Role of Pea LTPs and Abscisic Acid in Salt-Stressed Roots
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Preparation of Tissue Sections
2.3. Immunohistochemical Localization
2.4. Suberin Detection
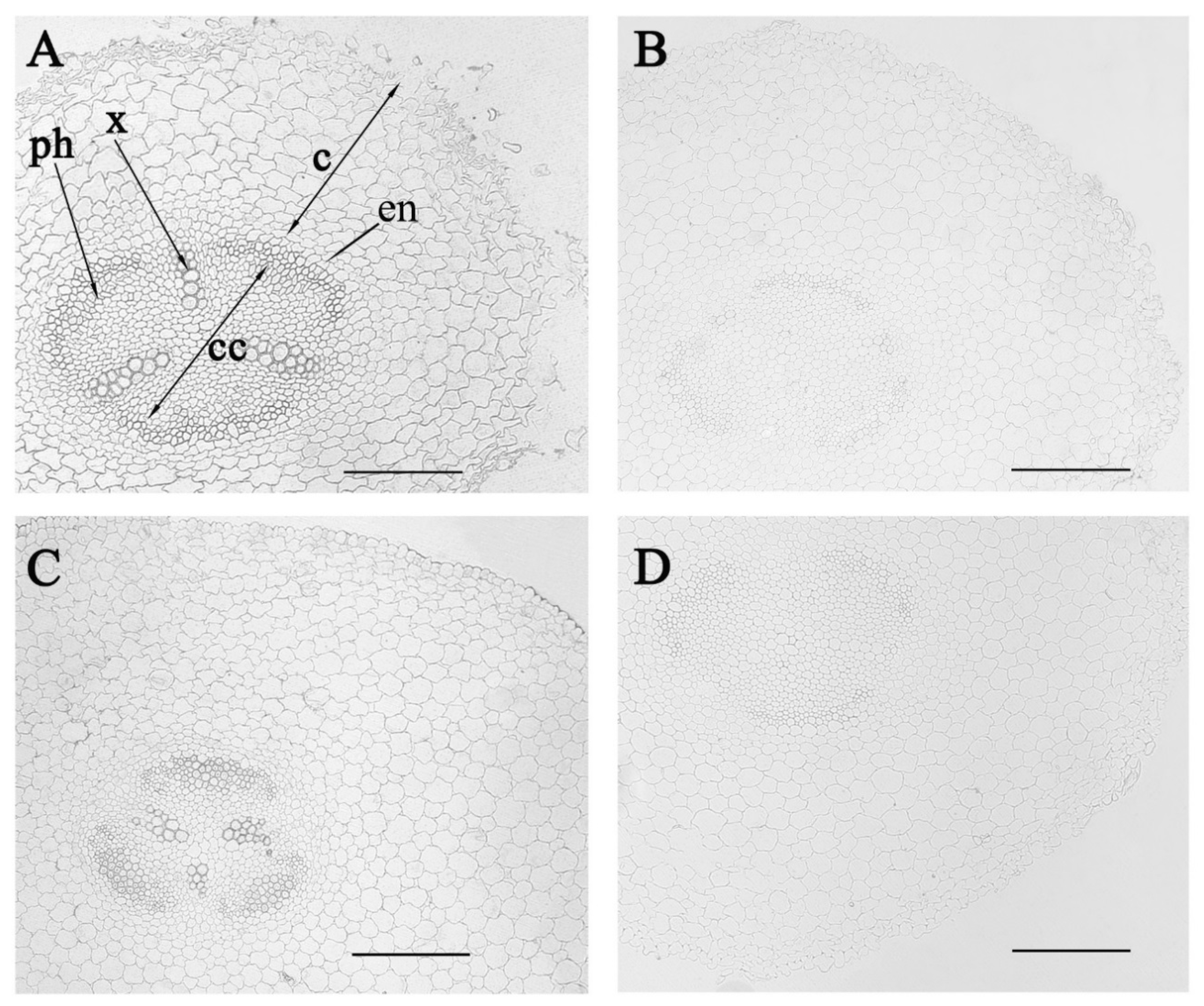
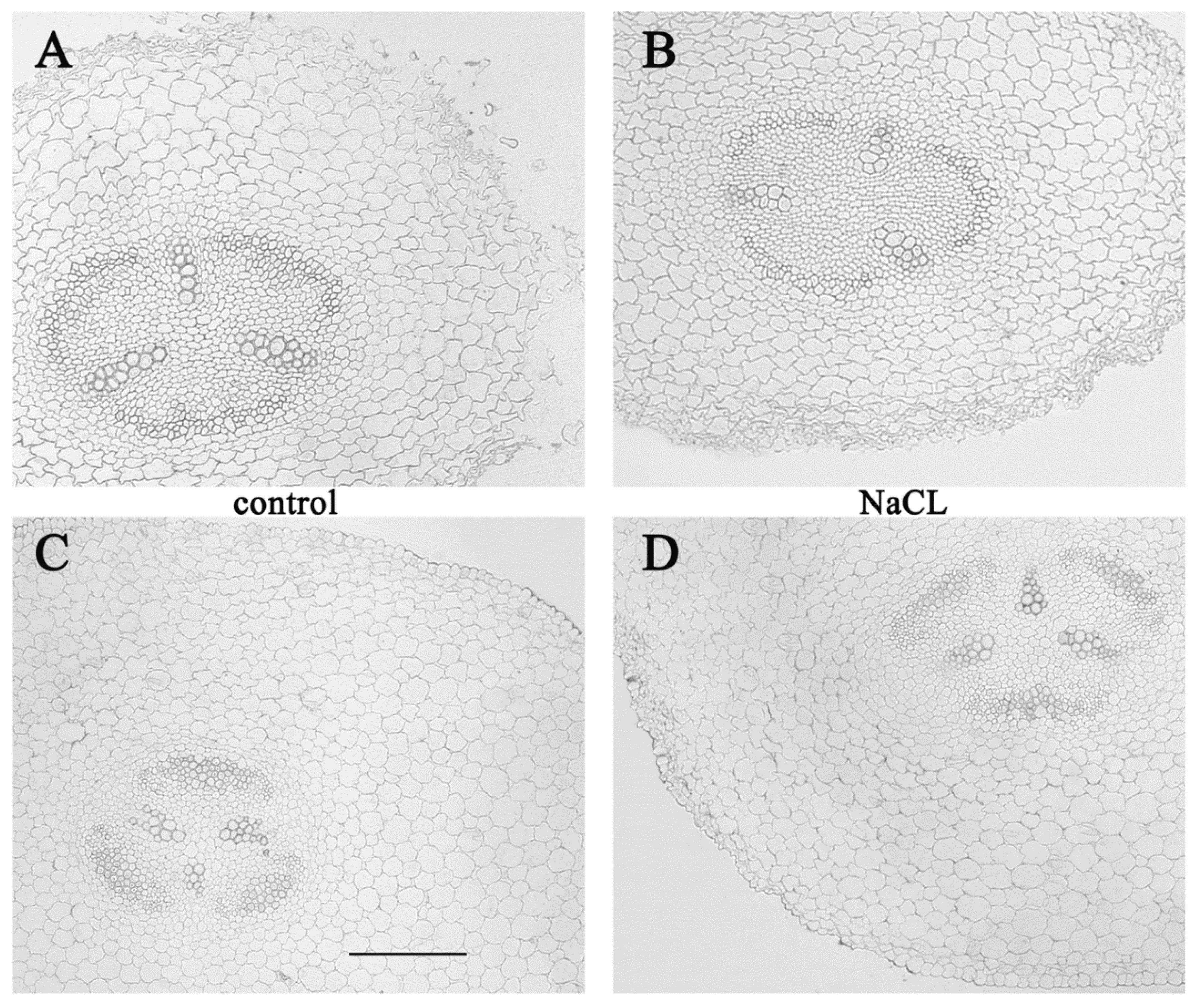
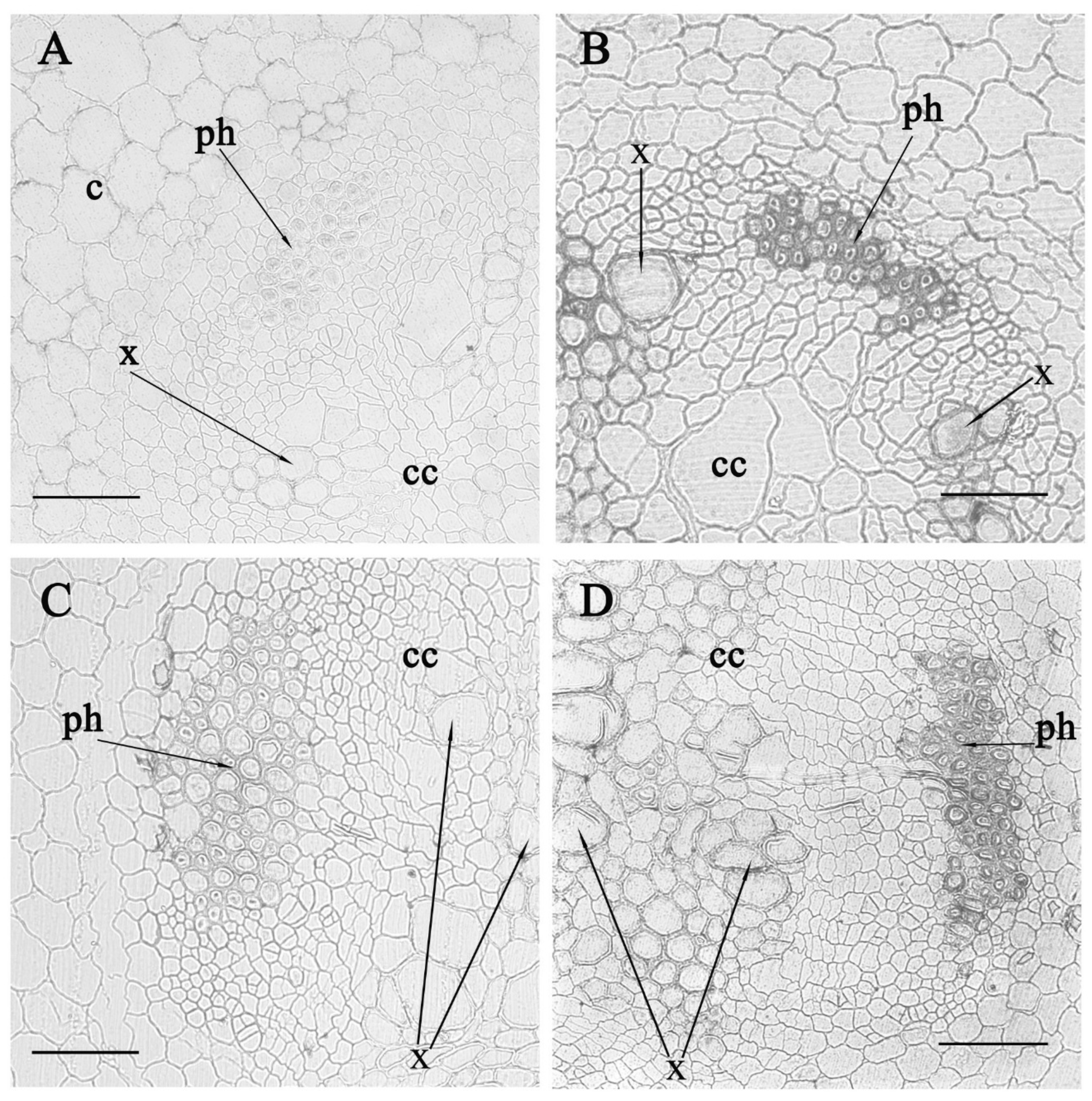
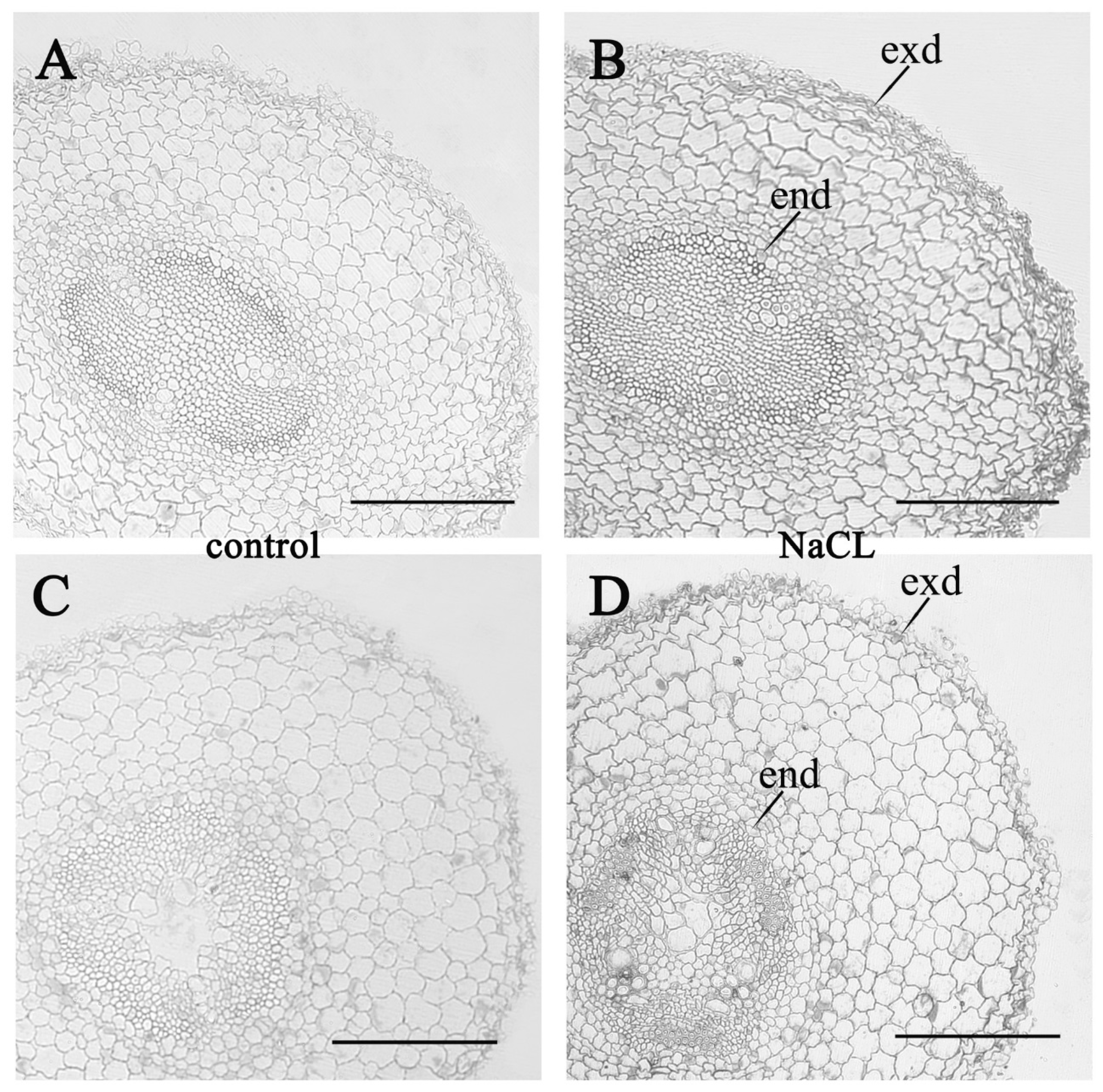
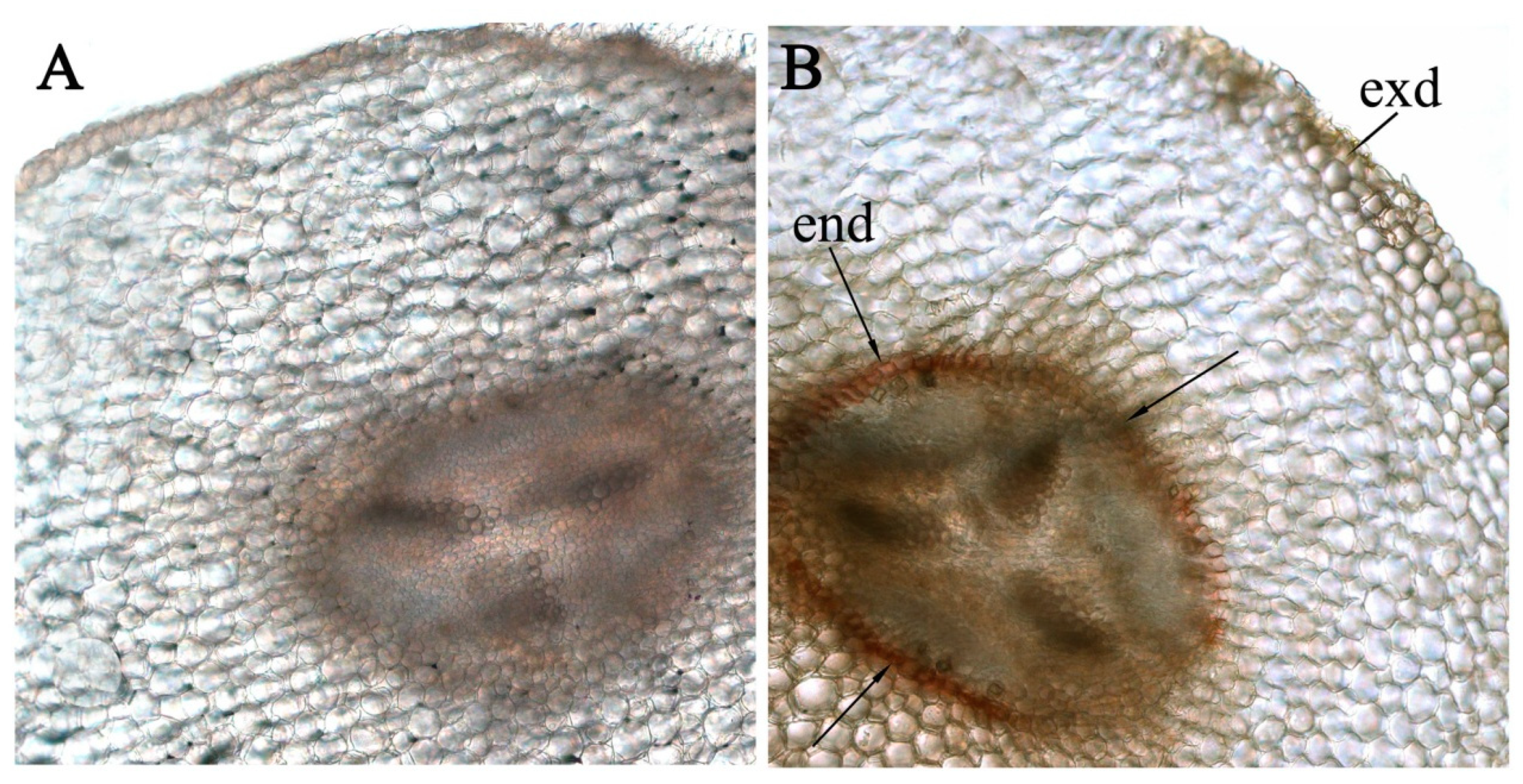
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Finkina, E.I.; Melnikova, D.N.; Bogdanov, I.V.; Ovchinnikova, T.V. Lipid transfer proteins as components of the plant innate immune system: Structure, functions, and applications. Acta Nat. 2016, 8, 47–61. [Google Scholar] [CrossRef]
- Melnikova, D.N.; Finkina, E.I.; Bogdanov, I.V.; Ovchinnikova, T.V. Plant pathogenesis-related proteins binding lipids and other hydrophobic ligands. Russ. J. Bioorganic Chem. 2018, 44, 586–594. [Google Scholar] [CrossRef]
- Simorre, J.; Caille, A.; Dominique, M.; Didier, M.; Ptak, M. Two- and three-dimensional 1H NMR studies of a wheat phospholipid transfer protein: Sequential resonance assignments and secondary structure. Biochemistry 1991, 30, 11600–11608. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.S.; Samuel, D.; Liu, Y.J.; Shyu, J.C.; Lai, S.M.; Lin, K.F.; Lyu, P.C. Binding mechanism of nonspecific lipid transfer proteins and their role in plant defense. Biochemistry 2004, 43, 13628–13636. [Google Scholar] [CrossRef]
- Pato, C.; Borgne, M.; Baut, G.; Papec, P.; Marion, D.; Douliez, J.P. Potential application of plant lipid transfer proteins for drug delivery. Biochem. Pharmacol. 2001, 62, 555–560. [Google Scholar] [CrossRef]
- Guerbette, F.; Grosbois, M.; Jolliot-Croquin, A.; Kader, J.C.; Zachowski, A. Comparison of lipid binding and transfer properties of two lipid transfer proteins from plants. Biochemistry 1999, 38, 14131–14137. [Google Scholar] [CrossRef]
- Edqvist, J.; Blomqvist, K.; Nieuwland, J.; Salminen, T.A. Plant lipid transfer proteins: Are we finally closing in on the roles of these enigmatic proteins? J. Lipid Res. 2018, 59, 1374–1382. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, X.; Lu, C.; Zeng, X.; Li, Y.; Fu, D.; Wu, G. Non-specific lipid transfer proteins in plants: Presenting new advances and an integrated functional analysis. J. Exp. Bot. 2015, 66, 5663–5681. [Google Scholar] [CrossRef]
- Edstam, M.M.; Edqvist, J. Involvement of GPI-anchored lipid transfer proteins in the development of seed coats and pollen in Arabidopsis thaliana. Physiol. Plant. 2014, 152, 32–42. [Google Scholar] [CrossRef]
- Ibort, P.; Imai, H.; Uemura, M.; Aroca, R. Proteomic analysis reveals that tomato interaction with plant growth promoting bacteria is highly determined by ethylene perception. J. Plant Physiol. 2018, 220, 43–59. [Google Scholar] [CrossRef]
- Pan, Y.; Li, J.; Jiao, L.; Li, C.; Zhu, D.; Yu, J. A Non-specific Setaria italica lipid transfer protein gene plays a critical role under abiotic stress. Front. Plant. Sci. 2016, 7, 1752. [Google Scholar] [CrossRef] [PubMed]
- Safi, H.; Saibi, W.; Alaoui, M.M.; Hmyene, A.; Masmoudi, K.; Hanin, M.; Brini, F. A wheat lipid transfer protein (TdLTP4) promotes tolerance to abiotic and biotic stress in Arabidopsis thaliana. Plant. Physiol. Biochem. 2015, 89, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Barberon, M.; Vermeer, J.E.M.; De Bellis, D.; Wang, P.; Naseer, S.; Andersen, T.G.; Humbel, B.M.; Nawrath, C.; Takano, J.; Salt, D.E.; et al. Adaptation of root function by nutrient-induced plasticity of endodermal differentiation. Cell 2016, 164, 447–459. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Lu, W.; Wei, X.; Li, L.; Mao, L.; Zhao, Y. Proteomics analysis to understand the ABA stimulation of wound suberization in kiwifruit. J. Proteom. 2018, 173, 42–51. [Google Scholar] [CrossRef]
- Deeken, R.; Saupe, S.; Klinkenberg, J.; Riedel, M.; Leide, J.; Hedrich, R.; Mueller, T.D. The nonspecific lipid transfer protein AtLtpI-4 is involved in suberin formation of Arabidopsis thaliana crown galls. Plant. Physiol. 2016, 172, 1911–1927. [Google Scholar] [CrossRef]
- Akhiyarova, G.R.; Fricke, W.; Veselov, D.S.; Kudoyarova, G.R.; Veselov, S.Y. ABA accumulation and distribution during the leaf tissues shows its role stomatal conductance regulation under short—Term salinity. Citologiya 2006, 48, 918–923. [Google Scholar]
- Veselov, D.S.; Sharipova, G.V.; Veselov, Y.S.; Dodd, I.C.; Ivanov, I.I.; Kudoyarova, G.R. Rapid changes in root HvPIP2;2 aquaporins abundance and ABA concentration are required to enhance root hydraulic conductivity and maintain leaf water potential in response to increased evaporative demand. Funct. Plant Biol. 2018, 45, 143–149. [Google Scholar] [CrossRef]
- Bogdanov, I.V.; Finkina, E.I.; Balandin, S.V.; Melnikova, D.N.; Stukacheva, E.A.; Ovchinnikova, T.V. Structural and functional characterization of recombinant isoforms of the lentil lipid transfer protein. Acta Nat. 2015, 7, 65–73. [Google Scholar] [CrossRef]
- Bogdanov, I.V.; Shenkarev, Z.O.; Finkina, E.I.; Melnikova, D.N.; Rumynskiy, E.I.; Arseniev, A.S.; Ovchinnikova, T.V. A novel lipid transfer protein from the pea Pisum sativum: Isolation, recombinant expression, solution structure, antifungal activity, lipid binding, and allergenic properties. BMC Plant Biol. 2016, 16, 107–124. [Google Scholar] [CrossRef]
- Sharipova, G.; Veselov, D.; Kudsoyarova, G.; Fricke, W.; Dodd, I.; Katsuhara, M.; Furuichi, T.; Ivanov, I.; Veselov, S. Exogenous application of abscisic acid (ABA) increases root and cell hydraulic conductivity and abundance of some aquaporin isoforms in the ABA deficient barley mutant Az34. Ann. Bot. 2016, 118, 777–785. [Google Scholar] [CrossRef]
- Efetova, M.; Zeier, J.; Riederer, M.; Lee, C.W.; Stingl, N.; Mueller, M.; Hartung, W.; Hedrich, R.; Deeken, R. A central role of abscisic acid in drought stress protection of Agrobacterium-induced tumors on Arabidopsis. Plant Physiol. 2007, 145, 853–862. [Google Scholar] [CrossRef] [PubMed]
- Chae, K.; Gonong, B.J.; Kim, S.C.; Kieslich, C.A.; Morikis, D.; Balasubramanian, S.; Lord, E.M. A multifaceted study of stigma/style cysteine-rich adhesin (SCA)-like Arabidopsis lipid transfer proteins (LTPs) suggests diversified roles for these LTPs in plant growth and reproduction. J. Exp. Bot. 2010, 61, 4277–4290. [Google Scholar] [CrossRef]
- Benning, U.F.; Tamot, B.; Guelette, B.S.; Hoffmann-Benning, S. New aspects of phloem-mediated long-distance lipid signaling in plants. Front. Plant Sci. 2012, 3, 53. [Google Scholar] [CrossRef] [PubMed]
- Guelette, B.S.; Benning, U.F.; Hoffman-Benning, S. Identification of lipids and lipid binding proteins in phloem exudate from Arabidopsis thaliana. J. Exp. Bot. 2012, 63, 3603–3616. [Google Scholar] [CrossRef] [PubMed]
- Carella, P.; Merl-Pham, J.; Wilson, D.C.; Dey, S.; Hauck, S.M.; Vlot, A.C.; Cameron, R.K. Comparative proteomics analysis of phloem exudates collected during the induction of systemic acquired resistance. Plant Physiol. 2016, 171, 1495–1510. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Melicherova, N.; Řemínek, R.; Foret, F. Application of capillary electrophoretic methods for the analysisof plant phloem and xylem saps composition: A review. J. Sep. Sci. 2019, 1–14. [Google Scholar]
- Djordjevic, M.A.; Oakes, M.; Li, D.X.; Hwang, C.H.; Hocart, C.H.; Gresshoff, P.M. The glycine max xylem sap and apoplast proteome. J. Proteome Res. 2007, 6, 3771–3779. [Google Scholar] [CrossRef]
- Rep, M.; Dekker, H.L.; Vossen, J.H.; de Boer, A.D.; Houterman, P.M.; de Koster, C.G.; Cornelissen, B.J.C. A tomato xylem sap protein represents a new family of small cysteine-rich proteins with structural similarity to lipid transfer proteins. FEBS Lett. 2003, 534, 82–86. [Google Scholar] [CrossRef]
- Buhtz, A.; Kolasa, A.; Arlt, K.; Walz, C.; Kehr, J. Xylem sap protein composition is conserved among different plant species. Planta 2004, 219, 610–618. [Google Scholar] [CrossRef]
- Perumalla, C.J.; Peterson, C.A. Deposition of Casparian bands and suberin lamellae in the exodermis and endodermis of young corn and onion roots. Can. J. Bot. 1986, 64, 1873–1878. [Google Scholar] [CrossRef]
- Hairat, S.; Baranwal, V.K.; Khurana, P. Identification of Triticum aestivum nsLTPs and functional validation of two members in development and stress mitigation roles. Plant Physiol. Biochem. 2018, 130, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Cai, X.; Wu, X.; Karahara, I.; Schreiber, L.; Lin, J. Casparian strip development and its potential function in salt tolerance. Plant Signal Behav. 2011, 6, 1499–1502. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zheng, X.; Song, Y.; Zhu, L.; Yu, Z.; Gan, L.; Zhou, S.; Liu, H.; Wen, F.; Zhu, C. NtLTP4, a lipid transfer protein that enhances salt and drought stresses tolerance in Nicotiana tabacum. Sci. Rep. 2018, 8, 8873. [Google Scholar] [CrossRef]
- Veselov, D.S.; Sharipova, G.V.; Veselov, Y.S.; Kudoyarova, G.R. The effects of NaCl treatment on water relations, growth and ABA content in barley cultivars differing in drought tolerance. J. Plant Growth Regul. 2008, 27, 380–386. [Google Scholar] [CrossRef]
- Edreva, A. Pathogenesis-related proteins: Research progress in the last 15 years. Gen. Appl. Plant Physiol. 2005, 31, 105–124. [Google Scholar]
- Wang, C.; Yang, C.; Gao, C.; Wang, Y. Cloning and expression analysis of 14 lipid transfer protein genes from Tamarix hispida responding to different abiotic stresses. Tree Physiol. 2009, 29, 1607–1619. [Google Scholar] [CrossRef]
- Gonzalez, L.E.; Keller, K.; Chan, K.X.; Gessel, M.M.; Thines, B.C. Transcriptome analysis uncovers Arabidopsis F-BOX STRESS INDUCED 1 as a regulator of jasmonic acid and abscisic acid stress gene expression. BMC Genom. 2017, 18, 533. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akhiyarova, G.R.; Finkina, E.I.; Ovchinnikova, T.V.; Veselov, D.S.; Kudoyarova, G.R. Role of Pea LTPs and Abscisic Acid in Salt-Stressed Roots. Biomolecules 2020, 10, 15. https://doi.org/10.3390/biom10010015
Akhiyarova GR, Finkina EI, Ovchinnikova TV, Veselov DS, Kudoyarova GR. Role of Pea LTPs and Abscisic Acid in Salt-Stressed Roots. Biomolecules. 2020; 10(1):15. https://doi.org/10.3390/biom10010015
Chicago/Turabian StyleAkhiyarova, Guzel R., Ekaterina I. Finkina, Tatiana V. Ovchinnikova, Dmitry S. Veselov, and Guzel R. Kudoyarova. 2020. "Role of Pea LTPs and Abscisic Acid in Salt-Stressed Roots" Biomolecules 10, no. 1: 15. https://doi.org/10.3390/biom10010015
APA StyleAkhiyarova, G. R., Finkina, E. I., Ovchinnikova, T. V., Veselov, D. S., & Kudoyarova, G. R. (2020). Role of Pea LTPs and Abscisic Acid in Salt-Stressed Roots. Biomolecules, 10(1), 15. https://doi.org/10.3390/biom10010015






