin-3 and claudin-4. Anticancer Res. 2010, 30, 4555–4562. [Google Scholar]
- Li, X.; Iida, M.; Tada, M.; Watari, A.; Kawahigashi, Y.; Kimura, Y.; Yamashita, T.; Ishii-Watabe, A.; Uno, T.; Fukasawa, M.; et al. Development of an anti-claudin-3 and -4 bispecific monoclonal antibody for cancer diagnosis and therapy. J. Pharmacol. Exp. Ther. 2014, 351, 206–213. [Google Scholar] [CrossRef]
- Winkler, L.; Gehring, C.; Wenzel, A.; Muller, S.L.; Piehl, C.; Krause, G.; Blasig, I.E.; Piontek, J. Molecular determinants of the interaction between clostridium perfringens enterotoxin fragments and claudin-3. J. Biol. Chem. 2009, 284, 18863–18872. [Google Scholar] [CrossRef]
- Kosmoliaptsis, V.; Goodman, R.S.; Dafforn, T.R.; Bradley, J.A.; Taylor, C.J. Structural implications of platelet endothelial cell adhesion molecule-1 polymorphisms. Transplantation 2008, 86, 484–485. [Google Scholar] [CrossRef]
- Jochems, C.; Hodge, J.W.; Fantini, M.; Fujii, R.; Morillon, Y.M., 2nd; Greiner, J.W.; Padget, M.R.; Tritsch, S.R.; Tsang, K.Y.; Campbell, K.S.; et al. An nk cell line (hank) expressing high levels of granzyme and engineered to express the high affinity cd16 allele. Oncotarget 2016, 7, 86359–86373. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kono, K.; Mizukami, Y.; Mimura, K.; Fujii, H. Mechanisms of escape from trastuzumab-mediated adcc in esophageal squamous cell carcinoma: Relation to susceptibility to perforin-granzyme. Anticancer Res. 2009, 29, 2137–2146. [Google Scholar]
- Lehmann, C.; Zeis, M.; Schmitz, N.; Uharek, L. Impaired binding of perforin on the surface of tumor cells is a cause of target cell resistance against cytotoxic effector cells. Blood 2000, 96, 594–600. [Google Scholar] [CrossRef]
- Hewitt, K.J.; Agarwal, R.; Morin, P.J. The claudin gene family: Expression in normal and neoplastic tissues. BMC Cancer 2006, 6, 186. [Google Scholar] [CrossRef]
- Santin, A.D.; Bellone, S.; Marizzoni, M.; Palmieri, M.; Siegel, E.R.; McKenney, J.K.; Hennings, L.; Comper, F.; Bandiera, E.; Pecorelli, S. Overexpression of claudin-3 and claudin-4 receptors in uterine serous papillary carcinoma. Cancer 2007, 109, 1312–1322. [Google Scholar] [CrossRef]
- Krause, G.; Protze, J.; Piontek, J. Assembly and function of claudins: Structure-function relationships based on homology models and crystal structures. In Seminars in Cell & Developmental Biology; Academic Press: London, UK, 2015; Volume 42, pp. 3–12. [Google Scholar]
- Chester, K.A.; Mayer, A.; Bhatia, J.; Robson, L.; Spencer, D.I.; Cooke, S.P.; Flynn, A.A.; Sharma, S.K.; Boxer, G.; Pedley, R.B. Recombinant anti-carcinoembryonic antigen antibodies for targeting cancer. Cancer Chemother. Pharmacol. 2000, 46, S8–S12. [Google Scholar] [CrossRef]
- Mayer, A.; Tsiompanou, E.; O’Malley, D.; Boxer, G.M.; Bhatia, J.; Flynn, A.A.; Chester, K.A.; Davidson, B.R.; Lewis, A.A.; Winslet, M.C. Radioimmunoguided surgery in colorectal cancer using a genetically engineered anti-cea single-chain fv antibody. Clin. Cancer Res. 2000, 6, 1711–1719. [Google Scholar]
- Steffens, M.G.; Boerman, O.C.; Oosterwijk-Wakka, J.C.; Oosterhof, G.O.; Witjes, J.A.; Koenders, E.B.; Oyen, W.J.; Buijs, W.C.; Debruyne, F.M.; Corstens, F.H.; et al. Targeting of renal cell carcinoma with iodine-131-labeled chimeric monoclonal antibody g250. J. Clin. Oncol 1997, 15, 1529–1537. [Google Scholar] [CrossRef]
- McLaughlin, P.M.; Harmsen, M.C.; Dokter, W.H.; Kroesen, B.-J.; van der Molen, H.; Brinker, M.G.; Hollema, H.; Ruiters, M.H.; Buys, C.H.; de Leij, L.F. The epithelial glycoprotein 2 (egp-2) promoter-driven epithelial-specific expression of egp-2 in transgenic mice: A new model to study carcinoma-directed immunotherapy. Cancer Res. 2001, 61, 4105–4111. [Google Scholar]
- Overdijk, M.B.; Verploegen, S.; Ortiz Buijsse, A.; Vink, T.; Leusen, J.H.; Bleeker, W.K.; Parren, P.W. Crosstalk between human igg isotypes and murine effector cells. J. Immunol. 2012, 189, 3430–3438. [Google Scholar] [CrossRef]
- Romani, C.; Cocco, E.; Bignotti, E.; Moratto, D.; Bugatti, A.; Todeschini, P.; Bandiera, E.; Tassi, R.; Zanotti, L.; Pecorelli, S.; et al. Evaluation of a novel human igg1 anti-claudin3 antibody that specifically recognizes its aberrantly localized antigen in ovarian cancer cells and that is suitable for selective drug delivery. Oncotarget 2015, 6, 34617–34628. [Google Scholar] [CrossRef][Green Version]
- Smaglo, B.G.; Aldeghaither, D.; Weiner, L.M. The development of immunoconjugates for targeted cancer therapy. Nat. Rev. Clin. Oncol. 2014, 11, 637–648. [Google Scholar] [CrossRef]
- Fleuren, E.D.; Versleijen-Jonkers, Y.M.; Heskamp, S.; van Herpen, C.M.; Oyen, W.J.; van der Graaf, W.T.; Boerman, O.C. Theranostic applications of antibodies in oncology. Mol. Oncol. 2014, 8, 799–812. [Google Scholar] [CrossRef]
- Moek, K.L.; Giesen, D.; Kok, I.C.; de Groot, D.J.A.; Jalving, M.; Fehrmann, R.S.N.; Lub-de Hooge, M.N.; Brouwers, A.H.; de Vries, E.G.E. Theranostics using antibodies and antibody-related therapeutics. J. Nucl. Med. 2017, 58, 83S–90S. [Google Scholar] [CrossRef]
- Pang, Y.; Hou, X.; Yang, C.; Liu, Y.; Jiang, G. Advances on chimeric antigen receptor-modified t-cell therapy for oncotherapy. Mol. Cancer 2018, 17, 91. [Google Scholar] [CrossRef]
- Rezvani, K.; Rouce, R.; Liu, E.; Shpall, E. Engineering natural killer cells for cancer immunotherapy. Mol. Ther. 2017, 25, 1769–1781. [Google Scholar] [CrossRef]
- Jiang, H.; Shi, Z.; Wang, P.; Wang, C.; Yang, L.; Du, G.; Zhang, H.; Shi, B.; Jia, J.; Li, Q.; et al. Claudin18.2-specific chimeric antigen receptor engineered t cells for the treatment of gastric cancer. J. Natl. Cancer Inst. 2018, 111, 409–418. [Google Scholar] [CrossRef]

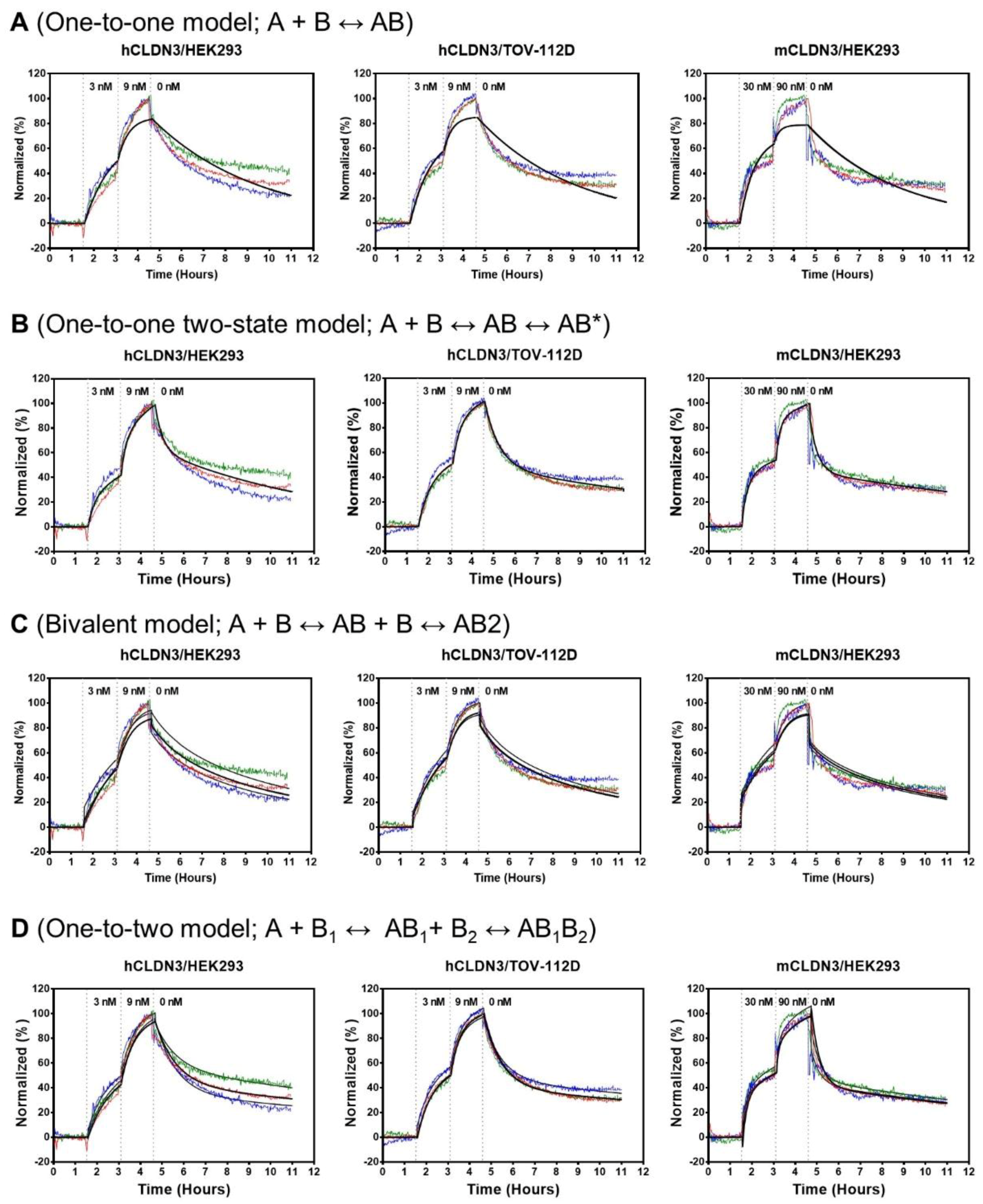
| CLDN3 Cell Lines | ka1 (M−1s−1) | kd1 (s−1) | ka2 (s−1) | kd2 (s−1) | KD (nM) |
|---|---|---|---|---|---|
| hCLDN3/HEK293 | 4.66 × 104 | 6.71 × 10−4 | 1.56 × 10−4 | 4.35 × 10−5 | 4.03 |
| hCLDN3/TOV-112D | 4.74 × 104 | 3.37 × 10−4 | 7.77 × 10−4 | 2.57 × 10−5 | 2.35 |
| mCLDN3/HEK293 | 8.69 × 103 | 7.07 × 10−4 | 8.63 × 10−5 | 2.27 × 10−5 | 20.4 |
| CLDN3 Cell Lines | ka1 (M−1s−1) | kd1 (s−1) | KD1 (nM) | ka2 (M−1s−1) | kd2 (s−1) | KD2 (nM) |
|---|---|---|---|---|---|---|
| hCLDN3/HEK293 | 3.57 × 104 | 2.98 × 10−4 | 8.33 | 3.03 × 104 | 1.53 × 10−5 | 0.504 |
| hCLDN3/TOV-112D | 6.66 × 104 | 3.43 × 10−4 | 5.15 | 2.90 × 104 | 1.26 × 10−5 | 0.434 |
| mCLDN3/HEK293 | 2.42 × 104 | 7.28 × 10−4 | 30.1 | 2.54 × 103 | 1.91 × 10−5 | 7.54 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, H.; Park, H.; Lee, Y.J.; Choi, J.Y.; Kim, T.; Rajasekaran, N.; Lee, S.; Song, K.; Hong, S.; Choi, J.-S.; et al. Development of Human Monoclonal Antibody for Claudin-3 Overexpressing Carcinoma Targeting. Biomolecules 2020, 10, 51. https://doi.org/10.3390/biom10010051
Yang H, Park H, Lee YJ, Choi JY, Kim T, Rajasekaran N, Lee S, Song K, Hong S, Choi J-S, et al. Development of Human Monoclonal Antibody for Claudin-3 Overexpressing Carcinoma Targeting. Biomolecules. 2020; 10(1):51. https://doi.org/10.3390/biom10010051
Chicago/Turabian StyleYang, Hobin, Hayeon Park, Yong Jin Lee, Jun Young Choi, TaeEun Kim, Nirmal Rajasekaran, Saehyung Lee, Kyoung Song, Sungyoul Hong, Joon-Seok Choi, and et al. 2020. "Development of Human Monoclonal Antibody for Claudin-3 Overexpressing Carcinoma Targeting" Biomolecules 10, no. 1: 51. https://doi.org/10.3390/biom10010051
APA StyleYang, H., Park, H., Lee, Y. J., Choi, J. Y., Kim, T., Rajasekaran, N., Lee, S., Song, K., Hong, S., Choi, J.-S., Shim, H., Kim, Y.-D., Hwang, S., Choi, Y.-L., & Shin, Y. K. (2020). Development of Human Monoclonal Antibody for Claudin-3 Overexpressing Carcinoma Targeting. Biomolecules, 10(1), 51. https://doi.org/10.3390/biom10010051






