Untargeted Metabolomics Toward Systematic Characterization of Antioxidant Compounds in Betulaceae Family Plant Extracts
Abstract
:1. Introduction
2. Results
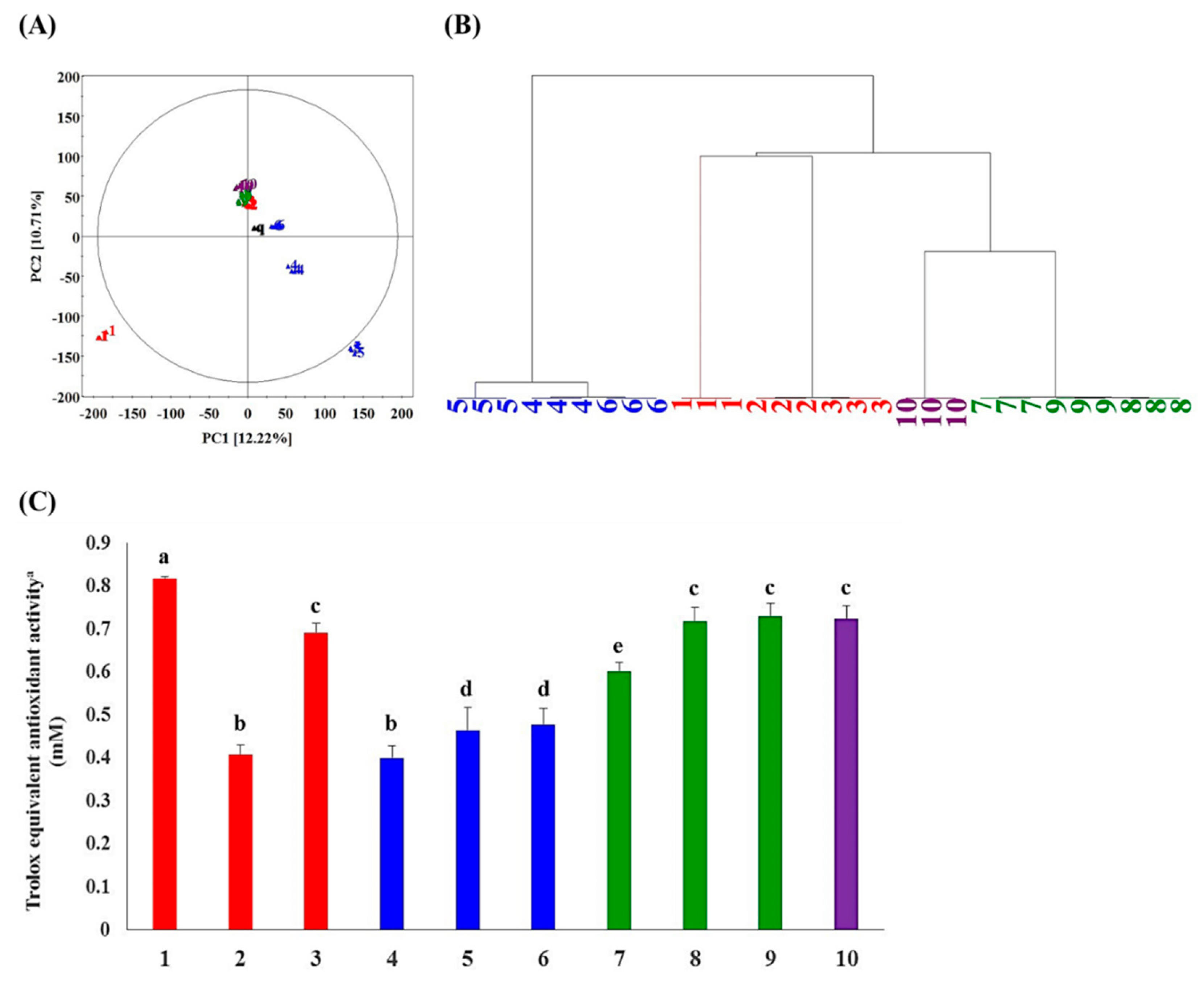
2.1. Metabolite Profiling and Antioxidant Activity Assay-Guided Screening of Plant Extracts
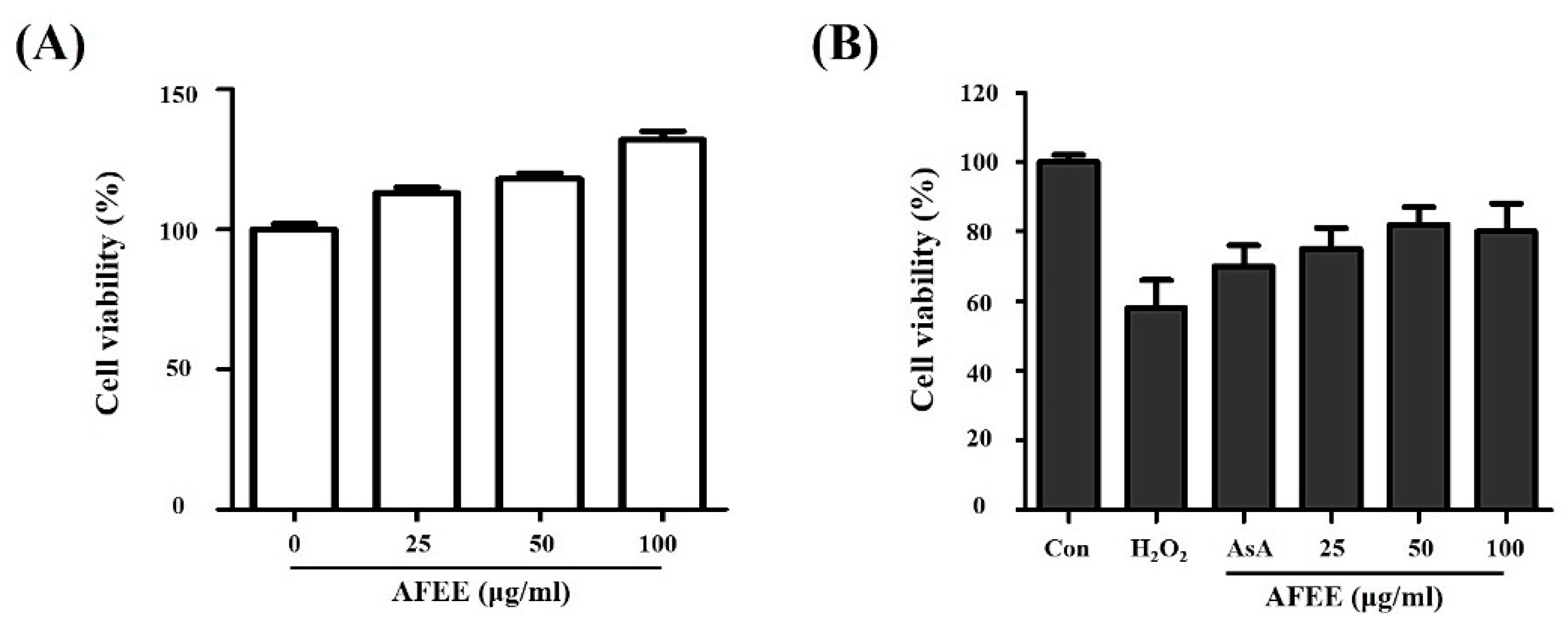
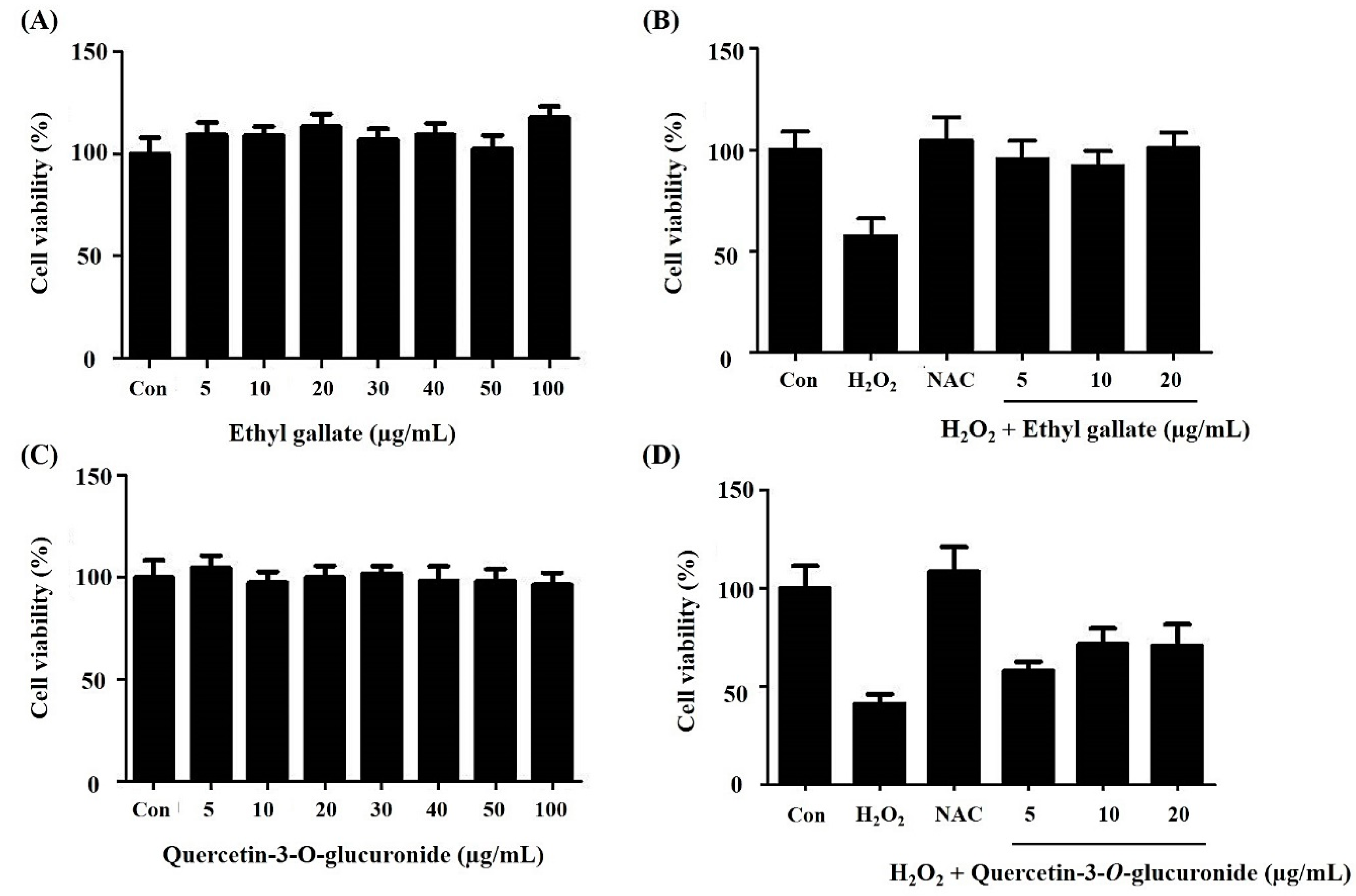
2.2. Effect of the Alnus firma Ethanol Extracts (AFEE) on H2O2-Induced HDF Cell Damage
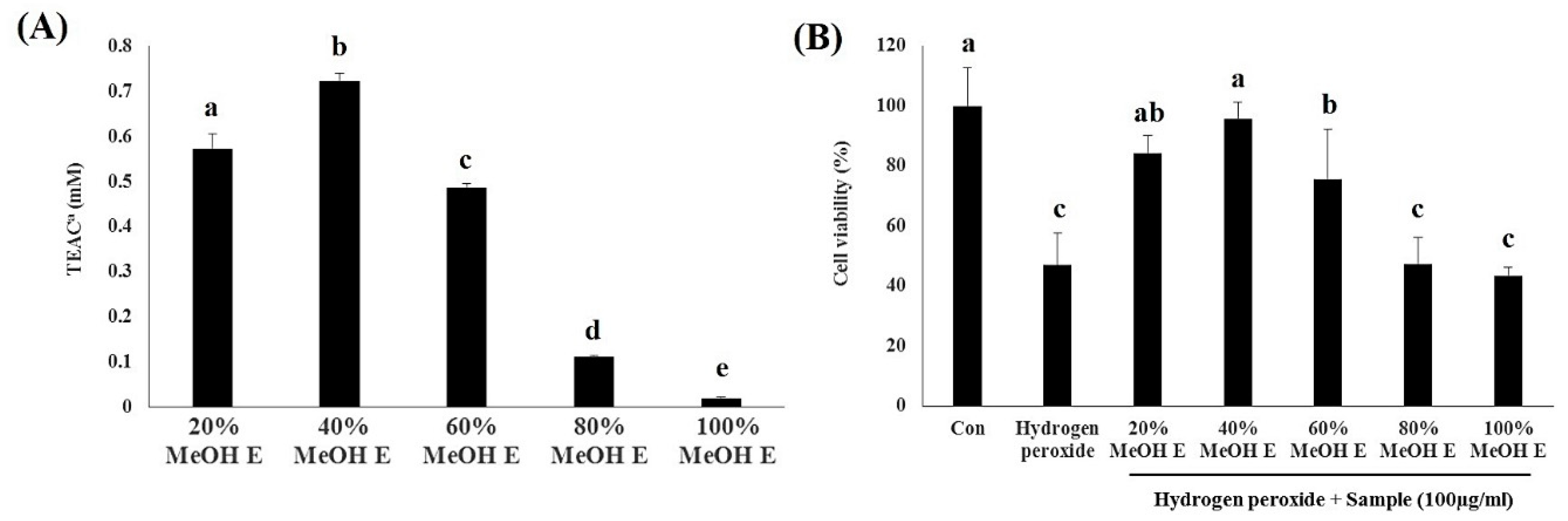
2.3. Bioactivity Assay for A. Firma SPE Fraction Eluates
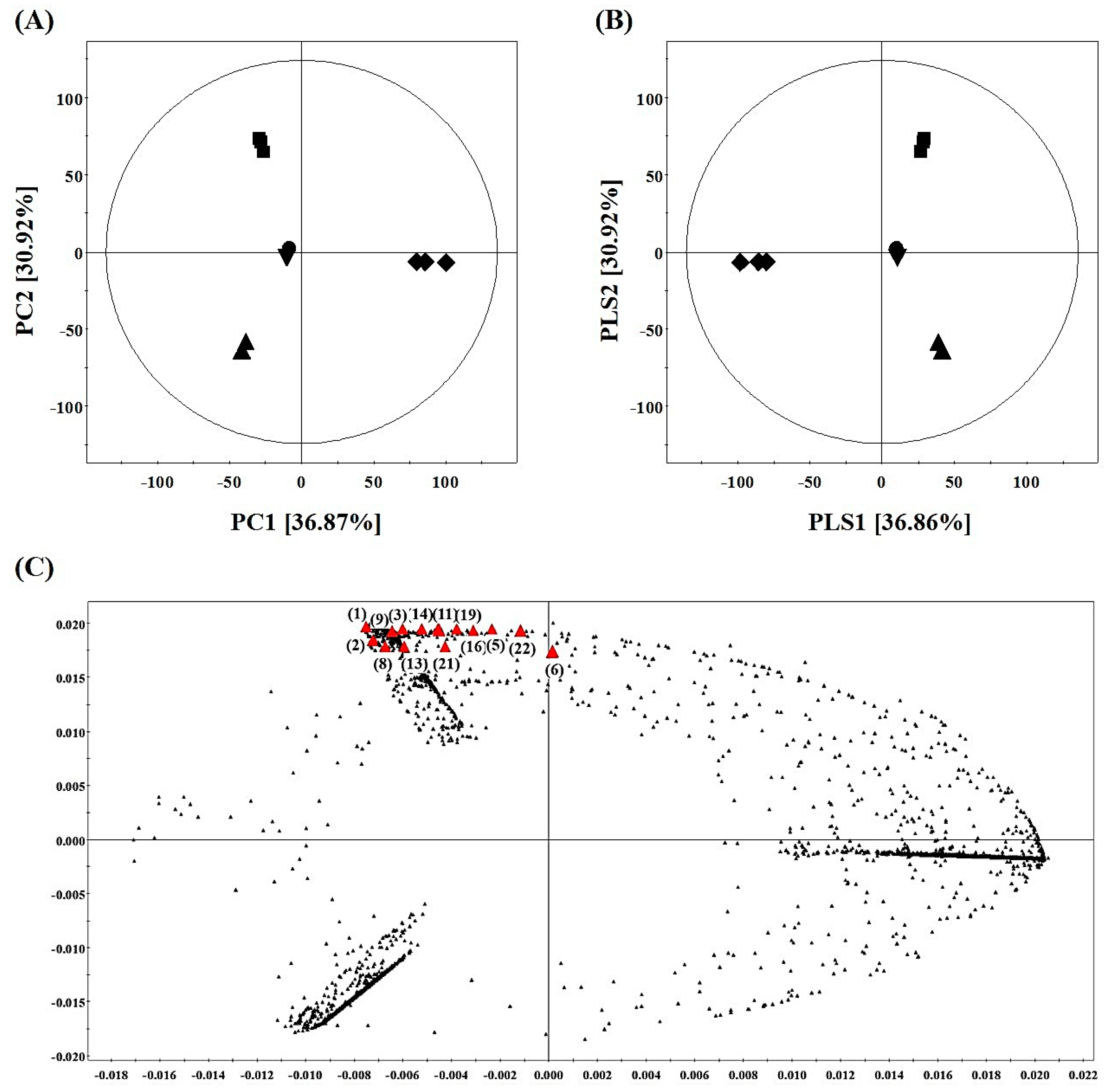
2.4. Metabolite Profiling of SPE MeOH Eluates of A. firma Extracts
2.5. Preparative HPLC Sub-Fractionation (15 and 31 Min) of 40% MeOH SPE Eluates and Antioxidant Assays
2.6. Bioactivity Validation of Proposed Metabolites
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Sample Information and Preparation
4.3. Metabolite Profiling of Plants from Betulaceae
4.4. Solid-Phase Extraction (SPE) for A. Firma Extracts
4.5. Metabolite Profiling and Multivariate Analysis of A. Firma SPE Samples
4.6. Bioactivity Assays
4.6.1. ABTS Radical Scavenging Assay of Plant Extracts Obtained from the Betulaceae Family
4.6.2. ABTS Radical Scavenging Assay for SPE Eluates of Extracts from A. firma
4.7. Cell Culture and Maintenance
XTT Cell Viability Assay using Extracts of A. firma Plants
4.8. Combined Preparative HPLC Analysis and ABTS Assay of 40% SPE Eluates of A. Firma Extracts
4.9. Analysis of Bioactive Prep-HPLC Sub-Fractions Using UHPLC-LTQ-IT-MS/MS and UHPLC-Q-Orbitrap-MS
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Chanwitheesuk, A.; Teerawutgulrag, A.; Rakariyatham, N. Screening of antioxidant activity and antioxidant compounds of some edible plants of Thailand. Food Chem. 2005, 92, 491–497. [Google Scholar] [CrossRef]
- Kasiotis, K.M.; Pratsinis, H.; Kletsas, D.; Haroutounian, S.A. Resveratrol and related stilbenes: Their anti-aging and anti-angiogenic properties. Food Chem. Toxicol. 2013, 61, 112–120. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.J.; Bao, J.L.; Wu, G.S.; Xu, W.S.; Huang, M.Q.; Chen, X.P.; Wang, Y.T. Quinones derived from plant secondary metabolites as anti-cancer agents. Anticancer Agents Med. Chem. 2013, 13, 456–463. [Google Scholar]
- Suh, D.H.; Lee, H.W.; Jung, E.S.; Singh, D.; Kim, S.H.; Lee, C.H. In vivo metabolomic interpretation of the anti-obesity effects of hyacinth bean (Dolichos lablab L.) administration in high-fat diet mice. Mol. Nutr. Food Res. 2017, 61, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Tagboto, S.; Townson, S. Antiparasitic properties of medicinal plants and other naturally occurring products. Adv. Parasitol. 2001, 50, 199–295. [Google Scholar] [PubMed]
- Han, J.S.; Lee, S.; Kim, H.Y.; Lee, C.H. MS-based metabolite profiling of aboveground and root components of Zingiber mioga and Officinale. Molecules 2015, 20, 16170–16185. [Google Scholar] [CrossRef]
- Kim, N.K.; Park, H.M.; Lee, J.; Ku, K.; Lee, C.H. Seasonal variations of metabolome and tyrosinase inhibitory activity of Lespedeza maximowiczii during growth periods. J. Agric. Food Chem. 2015, 63, 8631–8639. [Google Scholar] [CrossRef]
- Cuyckens, F.; Claeys, M. Mass spectrometry in the structural analysis of flavonoids. J. Mass Spectrom. 2004, 39, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yang, W.; Peng, Q.; He, Q.; Feng, Y.; Luo, S.; Yu, Z. Volatile phytochemical composition of rhizome of ginger after extraction by headspace solid-phase microextraction, petroleum ether extraction and steam distillation extraction. BDPS 2009, 4, 136–143. [Google Scholar]
- He, X.G.; Lin, L.Z.; Lian, L.Z. Electrospray High Performance Liquid Chromatography-Mass Spectrometry in Phytochemical Analysis of Kava (Piper methysticum) Extract. Planta Med. 1997, 63, 70–74. [Google Scholar] [CrossRef]
- Lee, S.; Oh, D.G.; Lee, S.; Kim, G.R.; Lee, J.S.; Son, Y.K.; Bae, C.H.; Yeo, J.; Lee, C.H. Chemotaxonomic metabolite profiling of 62 indigenous plant species and its correlation with bioactivities. Molecules 2015, 20, 19719–19734. [Google Scholar] [CrossRef] [PubMed]
- Son, S.Y.; Kim, N.K.; Lee, S.; Singh, D.; Kim, G.R.; Lee, J.S.; Yang, H.S.; Yeo, J.; Lee, S.; Lee, C.H. Metabolite fingerprinting, pathway analyses, and bioactivity correlations for plant species belonging to the Cornaceae, Fabaceae, and Rosaceae families. Plant Cell Rep. 2016, 35, 1917–1931. [Google Scholar] [CrossRef] [PubMed]
- He, X.G.; Lian, L.Z.; Lin, L.Z.; Bernart, M.W. High-performance liquid chromatography-electrospray mass spectrometry in phytochemical analysis of sour orange (Citrus aurantium L.). J. Chromatogr. A 1997, 791, 127–134. [Google Scholar] [CrossRef]
- Torre-Carbot, K.D.L.; Jauregui, O.; Gimeno, E.; Castellote, A.I.; Lamuela-Raventos, R.M.; Lopez-Sabater, M.C. Characterization and quantification of phenolic compounds in olive oils by solid-phase extraction, HPLC-DAD, and HPLC-MS/MS. J. Agric. Food Chem. 2005, 53, 4331–4340. [Google Scholar] [CrossRef] [PubMed]
- Kongstad, K.T.; Ozdemir, C.; Barzak, A.; Wubshet, S.G.; Staerk, D. Combined use of high-resolution α-glucosidase inhibition profiling and high-performance liquid chromatography-high-resolution mass spectrometry-solid-phase extraction-nuclear magnetic resonance spectroscopy for investigation of antidiabetic principles in crude plant extracts. J. Agric. Food Chem. 2015, 63, 2257–2263. [Google Scholar]
- Qureshi, R.A.; Ghufran, M.A.; Gilani, S.A.; Yousaf, Z.; Abbas, G.; Batool, A. Indigenous medicinal plants used by local women in southern Himalayan regions of Pakistan. Pak. J. Bot. 2009, 41, 19–25. [Google Scholar]
- Mariod, A.A.; Mohammed, N.M.F.; Nabag, F.O.; Hassan, A.A. Ethnobotanical study of three trees: Indigenous knowledge on trees used as cosmetic in Khartoum state, Sudan. AJPS 2014, 4, 178–182. [Google Scholar]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, S.; Pandey, M.M.; Rawat, A.K.S. Medicinal plants of the genus Betula-Traditional uses and a phytochemical-pharmacological review. J. Ethnopharmacol. 2015, 159, 62–83. [Google Scholar] [CrossRef]
- Ludwiczuk, A.; Saha, A.; Kuzuhara, T.; Asakawa, Y. Bioactivity guided isolation of anticancer constituents from leaves of Alnus sieboldiana (Betulaceae). Phytomedicine 2011, 18, 491–498. [Google Scholar] [CrossRef]
- Eom, H.J.; Kang, H.R.; Kim, H.K.; Jung, E.B.; Park, H.B.; Kang, K.S.; Kim, K.H. Bioactivity-guided isolation of antioxidant triterpenoids from Betula platyphylla var. japonica bark. Bioorg. Chem. 2016, 66, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.B.; Miyashiro, H.; Nakamura, N.; Hattori, M.; Park, J.C. Effects of triterpenoids and flavonoids isolated from Alnus firma on HIV-1 viral enzymes. Arch. Pharm. Res. 2006, 30, 820–826. [Google Scholar] [CrossRef]
- Sati, S.C.; Sati, N.; Sati, O.P. Bioactive constituents and medicinal importance of genus Alnus. Pharmacogn. Rev. 2011, 5, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Jung, Y.; Shin, J.H.; Kim, H.K.; Moon, B.C.; Ryu, D.H.; Hwang, G.S. Secondary metabolite profiling of Curcuma species grown at different locations using GC/TOF and UPLC/Q-TOF MS. Molecules 2014, 19, 9535–9551. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.D.; Manchester, S.R.; Sun, H.Y. Phylogeny and evolution of the Betulaceae as inferred from DNA sequences, morphology, and paleobotany. Am. Bot. 1999, 86, 1168–1181. [Google Scholar] [CrossRef]
- Yoo, K.O.; Wen, J. Phylogeny and biogeography of Carpinus and subfamily Coryloideae (Betulaceae). Int. J. Plant Sci. 2002, 163, 641–650. [Google Scholar] [CrossRef]
- del Rio, L.A. ROS and RNS in plant physiology: An overview. J. Exp. Bot. 2015, 66, 2827–2837. [Google Scholar] [CrossRef] [PubMed]
- Roselló-Soto, E.; Galanakis, C.M.; Brnčić, M.; Orlien, V.; Trujillo, F.J.; Mawson, R.; Knoerzer, K.; Tiwari, B.K.; Barba, F.J. Clean recovery of antioxidant compounds from plant foods, by-products and algae assisted by ultrasounds processing. Modeling approaches to optimize processing conditions. Trends Food Sci. Tech. 2015, 42, 134–149. [Google Scholar] [CrossRef]
- Romanik, G.; Gilgenast, E.; Przyjazny, A.; Kaminski, M. Techniques of preparing plant material for chromatographic separation and analysis. J. Biochem. Biophys. Methods 2007, 70, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Leopoldini, M.; Marino, T.; Russo, M.; Toscano, M. Antioxidant properties of phenolic compounds: H-atom versus electron transfer mechanism. J. Phys. Chem. 2004, 108, 4916–4922. [Google Scholar] [CrossRef]
- Badhani, B.; Sharma, N.; Kakkar, R. Gallic acid: A versatile antioxidant with promising therapeutic and industrial applications. RSC Adv. 2015, 5, 27540–27557. [Google Scholar] [CrossRef]
- Stojanovic, S.; Brede, O. Elementary reactions of the antioxidant actions of trans-stilbene derivatives: Resveratrol, pinosylvin and 4-hydroxystilbene. Phys. Chem. Chem. Phys. 2002, 4, 757–764. [Google Scholar] [CrossRef]
- Kuroyanagi, M.; Shimomae, M.; Nagashima, Y.; Muto, N.; Okuda, T.; Kawahara, N.; Nakane, T.; Sano, T. New diarylheptanoids from Alnus japonica and their antioxidative activity. Chem. Pharm. Bull. 2005, 53, 1519–1523. [Google Scholar] [CrossRef] [PubMed]
- Biskup, I.; Golonka, I.; Gamian, A.; Sroka, Z. Antioxidant activity of selected phenols estimated by ABTS and FRAP methods. Postepy Hig. Med. Dosw. 2013, 67, 958–963. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.H.; Tsushida, T.; Nakahara, K.; Terao, J. Identification of quercetin 3-O-β-d-glucuronide as an antioxidative metabolite in rat plasma after oral administration of quercetin. Free Radic. Biol. Med. 2001, 30, 1274–1285. [Google Scholar] [CrossRef]
- Kalaivani, T.; Rajasekaran, C.; Mathew, L. Free radical scavenging, cytotoxic, and hemolytic activities of an active antioxidant compound ethyl gallate from leaves of Acacia nilotica (L.) wild. Ex. Delile subsp. Indica (Benth.) Brenan. J. Food Sci. 2011, 76, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zuo, Z.; Lin, G. Intestinal and hepatic glucuronidation of flavonoids. Mol. Pharm. 2007, 4, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Re, R.; Pellegrini, N.; Proteggente, A.; Pannala, A.; Yang, M.; Rice-Evans, C. Antioxidant activity applying an improved ABTS radical cation decolorization assay. Free Radic. Biol. Med. 1999, 26, 1231–1237. [Google Scholar] [CrossRef]
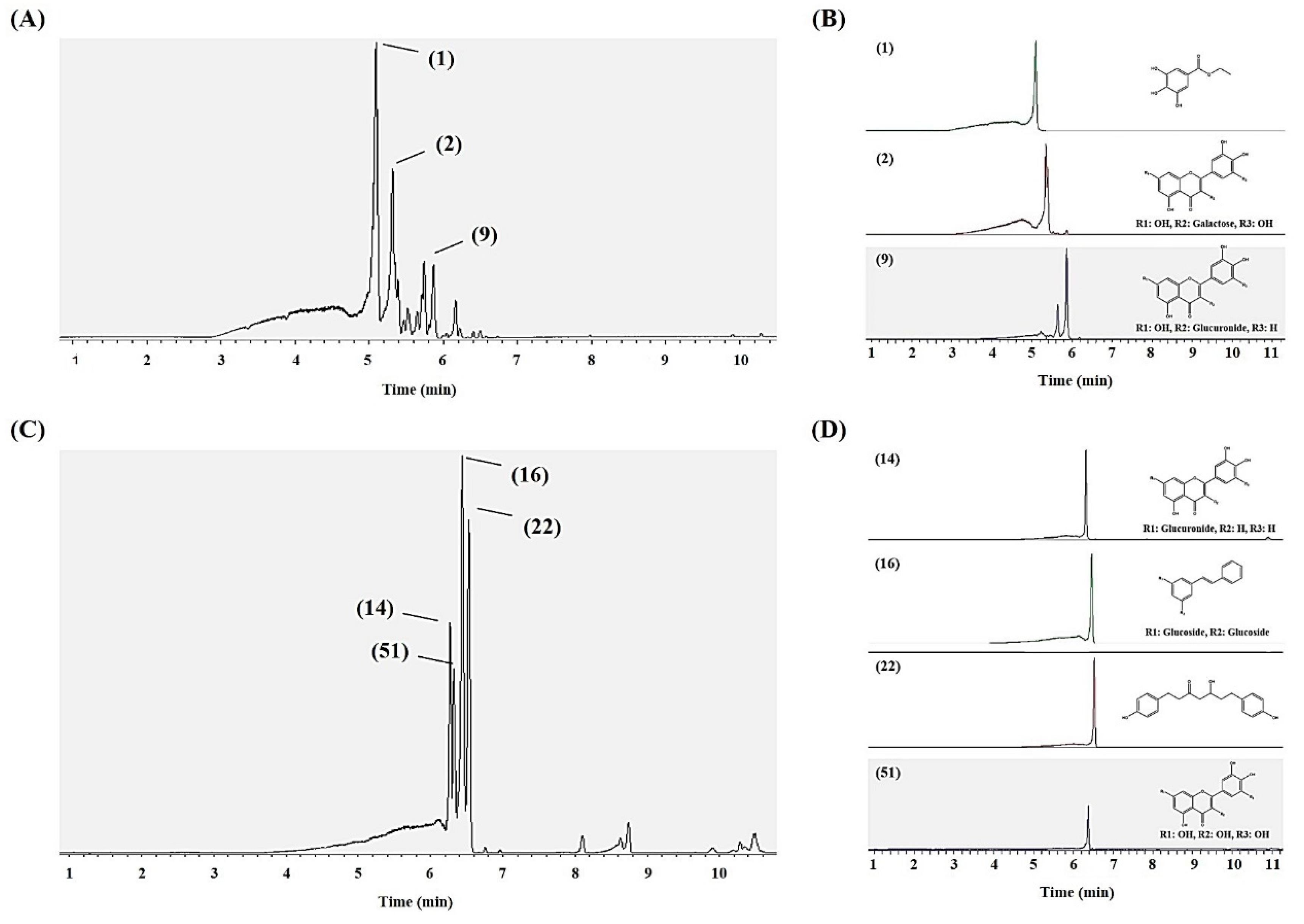
| No. a | Putative Identification b | UHPLC-LTQ-IT-MS/MS | |||||
|---|---|---|---|---|---|---|---|
| RT c (min) | m/z Nega d | m/z Posi e | M.W. f | MSn Fragment Pattern | UV(nm) | ||
| 1 | Ethyl gallate | 6.72 | 197 | 199 | 198 | 159 [posi] g | 217 |
| 2 | Myricetin-3-O-galactoside | 7.04 | 479 | 481 | 480 | 319 [posi] | 215,357 |
| 3 | N.I. 1 | 7.24 | 343 | 345 | 344 | 269 | 247 |
| 5 | Myricetin-3-O-pentoside | 7.44 | 449 | 451 | 450 | 319 [posi] | 218,355 |
| 6 | Quercetin-3-O-glucoside | 7.57 | 463 | 465 | 464 | 301 > 179, 151 | 207,255 |
| 8 | N.I. 2 | 7.72 | 413 | 415 | 414 | 311 | - |
| 9 | Quercetin-3-O-glucuronide | 7.73 | 477 | 479 | 478 | 301 | - |
| 11 | Hirsutoside | 8.02 | 491 | 493 | 492 | 311 | - |
| 13 | N.I. 3 | 8.03 | 537 | - | - | - | - |
| 14 | Luteolin-7-O-glucuronide | 8.22 | 461 | 463 | 462 | 285, 267 | - |
| 16 | Pinosylvin diglucoside | 8.27 | 581[M+FA]− | 537 | 536 | 375 > 213 [posi] | 214,274 |
| 19 | N.I. 6 | 8.47 | 325[M+FA]− | 303[M + Na]+ | 280 | 113 | - |
| 21 | N.I. 7 | 8.74 | 327 | 329 | 328 | - | - |
| 22 | Platyphyllonol | 8.79 | 313 | 315 | 314 | 297 [posi] | 205,366 |
| Fractions | No a | Putative Identification | UHPLC-LTQ-IT-MS/MS | UHPLC-Q-Orbitrap-MS | ID h | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RT b (min) | m/z Nega c | m/z Posi d | M.W. e | MSn Fragment Pattern | UV (nm) | RT (min) | m/z Nega | M.F.b g | Δppm | ||||
| 15 min | 1 | Ethyl gallate | 6.59 | 197 | 199 | 198 | 159 [posi] f | 217 | 5.10 | 197.0449 | C9H10O5 | 2.2 | STD |
| 2 | Myricetin-3-O-galactoside | 6.95 | 479 | 481 | 480 | 319 [posi] | 215,357 | 5.34 | 479.0837 | C21H20O13 | –0.3 | STD | |
| 3 | N.I. 1 | 7.17 | 343 | 345 | 344 | 269 | 247 | - | - | - | - | - | |
| 8 | N.I. 2 | 7.60 | 413 | 415 | 414 | 311 | - | - | - | - | - | - | |
| 9 | Quercetin-3-O-glucuronide | 7.60 | 477 | 479 | 478 | 301 | - | 5.87 | 477.0685 | C21H18O13 | –0.2 | STD | |
| 31 min | 14 | Luteolin-7-O-glucuronide | 8.08 | 461 | 463 | 462 | 285, 267 | - | 6.32 | 461.0734 | C21H18O12 | 0.2 | STD |
| 16 | Pinosylvin diglucoside | 8.13 | 581 [M+COOH]− | 537 | 536 | 375 > 213 [posi] | 214,274 | 6.44 | 581.1887 | C26H32O12 | –0.8 | CCD | |
| 21 | N.I. 7 | 8.61 | 327 | 329 | 328 | - | - | - | - | - | - | - | |
| 22 | Platyphyllonol | 8.64 | 313 | 315 | 314 | 297 [posi] | 205,366 | 6.53 | 313.1449 | C19H22O4 | 0.5 | CCD | |
| 49 | N.I. 17 | 8.36 | 447 | 449 | 448 | 317 | - | - | - | - | - | - | |
| 50 | N.I. 18 | 8.53 | 405 | 407 | 406 | 303 | 295 | - | - | - | - | - | |
| 51 | Myricetin | 8.97 | 317 | 319 | 318 | 289 | - | 6.38 | 317.0669 | C15H10O8 | –0.2 | STD | |
| 52 | N.I. 19 | 8.97 | 287 | 289 | 288 | 153 [posi] | - | - | - | - | - | - | |
| 53 | N.I. 20 | 7.94 | 542 | - | - | 466 | 215,247,362 | - | - | - | - | - | |
| Compounds | EC50 (µg/mL) | Regression Curve | R2 |
|---|---|---|---|
| Ethyl gallate (1) | 61.6 | y = –0.0048x + 0.6745 | 0.9872 |
| Quercetin-3-O-glucuronoide (9) | 64.6 | y = −0.0048x + 0.6745 | 0.9872 |
| Myricein-3-O-galactoside (2) | 144.9 | y = −0.0024x + 0.7120 | 0.9933 |
| Quercetin-3-O-glucoside (6) | 176.1 | y = −0.0019x + 0.6958 | 0.9953 |
| Luteolin-7-O-glucuronide (14) | 215.3 | y = −0.0016x + 0.6913 | 0.9959 |
| Myricetin (51) | 221.3 | y = −0.0016x + 0.7154 | 0.9997 |
| No. | Family | Genus | Species | Collection Area | Collection Date |
|---|---|---|---|---|---|
| 1 | Betulaceae | Alnus | firma | Sin-ri, Goryeong-eup, Goryeong-gun, Gyeongsangbuk-do | 2014-07-23 |
| 2 | Betulaceae | Alnus | hirsuta | Sangjung-ri, Geumgwang-myeon, Anseong-si, Gyeonggi-do | 2014-07-25 |
| 3 | Betulaceae | Alnus | japonica | Yonggi-ri, Gibuk-myeon, Buk-gu, Pohang-si, Gyeongsangbuk-do | 2014-07-30 |
| 4 | Betulaceae | Betula | schmidtii | Icheon-ri, Sangbuk-myeon, Ulju-gun, Ulsan | 2014-08-01 |
| 5 | Betulaceae | Betula | dahurica | Ungyo-ri, Bangnim-myeon, Pyeongchang-gun, Gangwon-do | 2014-08-08 |
| 6 | Betulaceae | Betula | pendula | Sogye-ri, Hwanggan-myeon, Yeongdong-gun, Chungcheongbuk-do | 2014-08-14 |
| 7 | Betulaceae | Carpinus | cordata | Apgok-ri, Bongsan-myeon, Hapcheon-gun, Gyeongsangnam-do | 2014-07-24 |
| 8 | Betulaceae | Carpinus | turczaninowii | Jiro-ri, Byeongyeong-myeon, Gangjin-gun, Jeollanam-do | 2014-08-12 |
| 9 | Betulaceae | Carpinus | laxiflora | Seonheul-ri, Jocheon-eup, Jeju-si, Jeju special self-governing province | 2014-08-24 |
| 10 | Betulaceae | Corylus | heterophylla | Apgok-ri, Bongsan-myeon, Hapcheon-gun, Gyeongsangnam-do | 2014-07-24 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Oh, D.-G.; Singh, D.; Lee, H.J.; Kim, G.R.; Lee, S.; Lee, J.S.; Lee, C.H. Untargeted Metabolomics Toward Systematic Characterization of Antioxidant Compounds in Betulaceae Family Plant Extracts. Metabolites 2019, 9, 186. https://doi.org/10.3390/metabo9090186
Lee S, Oh D-G, Singh D, Lee HJ, Kim GR, Lee S, Lee JS, Lee CH. Untargeted Metabolomics Toward Systematic Characterization of Antioxidant Compounds in Betulaceae Family Plant Extracts. Metabolites. 2019; 9(9):186. https://doi.org/10.3390/metabo9090186
Chicago/Turabian StyleLee, Sunmin, Dong-Gu Oh, Digar Singh, Hye Jin Lee, Ga Ryun Kim, Sarah Lee, Jong Seok Lee, and Choong Hwan Lee. 2019. "Untargeted Metabolomics Toward Systematic Characterization of Antioxidant Compounds in Betulaceae Family Plant Extracts" Metabolites 9, no. 9: 186. https://doi.org/10.3390/metabo9090186





