Multiplatform Urinary Metabolomics Profiling to Discriminate Cachectic from Non-Cachectic Colorectal Cancer Patients: Pilot Results from the ColoCare Study
Abstract
1. Introduction
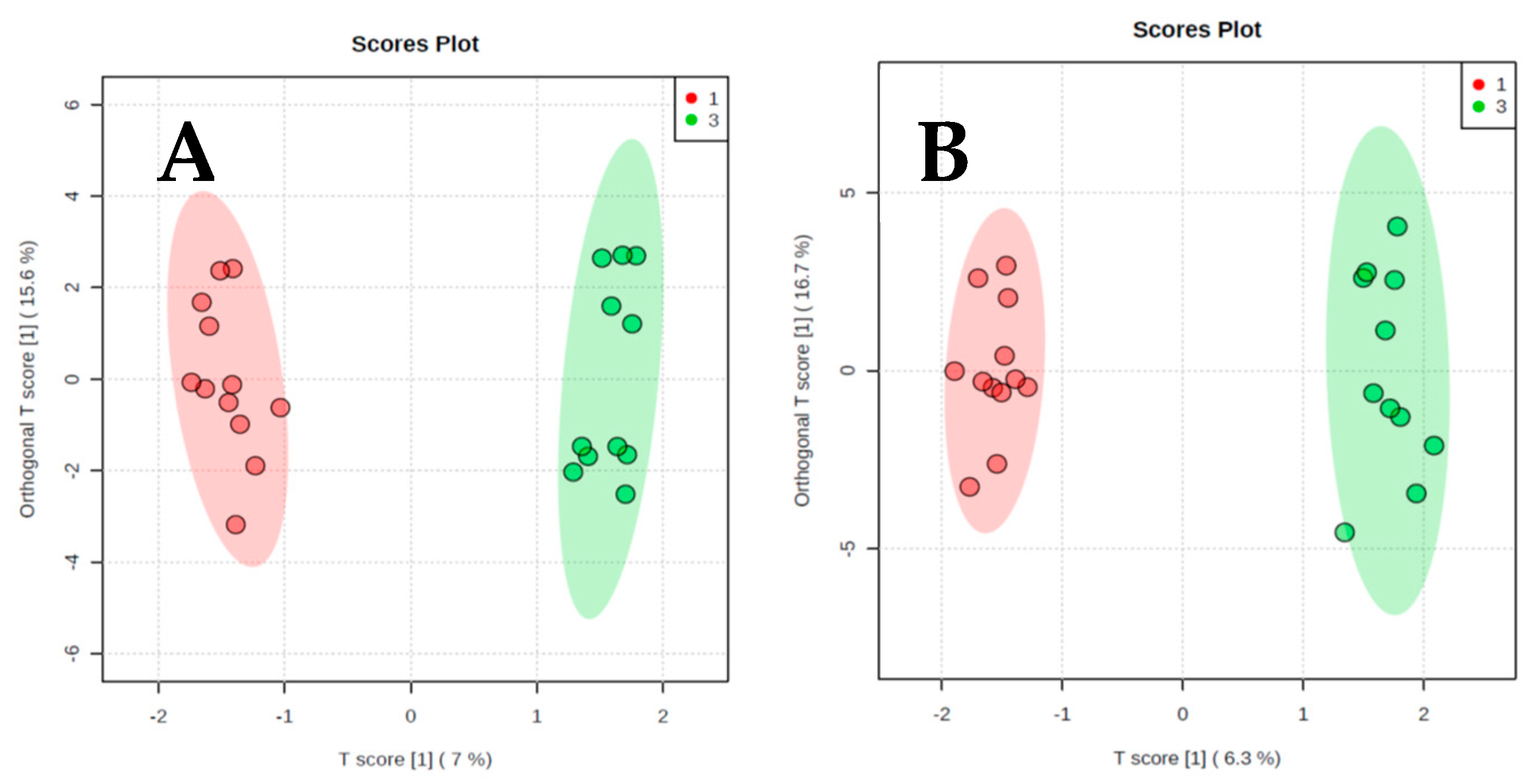
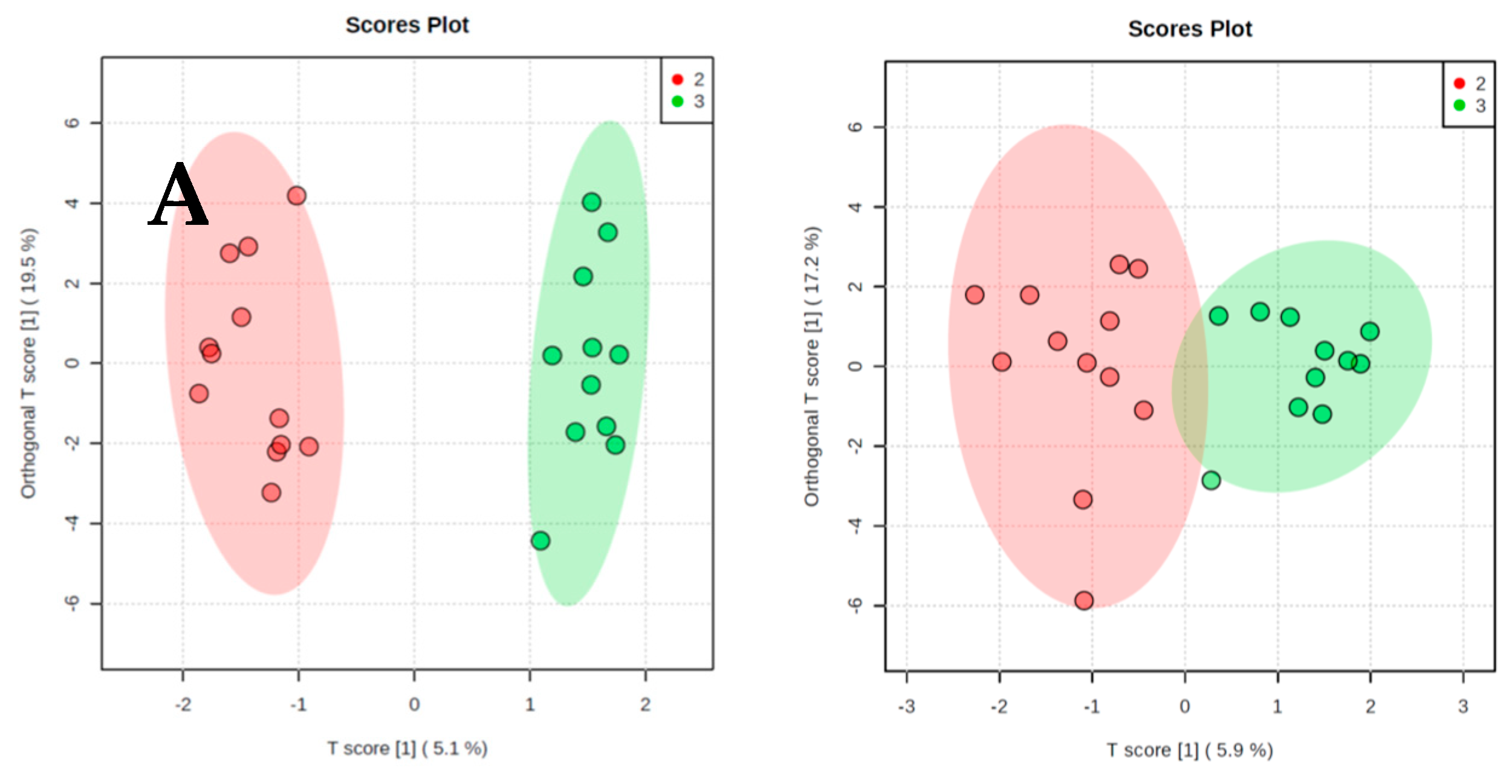
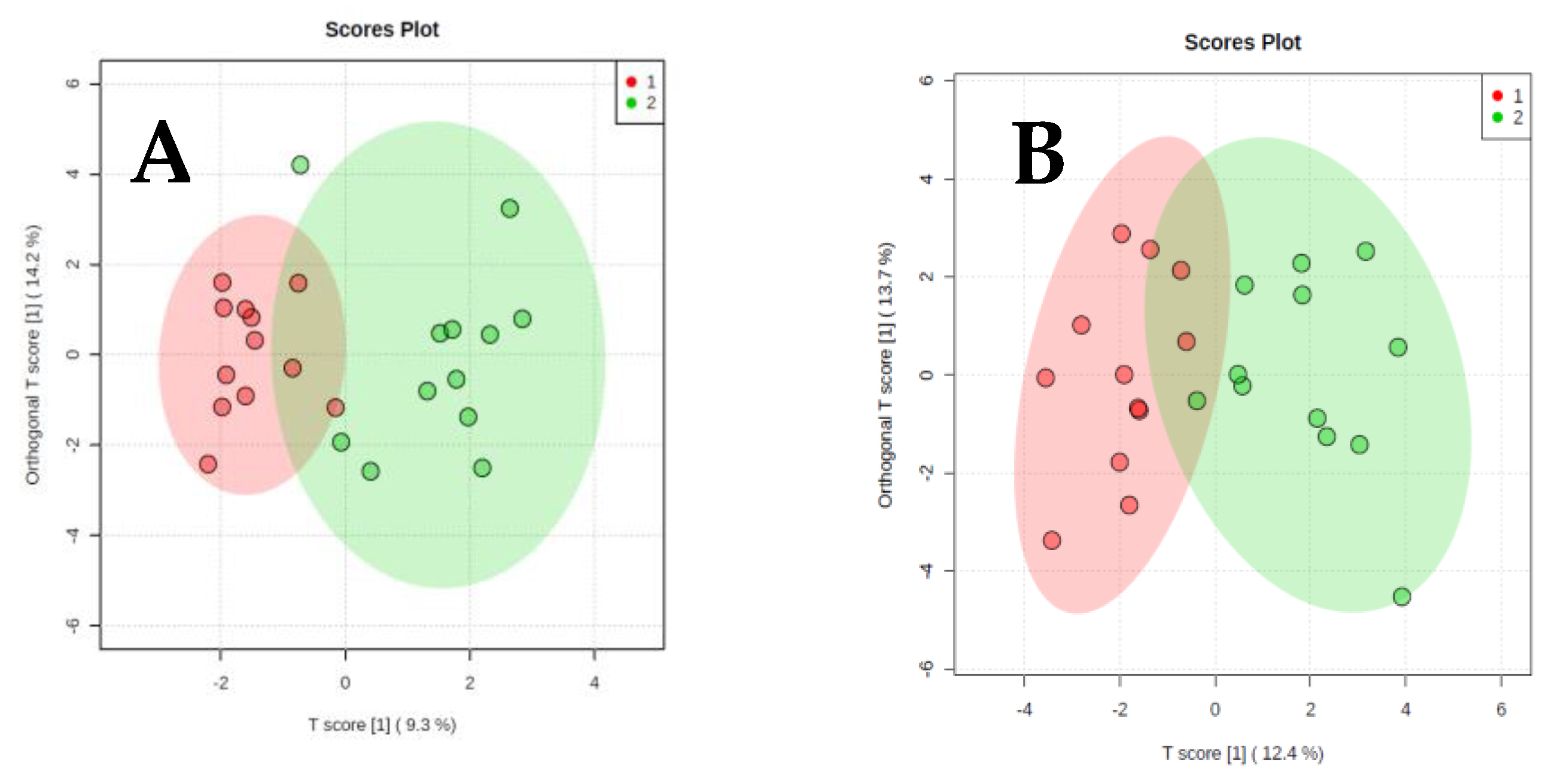
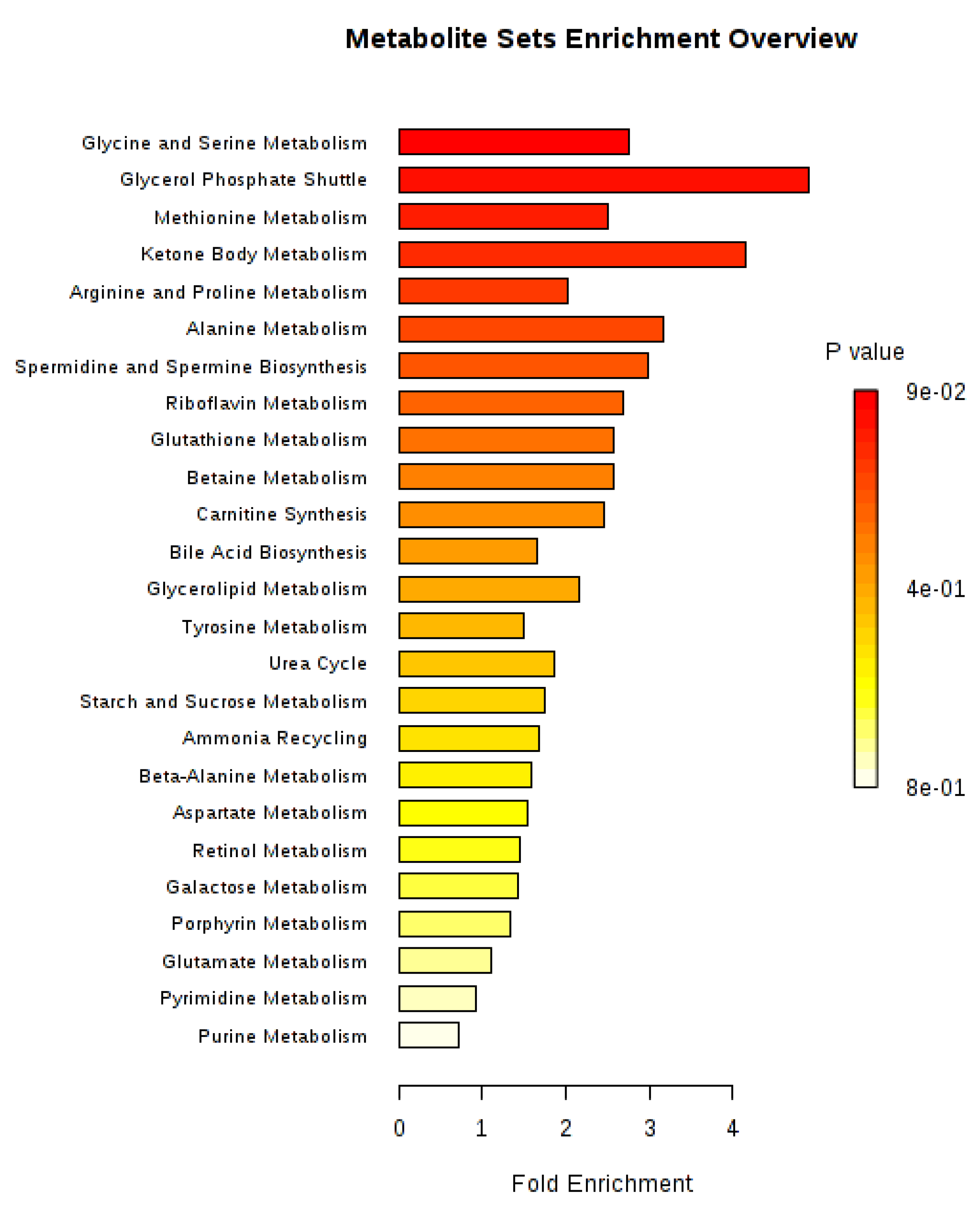
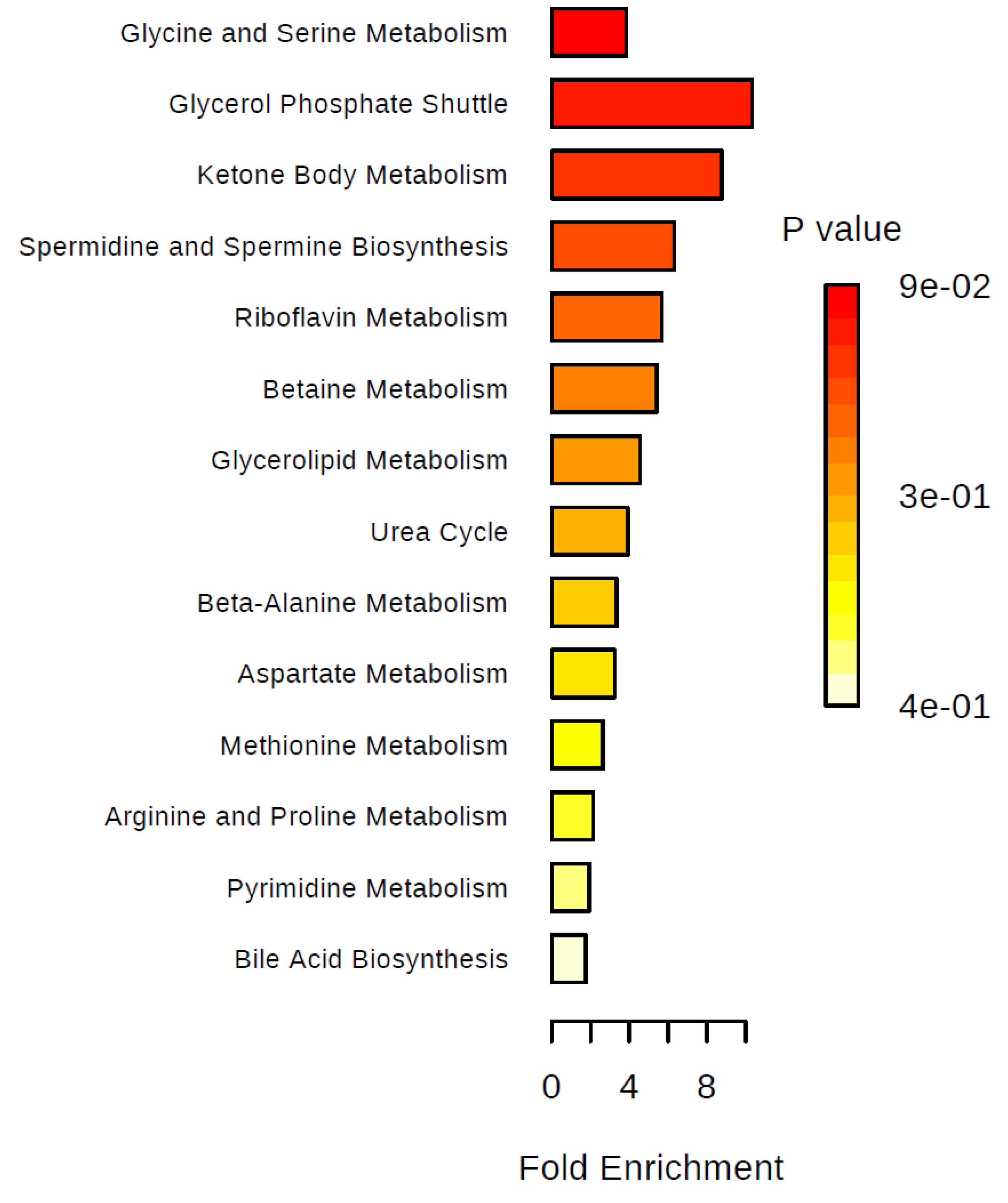
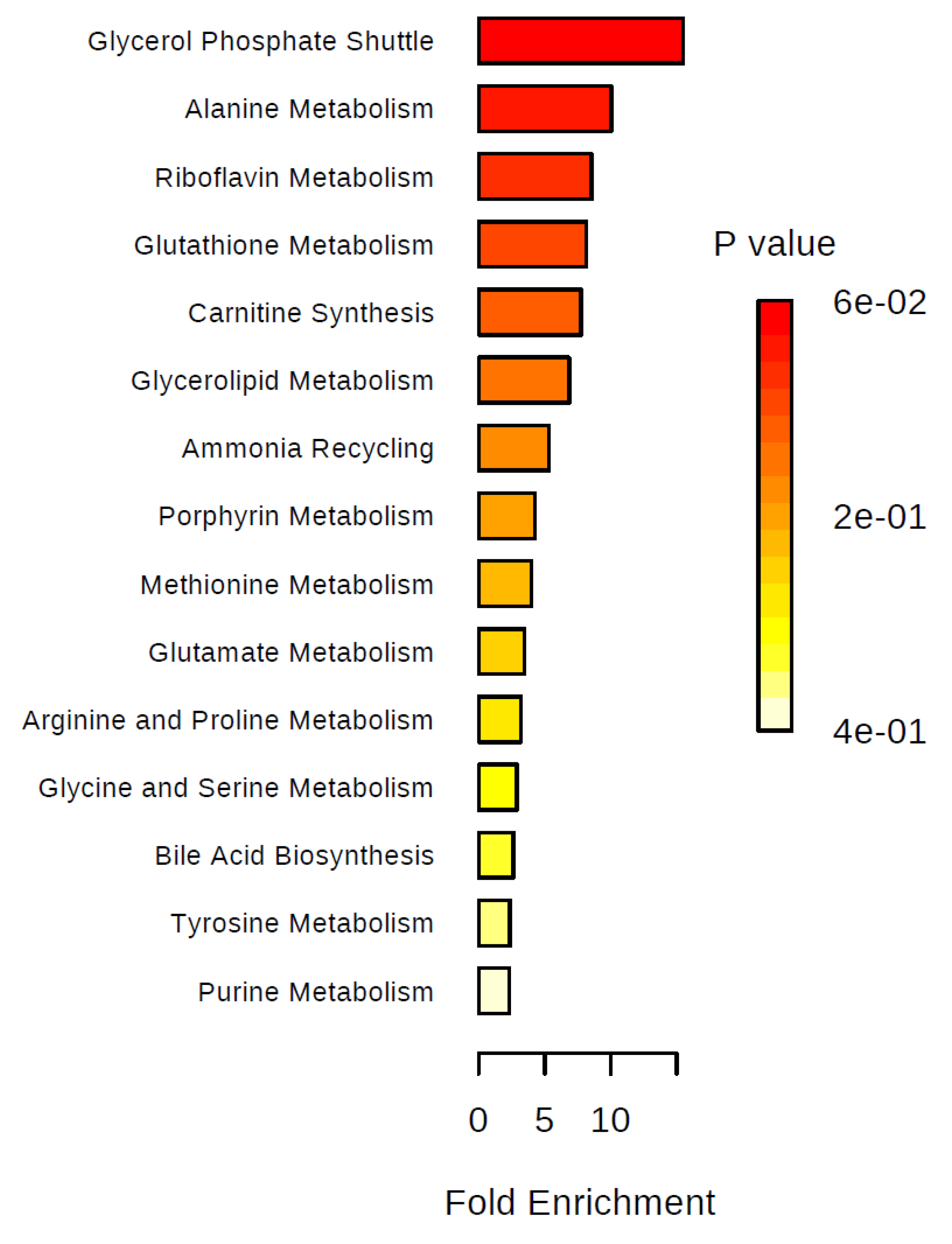
2. Results
3. Discussion
4. Materials and Methods
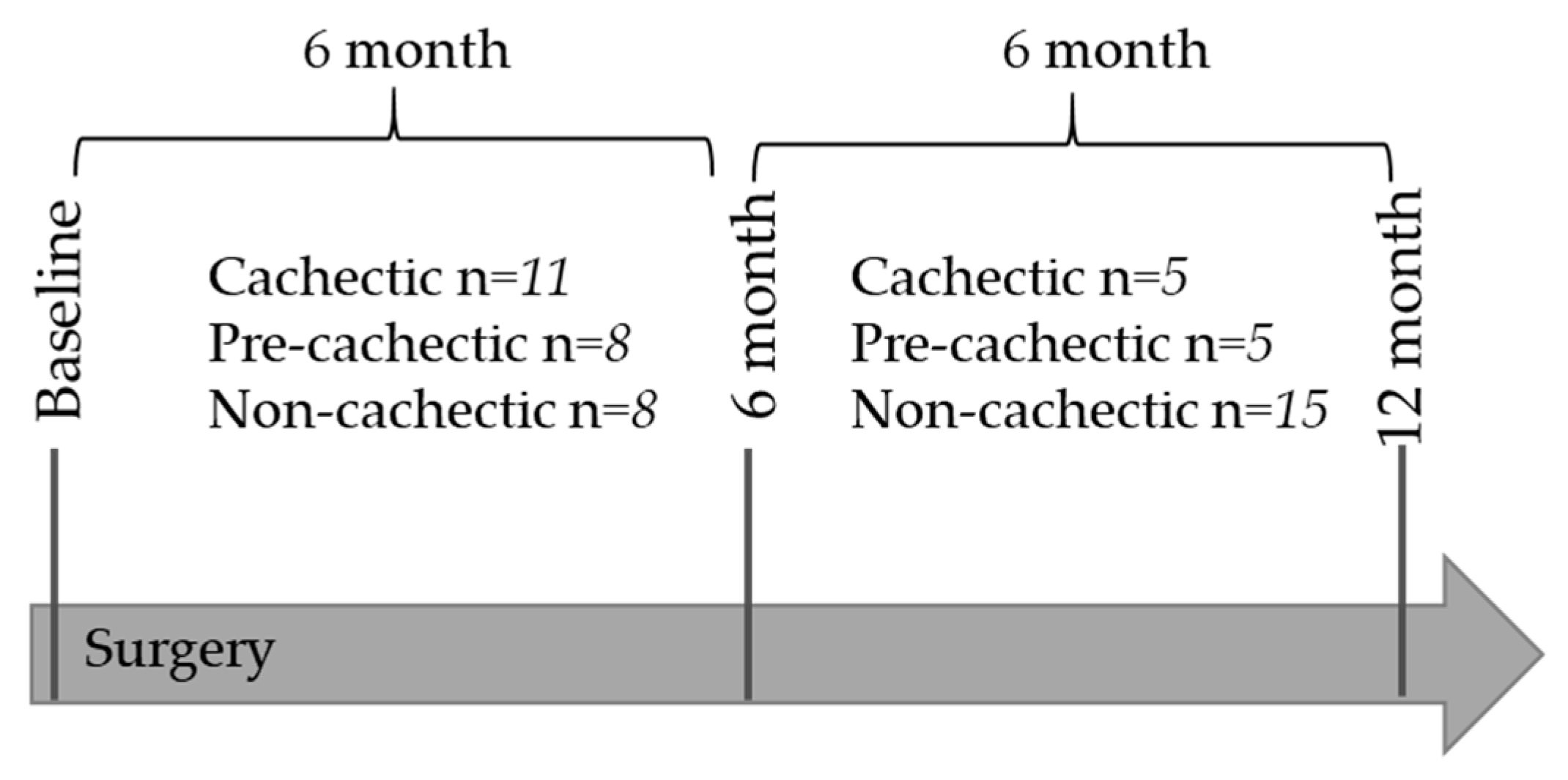
4.1. Patient Selection and Classification
4.2. Biospecimen Collection
4.3. Laboratory Analyses and Quality Control Measurements
4.4. Sample Preparation and Quality Control: GC–MS
4.5. Sample Preparation and Quality Control: 1H-NMR
4.6. Data Pre-Processing
4.7. Normalization of Metabolite Concentrations
4.8. Area-Based Computed Tomography (CT) Quantification of Abdominal Adipose Tissue and Muscle Tissue Area
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vagnildhaug, O.M.; Balstad, T.R.; Almberg, S.S.; Brunelli, C.; Knudsen, A.K.; Kaasa, S.; Thronæs, M.; Laird, B.; Solheim, T.S. A cross-sectional study examining the prevalence of cachexia and areas of unmet need in patients with cancer. Support. Care Cancer 2018, 26, 1871–1880. [Google Scholar] [CrossRef]
- Fearon, K.; Strasser, F.; Anker, S.D.; Bosaeus, I.; Bruera, E.; Fainsinger, R.L.; Jatoi, A.; Loprinzi, C.; Macdonald, N.; Mantovani, G.; et al. Definition and classification of cancer cachexia: An international consensus. Lancet Oncol. 2011, 12, 489–495. [Google Scholar] [CrossRef]
- Rohm, M.; Zeigerer, A.; Machado, J.; Herzig, S. Energy metabolism in cachexia. EMBO Rep. 2019, 20, e47258. [Google Scholar] [CrossRef]
- Schmidt, S.F.; Rohm, M.; Herzig, S.; Berriel Diaz, M. Cancer Cachexia: More Than Skeletal Muscle Wasting. Trends Cancer 2018, 4, 849–860. [Google Scholar] [CrossRef]
- Lee, J.; Lin, J.B.; Wu, M.H.; Jan, Y.T.; Chang, C.L.; Huang, C.Y.; Sun, F.; Chen, Y. Muscle radiodensity loss during cancer therapy is predictive for poor survival in advanced endometrial cancer. J. Cachexia Sarcopenia Muscle 2019, 10, 814–826. [Google Scholar] [CrossRef]
- Yoon, S.B.; Choi, M.H.; Song, M.; Lee, J.H.; Lee, I.S.; Lee, M.A.; Hong, T.H.; Jung, E.S.; Choi, M. Impact of preoperative body compositions on survival following resection of biliary tract cancer. J. Cachexia Sarcopenia Muscle 2019, 10, 794–802. [Google Scholar] [CrossRef]
- Koppe, L.; Fouque, D.; Kalantar-Zadeh, K. Kidney cachexia or protein-energy wasting in chronic kidney disease: Facts and numbers. J. Cachexia Sarcopenia Muscle 2019, 10, 479–484. [Google Scholar] [CrossRef]
- Kurk, S.; Peeters, P.; Stellato, R.; Dorresteijn, B.; de Jong, P.; Jourdan, M.; Creemers, G.; Erdkamp, F.; Jongh, F.; Kint, P.; et al. Skeletal muscle mass loss and dose-limiting toxicities in metastatic colorectal cancer patients. J. Cachexia Sarcopenia Muscle 2019, 10, 803–813. [Google Scholar] [CrossRef]
- Sasaki, S.; Oki, E.; Saeki, H.; Shimose, T.; Sakamoto, S.; Hu, Q.; Kudo, K.; Tsuda, Y.; Nakashima, Y.; Ando, K.; et al. Skeletal muscle loss during systemic chemotherapy for colorectal cancer indicates treatment response: A pooled analysis of a multicenter clinical trial (KSCC 1605-A). Int. J. Clin. Oncol. 2019. [Google Scholar] [CrossRef]
- Pigna, E.; Berardi, E.; Aulino, P.; Rizzuto, E.; Zampieri, S.; Carraro, U.; Kern, H.; Merigliano, S.; Gruppo, M.; Mericskay, M.; et al. Aerobic Exercise and Pharmacological Treatments Counteract Cachexia by Modulating Autophagy in Colon Cancer. Sci. Rep. 2016, 6, 26991. [Google Scholar] [CrossRef]
- Kasi, P.M.; Zafar, S.Y.; Grothey, A. Is obesity an advantage in patients with colorectal cancer? Expert Rev. Gastroenterol. Hepatol. 2015, 9, 1339–1342. [Google Scholar] [CrossRef]
- Ali, R.; Baracos, V.E.; Sawyer, M.B.; Bianchi, L.; Roberts, S.; Assenat, E.; Mollevi, C.; Senesse, P. Lean body mass as an independent determinant of dose-limiting toxicity and neuropathy in patients with colon cancer treated with FOLFOX regimens. Cancer Med. 2016, 5, 607–616. [Google Scholar] [CrossRef]
- Prado, C.M.; Cushen, S.J.; Orsso, C.E.; Ryan, A.M. Sarcopenia and cachexia in the era of obesity: Clinical and nutritional impact. Proc. Nutr. Soc. 2016, 75, 188–198. [Google Scholar] [CrossRef]
- Bruggeman, A.R.; Kamal, A.H.; Leblanc, T.W.; Ma, J.D.; Baracos, V.E.; Roeland, E.J. Cancer Cachexia: Beyond Weight Loss. J. Oncol. Pract. 2016, 12, 1163–1171. [Google Scholar] [CrossRef]
- Loumaye, A.; Thissen, J.P. Biomarkers of cancer cachexia. Clin. Biochem. 2017, 50, 1281–1288. [Google Scholar] [CrossRef]
- Liesenfeld, D.; Habermann, N.; Toth, R.; Owen, R.; Frei, E.; Böhm, J.; Schrotz-King, P.; Klika, K.D.; Ulrich, C.M. Changes in urinary metabolic profiles of colorectal cancer patients enrolled in a prospective cohort study (ColoCare). Metabolomics Off. J. Metabolomic Soc. 2015, 11, 998–1012. [Google Scholar] [CrossRef]
- Delphan, M.; Lin, T.D.; Liesenfeld, D.B.; Nattenmüller, J.; Böhm, J.T.; Gigic, B.; Habermann, N.; Zielske, L.; Schrotz-King, P.; Schneider, M.; et al. Associations of branched-chain amino acids with parameters of energy balance and survival in colorectal cancer patients: Results from the ColoCare Study. Metabolomics Off. J. Metabolomic Soci. 2018, 2018, 22. [Google Scholar] [CrossRef]
- Yang, Q.J.; Zhao, J.R.; Hao, J.; Li, B.; Huo, Y.; Han, Y.L.; Wan, L.L.; Li, J.; Huang, J.; Lu, J.; et al. Serum and urine metabolomics study reveals a distinct diagnostic model for cancer cachexia. J. Cachexia Sarcopenia Muscle 2018, 9, 71–85. [Google Scholar] [CrossRef]
- Cala, M.P.; Agulló-Ortuño, M.T.; Prieto-García, E.; González-Riano, C.; Parrilla-Rubio, L.; Barbas, C.; Díaz-García, C.V.; Garcia, A.; Pernaut, C.; Adeva, J.; et al. Multiplatform plasma fingerprinting in cancer cachexia: A pilot observational and translational study. J. Cachexia Sarcopenia Muscle 2018, 9, 348–357. [Google Scholar] [CrossRef]
- Fujiwara, Y.; Kobayashi, T.; Chayahara, N.; Imamura, Y.; Toyoda, M.; Kiyota, N.; Mukohara, T.; Nishiumi, S.; Azuma, T.; Yoshida, M.; et al. Metabolomics Evaluation of Serum Markers for Cachexia and Their Intra-Day Variation in Patients with Advanced Pancreatic Cancer. PLoS ONE 2014, 9, e113259. [Google Scholar] [CrossRef]
- Han, S.; Pan, Y.; Yang, X.; Da, M.; Wei, Q.; Gao, Y.; Qi, Q.; Ru, L. Intestinal microorganisms involved in colorectal cancer complicated with dyslipidosis. Cancer Biol. Ther. 2019, 20, 81–89. [Google Scholar] [CrossRef]
- Caldow, M.K.; Ham, D.J.; Godeassi, D.P.; Chee, A.; Lynch, G.S.; Koopman, R. Glycine supplementation during calorie restriction accelerates fat loss and protects against further muscle loss in obese mice. Clin. Nutr. 2016, 35, 1118–1126. [Google Scholar] [CrossRef]
- Koopman, R.; Caldow, M.K.; Ham, D.J.; Lynch, G.S. Glycine metabolism in skeletal muscle: Implications for metabolic homeostasis. Curr. Opin. Clin. Nutr. Metab. Care 2017, 20, 237–242. [Google Scholar] [CrossRef]
- Kremer, J.C.; Prudner, B.C.; Lange, S.E.S.; Bean, G.R.; Schultze, M.B.; Brashears, C.B.; Radyk, M.D.; Redlich, N.; Tzeng, S.-C.; Kami, K.; et al. Arginine Deprivation Inhibits the Warburg Effect and Upregulates Glutamine Anaplerosis and Serine Biosynthesis in ASS1-Deficient Cancers. Cell Rep. 2017, 18, 991–1004. [Google Scholar] [CrossRef]
- Albaugh, V.L.; Pinzon-Guzman, C.; Barbul, A. Arginine-Dual roles as an onconutrient and immunonutrient. J. Surg. Oncol. 2017, 115, 273–280. [Google Scholar] [CrossRef]
- Vynnytska-Myronovska, B.O.; Kurlishchuk, Y.; Chen, O.; Bobak, Y.; Dittfeld, C.; Hüther, M.; Kunz-Schughart, L.A.; Stasyk, O.V. Arginine starvation in colorectal carcinoma cells: Sensing, impact on translation control and cell cycle distribution. Exp. Cell Res. 2016, 341, 67–74. [Google Scholar] [CrossRef]
- Byeon, S.E.; Yi, Y.S.; Lee, J.; Yang, W.S.; Kim, J.H.; Kim, J.; Hong, S.; Kim, J.-H.; Cho, J.Y. Hydroquinone Exhibits In Vitro and In Vivo Anti-Cancer Activity in Cancer Cells and Mice. Int. J. Mol. Sci. 2018, 19, 903. [Google Scholar] [CrossRef]
- Mráček, T.; Drahota, Z.; Houštěk, J. The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim. Biophys. Acta 2013, 1827, 401–410. [Google Scholar] [CrossRef]
- Eto, K.; Tsubamoto, Y.; Terauchi, Y.; Sugiyama, T.; Kishimoto, T.; Takahashi, N.; Yamauchi, N.; Kubota, N.; Murayama, S.; Aizawa, T.; et al. Role of NADH shuttle system in glucose-induced activation of mitochondrial metabolism and insulin secretion. Science 1999, 283, 981–985. [Google Scholar] [CrossRef]
- Liu, X.; Qu, H.; Zheng, Y.; Liao, Q.; Zhang, L.; Liao, X.; Xiong, X.; Wang, Y.; Zhang, R.; Wang, H.; et al. Mitochondrial glycerol 3-phosphate dehydrogenase promotes skeletal muscle regeneration. EMBO Mol. Med. 2018, 10, e9390. [Google Scholar] [CrossRef]
- Koutnik, A.P.; D’Agostino, D.P.; Egan, B. Anticatabolic Effects of Ketone Bodies in Skeletal Muscle. Trends Endocrinol. Metab. 2019, 30, 227–229. [Google Scholar] [CrossRef]
- Altobelli, E.; Angeletti, P.M.; Latella, G. Role of Urinary Biomarkers in the Diagnosis of Adenoma and Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Cancer 2016, 7, 1984–2004. [Google Scholar] [CrossRef]
- Bingol, K.; Brüschweiler, R. Two elephants in the room: New hybrid nuclear magnetic resonance and mass spectrometry approaches for metabolomics. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 471–477. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; Kelley, D.E.; Thaete, F.L.; He, J.; Ross, R. Skeletal muscle attenuation determined by computed tomography is associated with skeletal muscle lipid content. J. Appl. Physiol. (1985) 2000, 89, 104–110. [Google Scholar] [CrossRef]
- Goodpaster, B.H.; Thaete, F.L.; Kelley, D.E. Composition of skeletal muscle evaluated with computed tomography. Ann. N. Y. Acad. Sci. 2000, 904, 18–24. [Google Scholar] [CrossRef]
- Skender, S.; Schrotz-King, P.; Böhm, J.; Abbenhardt, C.; Gigic, B.; Chang-Claude, J.; Siegel, E.M.; Steindorf, K.; Ulrich, C.M. Repeat physical activity measurement by accelerometry among colorectal cancer patients—feasibility and minimal number of days of monitoring. BMC Res. Notes 2015, 8, 3527. [Google Scholar] [CrossRef]
- Ulrich, C.M.; Gigic, B.; Bohm, J.; Ose, J.; Viskochil, R.; Schneider, M.; Colditz, G.A.; Figueiredo, J.C.; Grady, W.M.; Li, C.I.; et al. The ColoCare Study: A Paradigm of Transdisciplinary Science in Colorectal Cancer Outcomes. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 591–601. [Google Scholar] [CrossRef]
- Xiao, C.; Hao, F.; Qin, X.; Wang, Y.; Tang, H. An optimized buffer system for NMR-based urinary metabonomics with effective pH control, chemical shift consistency and dilution minimization. Analyst 2009, 134, 916–925. [Google Scholar] [CrossRef]
- Pluskal, T.; Castillo, S.; Villar-Briones, A.; Orešič, M. MZmine 2: Modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinform. 2010, 11, 395. [Google Scholar] [CrossRef]
- Wagner, B.D.; Accurso, F.J.; Laguna, T.A. The applicability of urinary creatinine as a method of specimen normalization in the cystic fibrosis population. J. Cyst. Fibros. 2010, 9, 212–216. [Google Scholar] [CrossRef]
- Himbert, C.; Ose, J.; Nattenmüller, J.; Warby, C.A.; Holowatyj, A.N.; Böhm, J.; Lin, T.; Haffa, M.; Gigic, B.; Hardikar, S.; et al. Body Fatness, Adipose Tissue Compartments, and Biomarkers of Inflammation and Angiogenesis in Colorectal Cancer: The ColoCare Study. Cancer Epidemiology Biomarkers Prev. 2018, 28, 76–82. [Google Scholar] [CrossRef]
- Nattenmueller, J.; Hoegenauer, H.; Boehm, J.; Scherer, D.; Paskow, M.; Gigic, B.; Schrotz-King, P.; Grenacher, L.; Ulrich, C.; Kauczor, H.-U. CT-based compartmental quantification of adipose tissue versus body metrics in colorectal cancer patients. Eur. Radiol. 2016, 26, 4131–4140. [Google Scholar] [CrossRef]
- Cheng, Y.; Xie, G.; Chen, T.; Qiu, Y.; Zou, X.; Zheng, M.; Tan, B.; Feng, B.; Dong, T.; He, P.; et al. Distinct urinary metabolic profile of human colorectal cancer. Journal of proteome research. 2012, 11, 1354–1363. [Google Scholar] [CrossRef]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]

| Category | Weight Loss | BMI |
|---|---|---|
| Non-Cachectic | weight loss ≤ 2% weight gain | - |
| Pre-Cachectic | weight loss * ≤ 5% | BMI ≤ 20 kg/m2 |
| Cachectic | weight loss * > 5% | |
| Cachectic | weight loss * > 2% | BMI < 20 kg/m2 |
| Cachectic (n = 16) | Pre-Cachectic (n = 13) | Non-Cachectic (n = 23) | p-Value | |
|---|---|---|---|---|
| Age at surgery + | 58.38 (±10.33) | 55.84 (±11.67) | 62.74 (±12.22) | 0.21 + |
| BMI 1 (kg/m2) | ||||
| at Baseline 2,+ | 25.60 (±2.71) | 29.39 (±3.35) | 26.48 (±4.26) | 0.02 + |
| 6 Months afterwards 3,+ | 23.37 (±3.08) | 28.11 (±3.42) | 26.54 (±4.41) | 0.004 + |
| Weight Change (kg) + | −7.12 (±3.63) | −2.54 (±1.27) | 2.78 (±2.68) | <0.001 + |
| Weight Change (%) | −0.09 (±0.05) | −0.02 (±0.02) | 0.03 (±0.03) | <0.001 + |
| Sex | 0.33 ++ | |||
| Male | 11 (69%) | 11 (85%) | 14 (61%) | |
| Female | 5 (31%) | 2 (15%) | 9 (39%) | |
| Stage | 0.11 ++ | |||
| I | 5 (31%) | 2 (15%) | 7 (30%) | |
| II | 1 (6%) | 5 (38%) | 9 (40%) | |
| III | 6 (38%) | 4 (31%) | 7 (30%) | |
| IV | 4 (25%) | 2 (15%) | 0 | |
| Site | 0.58 ++ | |||
| Colon | 6 (38%) | 5 (38%) | 12 (52%) | |
| Rectal | 10 (62%) | 8 (62%) | 11(48%) | |
| Adjuvant Therapy | 0.14 ++ | |||
| Yes | 10 (62%) | 8 (62%) | 7 (30%) | |
| No | 6 (38%) | 5 (38%) | 16 (70%) | |
| Neo-Adjuvant Therapy | 0.43 ++ | |||
| Yes | 6 (38%) | 4 (31%) | 6 (26%) | |
| No | 10 (62%) | 9 (70%) | 17 (74%) |
| Cachectic (n = 16) | Pre-Cachectic | Non-Cachectic | p-Value for Group Comparisons | ||||
|---|---|---|---|---|---|---|---|
| (n = 13) | (n = 23) | Three Group Comparison | Cachectic vs Non-Cachectic | Cachectic vs Pre-Cachectic | Pre-Cachectic vs Non-Cachectic | ||
| Visceral Fat Area | |||||||
| L3/L4 | 152.93 (±92.56) | 231.20 (±68.97) | 184.26 (±68.59) | 0.08 | 0.34 | 0.04 | 0.11 |
| L4/L5 | 134.90 (±74.86) | 204.51 (±57.86) | 154.96 (±51.81) | 0.04 | 0.44 | 0.03 | 0.04 |
| Subcutaneous Fat Area | |||||||
| L3/L4 | 204.58 (±101.90) | 301.70 (±125.96) | 255.47 (±130.21) | 0.20 | 0.30 | 0.06 | 0.39 |
| L4/L5 | 230.00 (±97.10) | 320.38 (±111.91) | 286.10 (±125.43) | 0.19 | 0.23 | 0.06 | 0.50 |
| Dorsal muscle Area | |||||||
| L3/L4 | 37.77 (±14.86) | 30.31 (16.34) | 35.43 (17.25) | 0.57 | 0.72 | 0.29 | 0.47 |
| L4/L5 | 29.44 (±10.26) | 21.84 (14.66) | 27.93 (11.34) | 0.32 | 0.73 | 0.18 | 0.26 |
| Psoas muscle Area | |||||||
| L3/L4 | 19.30 (±8.57) | 18.55 (±6.19) | 19.34 (±6.86) | 0.96 | 0.98 | 0.82 | 0.77 |
| L4/L5 | 28.81 (±24.44) | 19.29 (±6.58) | 22.30 (±7.93) | 0.34 | 0.36 | 0.25 | 0.34 |
| Abdominal muscle Area | |||||||
| L3/L4 | 42.88 (±17.46) | 26.02 (±14.07) | 34.25 (±15.12) | 0.06 | 0.20 | 0.03 | 0.19 |
| L4/L5 | 36.07 (±14.49) | 24.92 (±14.58) | 30.01 (±15.61) | 0.24 | 0.33 | 0.09 | 0.43 |
| Platform | Metabolite (mean ± std) | Cachectic | Non-Cachectic | Fold Change | pValue1 | pFDR2 | Identification |
|---|---|---|---|---|---|---|---|
| GC–MS 3 | 2.3-Butanediol | 0.2 ± 0.76 | 1.0 ± 0.73 | 0.45 | 0.01 | 0.93 | Level 2 |
| 2.3-Dihydroxybutyrate | 0.1 ± 0.59 | −0.5 ± 0.56 | 1.82 | 0.02 | 0.93 | Level 2 | |
| Sugar 15.798 min | 0.3 ± 1.02 | 1.1 ± 0.89 | 0.45 | 0.04 | 0.93 | Level 3 | |
| 4-Hydroxyphenylacetate | −0.2 ± 0.67 | 0.5 ± 0.98 | 0.50 | 0.04 | 0.93 | Level 2 | |
| Unknown Glucuronide 28.543 min | 0.3 ± 0.66 | 0.9 ± 0.98 | 0.55 | 0.04 | 0.93 | Level 3 | |
| 1H-NMR 4,5 | 3-Methylxanthine | 3.3 ± 1.81 | 5.8 ± 2.87 | 0.76 | 0.02 | 0.97 | NMR library |
| Acetone | 7.1 ± 7.37 | 1.7 ± 1.01 | 3.17 | 0.02 | 0.97 | NMR library | |
| Arginine | 25.8 ± 6.66 | 19.4 ± 8.03 | 0.33 | 0.04 | 0.97 | NMR library |
| Platform | Metabolite (Mean ± Std) | Cachectic | Pre-Cachectic | Fold Change | pValue1 | pFDR2 | Identification |
|---|---|---|---|---|---|---|---|
| GC-MS 3 | Hydroquinone | 0.6 ± 0.49 | −0.5 ± 1.20 | 3.00 | 0.006 | 0.59 | Level 1 |
| Unknown 13.271 min | 0.0 ± 0.86 | 0.8 ± 0.80 | 0.45 | 0.009 | 0.59 | Level 4 | |
| Aminomalonate | 0.4 ± 0.59 | −0.3 ± 0.77 | 2.01 | 0.01 | 0.59 | Level 2 | |
| Sugar Acid 15.429 min | 0.5 ± 0.74 | −0.1 ± 0.55 | 1.82 | 0.01 | 0.60 | Level 3 | |
| 4-Hydroxy-3-Methoxy-Mandelate | 0.6 ± 1.15 | −0.3 ± 0.83 | 2.46 | 0.03 | 0.79 | Level 2 | |
| Unknown Glucuronide 29.801 min | 0.7 ± 0.79 | 0.0 ± 1.06 | 2.01 | 0.03 | 0.79 | Level 3 | |
| Sugar Acid 15.750 min | 0.6 ± 0.79 | −0.1 ± 1.16 | 2.01 | 0.04 | 0.79 | Level 3 | |
| Unknown Sugar 14.575 min | 0.4 ± 0.94 | −0.4 ± 1.23 | 2.23 | 0.04 | 0.79 | Level 3 | |
| Unknown Dissacharide 29.943 min | 0.5 ± 0.76 | −0.2 ± 1.12 | 2.01 | 0.04 | 0.79 | Level 3 | |
| 1H-NMR 4,5 | Isobutyrate | 0.8 ± 0.78 | 1.7 ± 1.21 | 1.13 | 0.02 | 0.97 | NMR library |
| 0.79 | |||||||
| Glycine | 150.1 ± 118.51 | 72.5 ± 42.98 | 1.07E+33 | 0.04 | 0.97 | NMR library |
| Platform | Metabolite (Mean ± Std) | Pre-Cachectic | Non-Cachectic | Fold Change | pValue1 | pFDR2 | Identification |
|---|---|---|---|---|---|---|---|
| GC-MS 3 | Sugar Acid 15.429 min | −0.1 ± 0.55 | 0.8 ± 0.86 | 0.41 | 0.007 | 0.59 | Level 3 |
| 2-O-Glycerol-α-d-galactopyranoside | −0.4 ± 1.09 | 0.6 ± 0.49 | 0.37 | 0.01 | 0.59 | Level 2 | |
| Sugar 15.798 min | 0.0 ± 1.06 | 1.1 ± 0.89 | 0.33 | 0.01 | 0.59 | Level 3 | |
| Tartrate | 0.0 ± 0.85 | 1.0 ± 1.07 | 0.37 | 0.01 | 0.59 | Level 1 | |
| Hydroquinone | −0.5 ± 1.20 | 0.7 ± 1.14 | 0.30 | 0.02 | 0.59 | Level 1 | |
| Sugar Acid 15.750 min | −0.1 ± 1.16 | 0.8 ± 0.61 | 0.41 | 0.03 | 0.59 | Level 3 | |
| Unknown 28.636 min | 0.1 ± 1.14 | 1.1 ± 0.94 | 0.37 | 0.03 | 0.59 | Level 4 | |
| Unknown Glucuronide 27.116 min | 0.0 ± 0.83 | 0.7 ± 0.67 | 0.50 | 0.03 | 0.59 | Level 3 | |
| Unknown Glucuronide 28.543 min | 0.1 ± 0.89 | 0.9 ± 0.98 | 0.45 | 0.04 | 0.59 | Level 3 | |
| Sugar 14.052 min | −0.4 ± 1.34 | 0.7 ± 1.02 | 0.33 | 0.04 | 0.59 | Level 3 | |
| p-cresol-glucuronide | −0.3 ± 1.27 | 0.6 ± 0.68 | 0.41 | 0.04 | 0.59 | Level 1 | |
| Unknown Glucuronide 27.688 min | 0.1 ± 0.86 | 0.9 ± 0.75 | 0.45 | 0.04 | 0.59 | Level 3 | |
| Unknown Glucuronide 29.801 min | 0.0 ± 1.06 | 0.8 ± 0.90 | 0.45 | 0.049 | 0.59 | Level 3 | |
| 1H-NMR 4,5 | Uracil | 6.5 ± 2.10 | 3.9 ± 2.15 | 0.67 | 0.008 | 0.91 | NMR library |
| Cholate | 0.8 ± 0.12 | 1.6 ± 0.27 | 1.00 | 0.03 | 0.91 | NMR library | |
| Methionine | 4.4 ± 2.54 | 2.2 ± 1.37 | 1.00 | 0.03 | 0.91 | NMR library | |
| Acetone | 3.2 ± 1.83 | 1.7 ± 1.01 | 0.88 | 0.03 | 0.91 | NMR library | |
| 3-Phenylpropionate | 9.1 ± 3.97 | 14.4 ± 6.05 | 0.58 | 0.03 | 0.91 | NMR library | |
| Arginine | 28.7 ± 11.98 | 19.4 ± 8.03 | 0.48 | 0.045 | 0.91 | NMR library |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ose, J.; Gigic, B.; Lin, T.; Liesenfeld, D.B.; Böhm, J.; Nattenmüller, J.; Scherer, D.; Zielske, L.; Schrotz-King, P.; Habermann, N.; et al. Multiplatform Urinary Metabolomics Profiling to Discriminate Cachectic from Non-Cachectic Colorectal Cancer Patients: Pilot Results from the ColoCare Study. Metabolites 2019, 9, 178. https://doi.org/10.3390/metabo9090178
Ose J, Gigic B, Lin T, Liesenfeld DB, Böhm J, Nattenmüller J, Scherer D, Zielske L, Schrotz-King P, Habermann N, et al. Multiplatform Urinary Metabolomics Profiling to Discriminate Cachectic from Non-Cachectic Colorectal Cancer Patients: Pilot Results from the ColoCare Study. Metabolites. 2019; 9(9):178. https://doi.org/10.3390/metabo9090178
Chicago/Turabian StyleOse, Jennifer, Biljana Gigic, Tengda Lin, David B. Liesenfeld, Jürgen Böhm, Johanna Nattenmüller, Dominique Scherer, Lin Zielske, Petra Schrotz-King, Nina Habermann, and et al. 2019. "Multiplatform Urinary Metabolomics Profiling to Discriminate Cachectic from Non-Cachectic Colorectal Cancer Patients: Pilot Results from the ColoCare Study" Metabolites 9, no. 9: 178. https://doi.org/10.3390/metabo9090178
APA StyleOse, J., Gigic, B., Lin, T., Liesenfeld, D. B., Böhm, J., Nattenmüller, J., Scherer, D., Zielske, L., Schrotz-King, P., Habermann, N., Ochs-Balcom, H. M., Peoples, A. R., Hardikar, S., Li, C. I., Shibata, D., Figueiredo, J., Toriola, A. T., Siegel, E. M., Schmit, S., ... Ulrich, C. M. (2019). Multiplatform Urinary Metabolomics Profiling to Discriminate Cachectic from Non-Cachectic Colorectal Cancer Patients: Pilot Results from the ColoCare Study. Metabolites, 9(9), 178. https://doi.org/10.3390/metabo9090178





