Abstract
Serotonin N-acetyltransferase (SNAT) is the penultimate enzyme in the melatonin biosynthetic pathway, in which serotonin is converted into N-acetylserotonin (NAS) in plants. To date, two SNAT isogenes with low amino acid sequence homologies have been identified. Their single suppression in rice has been reported, but their double suppression in rice has not yet been attempted. Here, we generated double-suppression transgenic rice (snat1+2) using the RNA interference technique. The snat1+2 exhibited retarded seedling growths in conjunction with severe decreases in melatonin compared to wild-types and single-suppression rice plants (snat1 or snat2). The laminar angle was decreased in the snat1+2 rice compared to that of the wild-types and snat1, but was comparable to that of snat2. The reduced germination speed in the snat1+2 was comparable to that of snat2. Seed-aging testing revealed that snat1 was the most severely deteriorated, followed by snat1+2 and snat2, suggesting that melatonin is positively involved in seed longevity.
1. Introduction
Melatonin is a multifunctional regulator, serving as a signal molecule for various metabolic processes, and is also a potent antioxidant in plants [1,2,3,4]. Melatonin influences not only key plant hormones, such as auxin and cytokinin, which are essential for plant growth and development [5], but also plant redox signaling molecules, such as reactive oxygen species (ROS) and reactive nitrogen species (RNS), which play pivotal roles in numerous processes, including seed germination, senescence, and various stress responses [6]. Due to the large number of physiological roles played by melatonin, it has long been suspected that plants, like animals, possess melatonin receptors [7]. Recently, it was reported that Cand2 serves as a melatonin receptor involved in stomatal closure in Arabidopsis [8]. However, further in-depth studies are needed to understand the involvement of Cand2 in other melatonin-mediated plant functions, such as the induction of mitogen-activated protein kinase [9].
In plants, melatonin biosynthesis consists of four enzymatic steps, beginning with the aromatic amino acid tryptophan [10]. In plants, tryptophan decarboxylase (TDC) catalyzes the first committed step, in which tryptophan is converted into tryptamine, whereas, in animals, tryptophan hydroxylase (TPH) is the first enzyme, which catalyzes tryptophan conversion into 5‑hydroxytryptophan [2]. In plants, tryptamine is further hydroxylated into serotonin by the action of tryptamine 5-hydroxylase (T5H), a P450 enzyme localized in the endoplasmic reticulum. The penultimate enzyme is serotonin N-acetyltransferase (SNAT), which converts serotonin into N-acetylserotonin. The last enzyme is an O-methyltransferase that catalyzes the conversion of N-acetylserotonin into melatonin; both N-acetylserotonin O-methyltransferase (ASMT) and caffeic acid O-methyltransferase (COMT) are involved. The rate of serotonin synthesis is much greater than the rate of serotonin conversion into melatonin; thus, the latter half of the pathway plays a pivotal role in regulating melatonin levels in plants [10]. SNAT is believed to be the rate-limiting step enzyme, rather than ASMT, in view of the catalytic efficiencies (Vmax/Km) of the respective enzymes [10]. In 2013, the first SNAT gene (SNAT1) was cloned from rice [11]. The overexpression of SNAT1 conferred tolerance against abiotic stress [12], whereas its downregulation (snat1) led to a susceptibility to abiotic stress and retarded seedling growths, in conjunction with altered melatonin levels [13]. In marked contrast, downregulation of SNAT2 in rice (snat2) led to an enhanced tolerance to abiotic stress, a semidwarf phenotype with erect leaves, and a decrease in melatonin content [14,15]. The major difference between snat1 and snat2 rice was the level of brassinosteroid (BR), of which snat2 was more deficient than the wild-type, while snat1 had a BR content comparable to that of the wild-type. Both snat1 and snat2 rice had decreased melatonin levels, but they had different phenotypes and abiotic stress responses, depending on their BR levels [15,16]. These contradictory results prompted us to generate a double-suppression rice mutant (snat1+2) to observe the effects of the simultaneous downregulation of both genes.
2. Materials and Methods
2.1. Generation of a Double-Suppression Rice Mutant (snat1+2) Using RNA Interference
Rice (Oryza sativa) has two SNAT isogenes: SNAT1 and SNAT2, which share 27% amino acid identities [14]. SNAT1 is located on chromosome 5, whereas SNAT2 is located on chromosome 8 [17]. The full-length SNAT1 (GenBank accession number AK059369) and SNAT2 (GenBank accession number AK068156) genes from rice were provided by the National Institute of Agrobiological Sciences (NIAS, Tsukuba, Japan) [17]. To simultaneously knock down SNAT1 and SNAT2 gene expressions in rice, a chimeric gene composed of SNAT1 and SNAT2 was constructed as follows: The SNAT1 insert, positioned in the middle of the cDNA, was PCR-amplified with forward (5′-ACT AGT ACT CCT AGA AAG-3′ [SpeI site underlined]) and reverse (5′-CTC GAG AGG CAT AGT AAC TGA-3′ [XhoI site underlined]) primers. The SNAT2 insert, positioned in the middle of the cDNA, was PCR-amplified with forward (5′-CTC GAG GCG GGG GAC GGC GTG-3′ [XhoI site underlined]) and reverse (5′-GAG CTC GGG GTC CAT GGC GAA-3′ [SacI site underlined]) primers. Both PCR fragments were digested by XhoI and ligated together (RBC Bioscience, New Taipei City, Taiwan). Using the ligated product as a template, PCR was carried out to obtain a chimeric gene (SNAT1 + SNAT2) with a primer set (forward 5′-ACT AGT ACT CCT AGA AAG-3′ [SpeI site underlined] and reverse 5′-GAG CTC GGG GTC CAT GGC GAA-3′ [SacI site underlined]). The resulting 393-bp chimeric fragment was cloned into the T&A cloning vector (T&A:OsSNAT1+2; RBC Bioscience), and the antisense SNAT1+2 insert was obtained by SacI and SpeI double-digestion and ligated into the pTCK303 binary vector [18], which had been digested by the SacI and SpeI restriction enzymes. Thereafter, the sense fragment of the SNAT1+2 insert was from KpnI and BamHI digestion from the T&A:OsSNAT1+2 plasmid and purified on DE81 ion exchange paper (Whatman, Maidstone, UK). The purified KpnI and BamHI insert was further ligated into the pTCK303 vector harboring the corresponding antisense SNAT1+2 fragments, which were predigested with KpnI and BamHI.
The pTCK303:OsSNAT1+2 RNAi binary vector (Figure 1A) was transformed into Agrobacterium tumefaciens LBA4404 using the freeze-and-thaw method, followed by transformation into rice, as described previously [19].
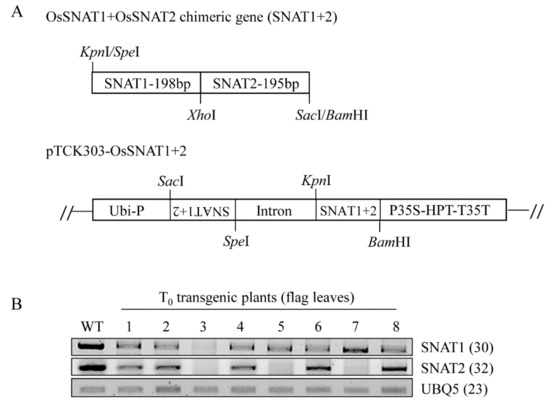
Figure 1.
Schematic diagram of the SNAT1+2 chimeric gene construct and reverse transcription–polymerase chain reaction (RT-PCR) analyses of T0 transgenic rice plants. (A) Construction of chimeric gene containing SNAT1 and SNAT2 and the binary vector used for SNAT1+2 suppression. (B) RT-PCR analyses of independent T0 transgenic lines grown for 15 weeks in a paddy field. SNAT = serotonin N-acetyltransferase, Ubi-P = maize ubiquitin promoter, HPT = hygromycin phosphotransferase, WT = wild-type, UBQ5 = rice ubiquitin5 gene, and 1–8 = SNAT1+2‑underexpression line. The GenBank accession numbers of SNAT1, SNAT2, and UBQ5 are AK059369, AK068156, and Os03g13170.
2.2. Plant Growth Conditions
Seeds of wild-type (Oryza sativa cv. Dongjin) and transgenic rice were soaked in sterile distilled water for three days at 28 °C, and the germinated seeds were transplanted into soil for seed production. The plants were grown in a culture room at 28 °C/24 °C (day/night) with a 14 h-light/10 h-dark cycle or in a paddy field at the Chonnam National University (53 m a.s.l.; 35°09′ N and 126°54′ W), Gwangju, Korea. The paddy field was a controlled area for growing the transgenic rice plants permitted by the Rural Development Administration of Korea. The angles of the lamina joints of the second leaves were measured in 14-day-old rice seedlings. The RNAi lines of rice SNAT1 (s1) and SNAT2 (s2) were obtained from previous reports [13,14]. Fertilizer was applied at 70 N/40 P/70 K kg/ha. The rice seeds were harvested by cutting mature panicles with a scissor, and spikelets were hand-threshed.
2.3. Semi-Quantitative Reverse Transcription–Polymerase Chain Reaction (RT-PCR) Analysis and Quantitative Real-Time (qRT)-PCR Analyses
The total RNA of the rice plants was isolated using a NucleoSpin RNA Plant Kit (Macherey-Nagel, Düren, Germany). First-strand cDNA was synthesized from 2 μg of total RNA using MG MMLV Reverse Transcriptase (MGmed, Inc.; Seoul; Korea) and an oligo dT18 primer at 42 °C for 1 h. Semi-quantitative RT-PCR was performed, as described previously [14]. Real-time PCR was performed in a Mic qPCR cycler system (Biomolecular Systems, Queensland, VIC, Australia) with specific primers and the Luna Universal qPCR Master Mix (New England Biolabs, Ipswich, MA, USA). The expressions of genes were analyzed using Mic’s RQ software (Biomolecular Systems) and normalized to UBQ5. Semi-quantitative RT-PCR and real-time PCR were performed with the following primer set: SNAT1 forward 5′-CAG TAG AGC CAC CAT CAG CA-3′, SNAT1 reverse 5′-ATC CCA CCT TGT CGC ATA AA-3′, SNAT2 forward 5′-GTC TGG GAC GTG GTC GTG-3′, SNAT2 reverse 5′-GTT GCC TTG AGC GGT AGA AG-3′, DWARF4 forward 5′-GTG CTG CCA TTC TCG GAG TAA TAG-3′, DWARF4 reverse 5′-CTC AGC AAG AGG TCC AGG ATT TGC-3′, UBQ5 forward 5′-CCG ACT ACA ACA TCC AGA AGG AG-3′, and UBQ5 reverse 5′-AAC AGG AGC CTA CGC CTA AGC-3′.
2.4. Quantification of Melatonin
Frozen rice samples (0.1 g) were ground into powder in liquid nitrogen using the TissueLyser II (Qiagen, Tokyo, Japan) and extracted with 1 mL of chloroform for 24 h at 4 °C. The chloroform extracts (200 µL) were completely evaporated and dissolved in 0.1 mL of 40% methanol, and 20 µL aliquots were subjected to high-performance liquid chromatography using a fluorescence detector system (Waters, Milford, MA, USA). Melatonin was detected by emissions at 348 nm, using 280 nm excitation. All measurements were taken in triplicate.
2.5. Germination Assay
Seeds of wild-type and transgenic rice (n = 25) were surface-sterilized and germinated in 3 mL distilled water in six-well plates in a culture room at 28 °C/24 °C (day/night) with a 14 h‑light/10 h-dark cycle. A seed was considered to have germinated if the seed coat was ruptured and a radicle of >1 mm in length had emerged. The germination speed index (GSI) was calculated using the formula GSI = (G1/N1) + (G2/N2) + … +(Gn/Nn) reported by Maguire [20]. Each treatment was replicated three times.
2.6. Giberellic Acid (GA) Treatment for Measuring Third-Leaf Sheath Elongations
Rice seeds were surface-sterilized and placed on half-strength Murashige and Skoog (MS) mediums (MB Cell, Seoul, Korea) at 28 °C/24 °C (day/night) with a 14 h-light/10 h-dark cycle for four days and then transferred to half-strength MS mediums containing 10 μM GA3. The length of the third-leaf sheaths (n = 15) were measured four days after applications of GA3 [21]. All measurements were taken in triplicate.
2.7. Accelerated Aging Treatment and Seed Germination Determination
An accelerated aging treatment was performed, as described previously [22]. To accelerate aging, rice seeds were incubated at 42 °C and 100% relative humidity in a growth cabinet for four days. These aged seeds were then surface-sterilized and soaked in sterile distilled water. Germination tests (n = 25) were carried out in a culture room at 28 °C/24 °C (day/night) with a 14 h‑light/10 h-dark cycle for six days. All measurements were taken in triplicate.
2.8. Statistical Analysis
Data were analyzed by analysis of variance using IBM SPSS Statistics 23 software (IBM Corp. Armonk, NY, USA). Means with different letters or asterisks indicate significantly different values at p < 0.05, according to Tukey’s post-hoc honestly significant difference (HSD) test. Data are presented as means ± standard deviations.
3. Results and Discussion
3.1. Generation of SNAT1+2 Double-Suppression Transgenic Rice Plants
Due to a low amino acid sequence identity between rice SNAT1 and rice SNAT2 (>27%), there was no highly homologous DNA region common to both of them. Thus, a chimeric gene comprising SNAT1 and SNAT2 was constructed and used as a single-transgene construct for downregulation of both SNAT1 and SNAT2 genes (Figure 1A). This approach was successful in the suppression of three members of the OsRac gene family by a single chimeric construct [23].
Through the scutellum-derived calli transformation by Agrobacterium infections, we first generated eight independent T0 transgenic rice plants (snat1+2), in which both SNAT1 and SNAT2 genes were downregulated, as indicated by RT-PCR analyses (Figure 1B). By self-crossing T1 snat1+2 rice, we selected and generated three independent homozygous lines of T2 snat1+2 rice: lines 3, 5, and 8. Although the expression levels of both SNAT1 and SNAT2 in the T2 rice were diminished, compared with the wild-type, the degree of suppression was significantly greater for SNAT1 than SNAT2 when measured by qRT-PCR (Figure 2A). This result indicates that the simultaneous suppression of both SNAT1 and SNAT2 was stably transmitted from the T0 to T2 generation. The variations of SNAT1 and SNAT2 expressions between the snat1+2 lines were attributed to the positional effects, indicating that the T-DNA in the three independent homozygous lines was inserted in different loci of rice chromosomes. Collectively, we successfully generated three T2 homozygous snat1+2 transgenic rice lines exhibiting suppression of both SNAT1 and SNAT2 mRNA compared to the wild-types (Figure 2A). When grown in MS mediums, the snat1+2 rice exhibited retarded seedling growths and inhibited growths of both the shoots and roots (Figure 2B,C). Compared to single-suppression rice, such as snat1 (s1) and snat2 (s2), the shoot lengths of snat1+2 were comparable to those of s1 and s2, and the root lengths of snat1+2 were equal to those of s2. Based on the seedling phenotypes, the snat1+2 double-suppression rice was similar to s2.
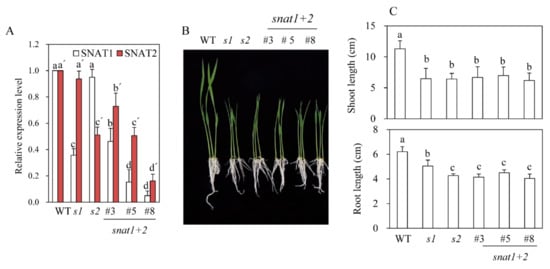
Figure 2.
Expressions of SNAT isogenes and growths of snat1+2 rice seedlings. (A) qRT-PCR analysis of SNAT1 and SNAT2 messenger RNA (mRNA) in snat1+2 rice. (B) Seedling phenotypes of snat1+2 rice grown for one week. (C) Shoot and root lengths of snat1+2 (n = 20). s1 = snat1 rice, s2 = snat2 rice, and snat1+2 = double-suppression rice. Different letters indicate significant differences from the wild-types (Tukey’s post-hoc honest significant difference (HSD) test; p <0.05).
3.2. Quantification of Melatonin in snat1+2 Rice
To determine whether the suppression of SNAT was closely associated with melatonin synthesis, we first measured melatonin content in dehulled seeds that had imbibed water for 9 h. As shown in Figure 3A, wild-type seeds contained 0.6 ng/g fresh weight (FW) melatonin, whereas the s1 and s2 mutants contained approximately half as much melatonin. Furthermore, the snat1+2 rice contained significantly less melatonin than either the s1 or s2 lines. Melatonin was also measured in seven-day-old rice seedlings that had been rhizospherically challenged with 0.5 mM cadmium chloride for three days to induce melatonin biosynthesis under continuous light. Under these conditions, the wild-types had melatonin levels of 100 ng/g FW, whereas the s1 and s2 lines produced 38 ng/g FW and 29 ng/g FW, respectively. Not surprisingly, the melatonin content was smallest in the snat1+2 rice (20 ng/g FW). These data suggest that SNAT1 and SNAT2 are synergistically involved in melatonin synthesis in rice.
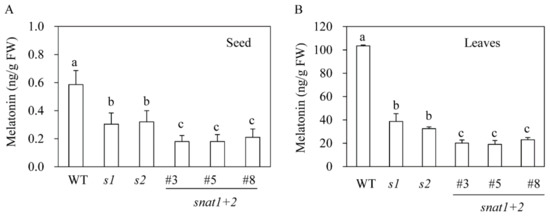
Figure 3.
Melatonin content of seeds and leaves. (A) Melatonin content of seeds. (B) Melatonin content of rice leaves upon cadmium treatment. Seeds imbibed water for 9 h and were then subjected to high-performance liquid chromatography (HPLC) analysis for melatonin quantification. Seven-day-old rice seedlings were rhizospherically challenged with 500 μM CdCl2 for three days to induce melatonin production, and leaves were subjected to melatonin analysis.
3.3. Leaf Angle Phenotypes
Previous research has revealed that the s2 rice exhibits a phenotype of dwarf plants with erect leaves [14], whereas the s1 phenotype is a dwarf without erect leaves [13,15]. The erect leaves in s2 rice were attributed to a decrease in BR content [14]. As shown in Figure 4, the snat1+2 rice was phenotypically similar to s2 plants, with a decreased leaf angle, while s1 plants were phenotypically similar to the wild-types with respect to leaf angle. DWARF4, a key BR biosynthetic gene, was significantly reduced in s2 and snat1+2, compared to s1 (Figure 4C), indicative of severe BR decreases in both s2 and snat1+2, compared to s1 and wild-types. DWARF4 is a rate-limiting step enzyme that catalyzes C22-hydroxylation from 6-oxo-campestanol into cathasterone. The failure or reduction of DWARF4 expression leads to dwarf, erect leaf, and aberrant skotomorphogenic phenotypes in plants [14,15]. These observations suggest different physiological roles for the SNAT1 and SNAT2 genes, although they are both involved in melatonin biosynthesis in rice plants. Leaf angle is affected by plant hormones, such as BR and GA [24]. Thus, it is likely that melatonin interacts with these plant hormones to orchestrate plant growths and developments in much the same way that melatonin interacts with key molecules in animals [2,25,26].
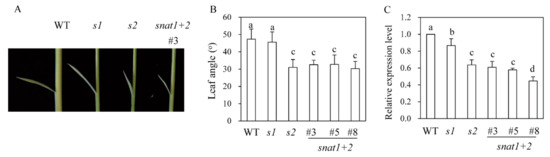
Figure 4.
Measurements of second leaf angles and DWARF4 expressions. (A) Photograph of representative transgenic rice plants. (B) Leaf angle measurements (n = 10). (C) Relative expression levels of DWARF4.
3.4. Germination Speed and GA Response to Leaf Sheath Growths
All of the rice cultivars (wild-type, s1, s2, and snat1+2) had the same germination rates of approximately 90% after six days (Figure 5A). However, when assessed at three days, the degree of germination successes differed significantly between the wild-types and RNAi lines. The s1 rice have three-day germination rates similar to that of the wild-types, whereas s2 have three-day germination rates approximately half that of the wild-types. In contrast, snat1+2 rice have three-day germination rates intermediate between the wild-types and s1. Consequently, the germination speed index was 2.0, 1.8, 1.1, and 1.3 for wild-type, s1, s2, and snat1+2 rice, respectively (Figure 5B). These data suggest an antagonistic effect on germination speeds between SNAT1 and SNAT2. Since germination and germination speeds are closely associated with GA, we measured the elongation patterns of third-leaf sheaths in response to GA3 treatments. The leaf sheath elongations assay is simple and more precise than the germination assay as a measure of GA involvement in certain physiological processes [27]. As shown in Figure 5C, treatments with GA3 (10 μM) caused a 1.5-fold increase in leaf sheath lengths of wild-type plants. Similarly, GA3 treatments of s1, s2, and snat1+2 caused a similar 1.5-fold increase in leaf sheath lengths, although the total leaf sheath lengths were still shorter than those of the wild-types, indicating that the shorter leaf sheath lengths in the RNAi lines were not rescued by GA treatments. These data are consistent with a previous report that s2 shoot lengths are insensitive to GA3 after pretreatments with the GA-inhibitor paclobutrazol [15].

Figure 5.
Germination rates, speeds, and increases in leaf sheaths after GA3 treatments. (A) Time–course experiment of germination rates. (B) Germination speed index. (C) Length of third-leaf sheaths in response to 10 μM GA3. Germination rates were measured at the indicated number of days following imbibition.
3.5. Seed Viability
To test seed viability, rice seeds were exposed to a high temperature (42 °C) under 100% humidity for four days to accelerate seed aging [28]. After the accelerated aging treatment, seeds were allowed to germinate for six days at 28 °C. As shown in Figure 6, all of the genotypes (wild-types, s1, s2, and snat1+2) had 92% six-day germination rates in the absence of accelerated aging, but after accelerated aging, the germination rates declined to 24%, 62%, and 42% for s1, s2, and snat1+2, respectively. Interestingly, the germination rates of snat1+2 after accelerated aging were intermediate between s1 and s2, indicating that suppression of SNAT2 is relatively protective of seed deterioration by accelerated aging. This observation suggests antagonistic physiological effects of SNAT1 versus SNAT2 in terms of seed viability.
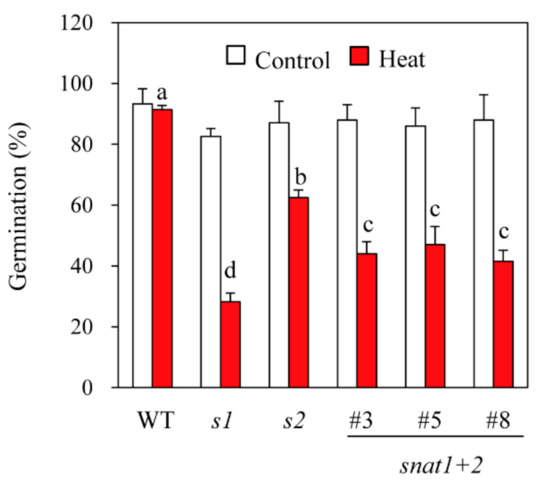
Figure 6.
Seed germination in response to accelerated aging treatments. Rice seeds were exposed to heat (42 °C) under high humidity for four days, followed by germination at 28 °C for six days.
4. Conclusions
Although the role of melatonin in plants is a relatively new field of study, compared with that in animals [2], much progress has been achieved since the first pharmacological study was reported in Scadoxus multiforus in 1969 [29]. The basic function of melatonin in plants is believed to act as a potent antioxidant, as it does in animals [4,30,31]. Thus, melatonin confers tolerance against various oxidative stresses caused by cadmium [32,33], herbicide [34,35], toxic compounds [9,36,37], and various abiotic stresses [38,39] in both plants and animals [2,40,41]. In addition to conferring tolerance against various abiotic stresses, melatonin is also implicated in several physiological and developmental processes, including maintenance of the endoplasmic reticulum [9]; sugar synthesis [42]; secondary metabolite synthesis [43,44]; growth [15]; somatic embryogenesis [45]; senescence [46,47]; root growth [48]; stomatal closure [8]; and flowering [49,50]. These diverse effects of melatonin are mediated by signaling molecules, such as H2O2 and NO, in combination with phytomelatonin receptors [6,8]. These data suggest that melatonin is multifunctional as both a hormone and biostimulator [1].
All genes responsible for melatonin biosynthesis in plants exist as a small gene family, except T5H. For example, TDC and ASMT exist in at least three copies in rice genomes [51,52], whereas SNAT has two copies with low amino acid homologies [53]. Among these four genes, SNAT seems to play an important role in synthesizing melatonin because it acts as a rate-limiting enzyme, and two SNAT isogenes play different physiological roles in rice plants. For example, SNAT1 is positively associated with abiotic tolerance [12], whereas SNAT2 is negatively associated with abiotic stress tolerance [15]. These different physiological roles of the SNAT isogenes may result from their interactions with BR. SNAT1 functions independently of BR, whereas SNAT2 functions in a manner that is dependent on BR [16].
Seeds lose viability during storage for a variety of reasons, including genetic damage, lipid peroxidation, and loss of membrane integrity [22]. Among these, lipid peroxidation is the major cause of seed deterioration. The relevance of melatonin to seed viability was first proposed by Manchester et al. [54], because melatonin is abundant in many edible seeds that are highly vulnerable to oxidative stress and storage. However, to date, no direct evidence has demonstrated that melatonin affects seed viability, although exogenous melatonin treatments do enhance germination rates in response to heat stress [55]. In this report, we have confirmed for the first time that endogenous melatonin levels are closely coupled with seed viability in rice. The largest decreases in germination rates after accelerated aging were observed in snat1 rice, followed by snat1+2 and snat2. The relatively mild suppression in snat2 may be due to the simultaneous reduction of BR, which confers tolerance against various abiotic stresses [15]. Collectively, these results confirm and extend many previous reports that melatonin acts as a potent antioxidant that effectively prevents lipid peroxidation in living cells, as well as seeds in storage.
Author Contributions
Conceptualization, K.B.; data curation, O.J.H.; formal analysis, O.J.H.; funding, K.B.; writing—original draft, K.B.; and writing—review and editing, K.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Next-Generation BioGreen 21 Program (SSAC Project No. PJ1325501) and the Basic Science Research Program of the National Research Foundation of Korea (2017R1A2A2A05069253) funded by the Ministry of Education, Republic of Korea.
Conflicts of Interest
The authors declared no conflicts of interest.
References
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Yu, Y.; Shen, Y.; Liu, Q.; Zhao, Z.; Sharma, R.; Reiter, R.J. Melatonin synthesis and function: Evolutionary history in animals and plants. Front. Endocrinol. 2019, 10, 249. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R. Melatonin in the evolution of plants and other phototrophs. Melatonin Res. 2019, 2, 10–36. [Google Scholar] [CrossRef]
- Reina, M.; Castañeda-Arriaga, R.; Perez-Gonzalez, A.; Guzman-Lopez, E.G.; Tan, D.X.; Reiter, R.J.; Galano, A. A computer-assisted systematic search for melatonin derivatives with high potential as antioxidants. Melatonin Res. 2018, 1, 27–58. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and its relationship to pant hormones. Ann. Bot. 2018, 121, 195–207. [Google Scholar] [CrossRef]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin and reactive oxygen and nitrogen species: A model for the plant redox network. Melatonin Res. 2019, 2, 152–168. [Google Scholar] [CrossRef]
- Tan, D.-X.; Reiter, R.J. Mitochondria: The birth place, battle ground and site of melatonin metabolism in cells. Melatonin Res. 2019, 2, 44–66. [Google Scholar] [CrossRef]
- Wei, J.; Li, D.X.; Zhang, J.R.; Shan, C.; Rengel, Z.; Song, Z.B.; Chen, Q. Phytomelatonin receptor PMTR1-mediated signaling regulates stomatal closure in Arabidopsis thaliana. J. Pineal Res. 2018, 65, e12500. [Google Scholar] [CrossRef]
- Lee, H.Y.; Back, K. Melatonin plays a pivotal role in conferring tolerance against endoplasmic reticulum stress via mitogen-activated protein kinases and bZIP60 in Arabidopsis thaliana. Melatonin Res. 2018, 1, 93–107. [Google Scholar] [CrossRef]
- Back, K.; Tan, D.-X.; Reiter, R.J. Melatonin biosynthesis in plants: Multiple pathways catalyze tryptophan to melatonin in the cytoplasm or chloroplasts. J. Pineal Res. 2016, 61, 426–437. [Google Scholar] [CrossRef]
- Kang, K.; Lee, K.; Park, S.; Byeon, Y.; Back, K. Molecular cloning of rice serotonin N-acetyltransferase, the penultimate gene in plant melatonin biosynthesis. J Pineal Res. 2013, 55, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Back, K. Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62, e12392. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.; Back, K. Low melatonin production by suppression of either serotonin N-acetyltransferase or N-acetylserotonin methyltransferase in rice causes seedling growth retardation with yield penalty, abiotic stress susceptibility, and enhanced coleoptile growth under anoxic conditions. J. Pineal Res. 2016, 60, 348–359. [Google Scholar] [PubMed]
- Hwang, O.J.; Back, K. Melatonin is involved in skotomorphogenesis by regulating brassinosteroid biosynthesis in plants. J. Pineal Res. 2018, 65, e12495. [Google Scholar] [CrossRef] [PubMed]
- Hwang, O.J.; Back, K. Melatonin deficiency confers tolerance to multiple abiotic stresses in rice via decreased brassinosteroid levels. Int. J. Mol. Sci. 2019, 20, 5173. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Back, K. Melatonin-deficient rice plants show a common semidwarf phenotype either dependent or independent of brassinosteroid biosynthesis. J. Pineal Res. 2019, 66, e12537. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, S.; Satoh, K.; Nagata, T.; Kawagashira, N.; Doi, K.; Kishimoto, N.; Yazaki, J.; Ishikawa, M.; Yamada, H.; Ooka, H.; et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science 2003, 301, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, C.; Xu, Y.; Jiang, R.; Han, Y.; Xu, Z.; Chong, K. A practical vector for efficient knockdown of gene expression in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 2004, 22, 409–417. [Google Scholar] [CrossRef]
- Lee, H.J.; Lee, S.B.; Chung, J.S.; Han, S.U.; Han, O.; Guh, J.O.; Jeon, J.S.; An, G.; Back, K. Transgenic rice plants expressing a Bacillus subtilis protoporphyrinogen oxidase gene are resistant to diphenyl ether herbicide oxyfluorfen. Plant Cell Physiol. 2000, 41, 743–749. [Google Scholar] [CrossRef]
- Maguire, J.D. Speed of germination-aid in selection and evaluation for seedling emergence and vigor. Crop Sci. 1962, 2, 176–177. [Google Scholar] [CrossRef]
- Komorisono, M.; Ueguchi-Tanaka, M.; Aichi, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M.; Sazuka, T. Analysis of the rice mutant dwarf and gladius leaf 1. Aberrant katanin-mediated microtubule organization causes up-regulation of gibberellin biosynthetic genes independently of gibberellin signaling. Plant Physiol. 2005, 138, 1982–1993. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, R.; Jing, W.; Zhang, W. Quantitative dissection of lipid degradation in rice seeds during accelerated aging. Plant Growth Regul. 2012, 66, 49–58. [Google Scholar] [CrossRef]
- Miki, D.; Itoh, R.; Shimamoto, K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005, 138, 1903–1913. [Google Scholar] [CrossRef] [PubMed]
- Unterholzner, S.J.; Rozhon, W.; Papacek, M.; Ciomas, J.; Lange, T.; Kugler, K.G.; Mayer, K.F.; Sieberer, T.; Poppenberger, B. Brassinosteroids are master regulators of gibberellin biosynthesis in Arabidopsis. Plant Cell 2015, 27, 2261–2272. [Google Scholar] [CrossRef] [PubMed]
- Hardeland, R. Melatonin and retinoid orphan receptors: Demand for new interpretations after their exclusion as nuclear melatonin receptors. Melatonin Res. 2018, 1, 78–93. [Google Scholar] [CrossRef]
- Potes, Y.; de Luxan-Delgado, B.; Rubio-González, A.; Reiter, R.J.; Coto-Montes, A. Dose-dependent beneficial effect of melatonin on obesity; interaction of melatonin and leptin. Melatonin Res. 2019, 2, 1–8. [Google Scholar] [CrossRef]
- Li, R.; Xia, J.; Xu, Y.; Zhao, X.; Liu, Y.-G.; Chen, Y. Characterization and genetic mapping of a Photoperiod-sensitive dwarf1 locus in rice (Oryza sativa L.). Theor. Appl. Genet. 2014, 127, 241–250. [Google Scholar] [CrossRef]
- Powell, A.M.; Matthews, S. Application of the controlled deterioration vigor test to detect seed lots of Brussels sprouts with low potential for storage under commercial conditions. Seed Sci. Technol. 1984, 12, 649–657. [Google Scholar]
- Jackson, W.T. Regulation of mitosis. II. Interaction of isopropyl N-phenylcarbamate and melatonin. J. Cell Sci. 1969, 5, 745–755. [Google Scholar]
- Yu, Y.; Lv, Y.; Shi, Y.; Li, T.; Chen, Y.; Zhao, D.; Zhao, Z. The role of phyto-melatonin and related metabolites in response to stress. Molecules 2018, 23, 1887. [Google Scholar] [CrossRef]
- Salehi, B.; Sharopov, F.; Fokou, P.V.T.; Kobylinska, A.; de Jonge, L.; Tadio, K.; Sharifi-Rad, J.; Posmyk, M.M.; Martorell, M.; Martins, N.; et al. Melatonin in medicinal and food plants: Occurrence, bioavailability, and health potential for humans. Cells 2019, 8, 681. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.-H.; Back, K. Suppression of melatonin 2-hydroxylase increases melatonin production leading to the enhanced abiotic stress tolerance against cadmium, senescence, salt, and tunicamycin in rice plants. Biomolecules 2019, 9, 589. [Google Scholar] [CrossRef] [PubMed]
- Mitra, E.; Bhattacharjee, B.; Pal, P.K.; Ghosh, A.K.; Mishra, S.; Chattopadhyay, A.; Bandyopadhyay, D. Melatonin protects against cadmium-induced oxidative damage in different tissues of rat: A mechanistic insight. Melatonin Res. 2019, 2, 1–21. [Google Scholar] [CrossRef]
- Daniela, M.; Russel, R.J.; Ahmed, A.M.; Masayuki, H.; Alejandro, B.; Giuseppe, N. Potent protective effect of melatonin on in vivo paraquat-induced oxidative damage in rats. Life Sci. 1994, 56, 83–89. [Google Scholar]
- Szafrańska, K.; Reiter, R.J.; Posmyk, M.M. Melatonin application to Pisum sativum L. seeds positively influences the function of the photosynthetic apparatus in growing seedlings during paraquat-induced oxidative stress. Front Plant Sci. 2016, 7, 1663. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, Y.; Tian, Y.; Qu, M.; Zhang, W.; Gao, L. Melatonin has the potential to alleviate cinnamic acid stress in cucumber seedlings. Front. Plant Sci. 2017, 8, 1193. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Naaz, S.; Ghosh, A.K.; Mishra, S.; Chattopadhyay, A.; Bandyopadhyay, D. Melatonin chelates iron and binds directly with phenylhydrazine to provide protection against phenylhydrazine induced oxidative damage in red blood cells along with its antioxidant mechanisms: An in vitro study. Melatonin Res. 2018, 1, 1–20. [Google Scholar] [CrossRef]
- Manda, K.; Anzai, K.; Kumari, S.; Bhatia, A.L. Melatonin attenuates radiation-induced learning deficit and brain oxidative stress in mice. Acta Neurobiol. Exp. 2007, 67, 63–70. [Google Scholar]
- Huang, B.; Chen, Y.-E.; Zhao, Y.-Q.; Ding, C.-B.; Liao, J.-Q.; Hu, C.; Zhou, L.-J.; Zhang, Z.-W.; Yuan, S.; Yuan, M. Exogenous melatonin alleviates oxidative damages and protects photosystem II in maize seedlings under drought stress. Front. Plant Sci. 2019, 10, 677. [Google Scholar] [CrossRef]
- Wang, Y.; Reiter, R.J.; Chan, Z. Phytomelatonin: A universal abiotic stress regulator. J. Exp. Bot. 2018, 69, 963–974. [Google Scholar] [CrossRef]
- Pal, P.K.; Bhattacharjee, B.; Chattopadhyay, A.; Bandyopadhyay, D. Pleiotropic roles of melatonin against oxidative stress mediated tissue injury in the gastrointestinal tract: An overview. Melatonin Res. 2019, 2, 158–184. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, C.; Wang, Z.; Sun, S.; Zhan, R.; Zhao, Y.; Ma, B.; Ma, F.; Li, M. Melatonin-mediated sugar accumulation and growth inhibition in apple plants involves down-regulation of fructokinase 2 expression and activity. Front. Plant Sci. 2019, 10, 150. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yue, Q.; Bian, F.; Sun, H.; Zhai, H.; Yao, Y. Melatonin enhances phenolic accumulation partially via ethylene signaling and resulted in high antioxidant capacity in grape berries. Front. Plant Sci. 2017, 8, 1426. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Hao, P.; Peng, J.; Zhao, Y.; Xu, J.-W.; Li, T.; Reiter, R.J.; Ma, H.; Yu, X. Melatonin enhances astaxanthin accumulation in the green microalga Haematococcus pluvialis by mechanisms possibly related to abiotic stress tolerance. Algal Res. 2018, 33, 256–265. [Google Scholar] [CrossRef]
- Ramakrishna, A.; Giridhar, P.; Jobin, M.; Paulose, C.S.; Ravishankar, G.A. Indoleamines and calcium enhance somatic embryogenesis in Coffea canephora P ex Fr. Plant Cell Tissue Organ Cult. 2012, 108, 267–278. [Google Scholar] [CrossRef]
- Hong, Y.; Zhang, Y.; Sinumporn, S.; Yu, N.; Zhan, X.; Shen, X.; Chen, D.; Yu, P.; Wu, W.; Liu, Q.; et al. Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. 2018, 95, 877–891. [Google Scholar] [CrossRef]
- Liang, D.; Shen, Y.; Ni, Z.; Wang, Q.; Lei, Z.; Xu, N.; Deng, Q.; Lin, L.; Wang, J.; Lv, X.; et al. Exogenous melatonin application delays senescence of kiwifruit leaves by regulating the antioxidant capacity and biosynthesis of flavonoids. Front. Plant Sci. 2018, 9, 426. [Google Scholar] [CrossRef]
- Liang, C.; Li, A.; Yu, H.; Li, W.; Liang, C.; Guo, S.; Zhang, R.; Chu, C. Melatonin regulates root architecture by modulating auxin response in rice. Front. Plant Sci. 2017, 8, 134. [Google Scholar] [CrossRef]
- Shi, H.; Wei, Y.; Wang, Q.; Reiter, R.J.; He, C. Melatonin mediates the stabilization of DELLA proteins to repress the floral transition in Arabidopsis. J Pineal Res. 2016, 60, 373–379. [Google Scholar] [CrossRef]
- Lee, H.Y.; Lee, K.; Back, K. Knockout of Arabidopsis serotonin N-acetyltransferase-2 reduces melatonin levels and delays flowering. Biomolecules 2019, 9, 712. [Google Scholar] [CrossRef]
- Byeon, Y.; Park, S.; Lee, H.Y.; Kim, Y.S.; Back, K. Elevated production of melatonin in transgenic rice seeds expressing rice tryptophan decarboxylase. J Pineal Res. 2014, 56, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Byeon, Y.; Back, K. Functional analysis of three ASMT gene family members in rice plants. J. Pineal Res. 2013, 55, 409–415. [Google Scholar] [PubMed]
- Byeon, Y.; Lee, H.Y.; Back, K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa). J Pineal Res. 2016, 61, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Manchester, L.C.; Tan, D.X.; Reiter, R.J.; Park, W.; Monis, K.; Qi, W. High levels of melatonin in the seeds of edible plants: Possible function in germ cell protection. Life Sci. 2000, 67, 3023–3029. [Google Scholar] [CrossRef]
- Hernández, I.G.; Gomez, F.J.V.; Cerutti, S.; Arana, M.V.; Silva, M.F. Melatonin in Arabidopsis thaliana acts as plant growth regulator at low concentration and preserves seed viability at high concentrations. Plant Physiol. Biochem. 2015, 94, 191–196. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).