Metabolic Regulation of Glycolysis and AMP Activated Protein Kinase Pathways during Black Raspberry-Mediated Oral Cancer Chemoprevention
Abstract
1. Introduction
2. Results
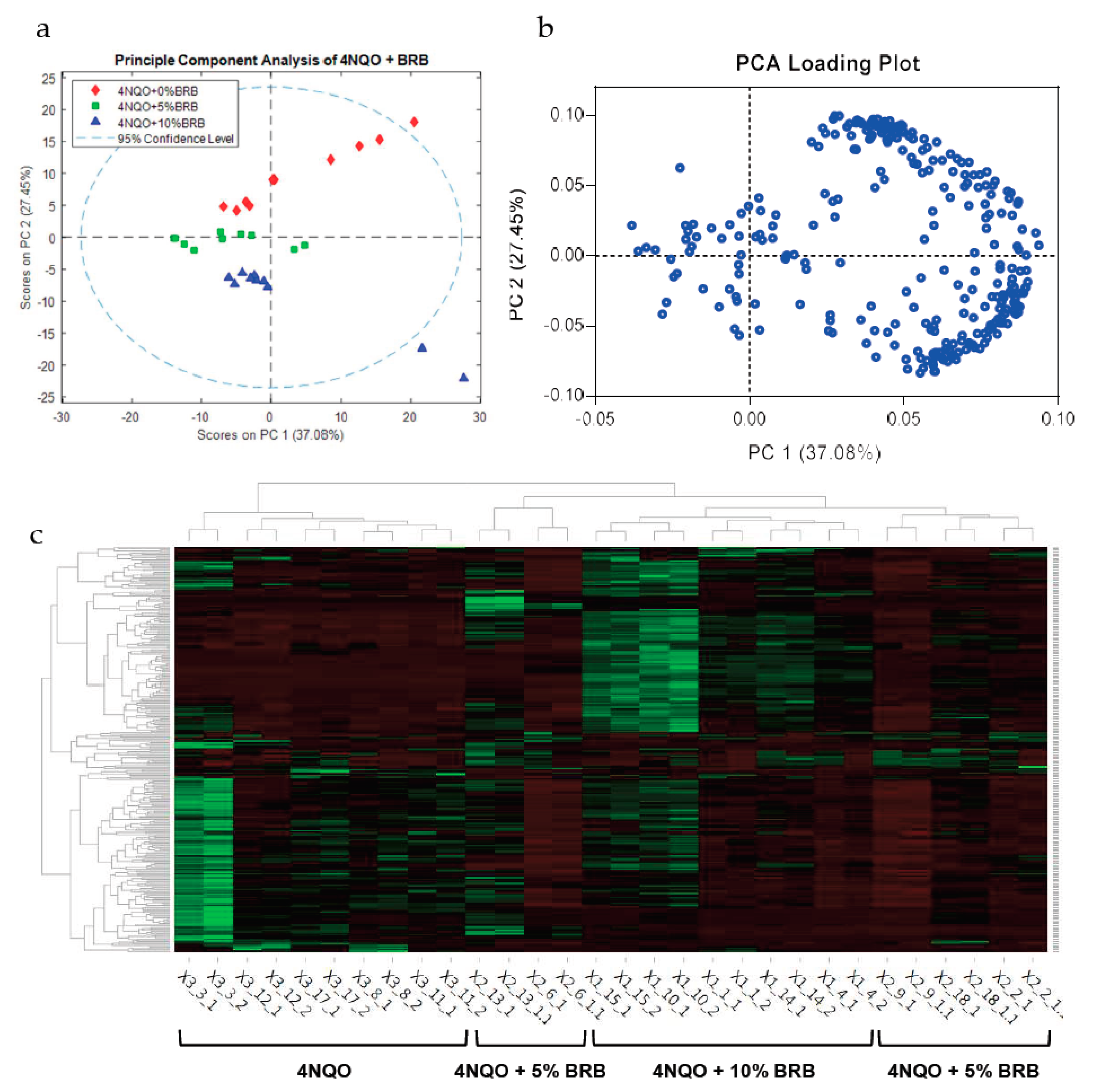
2.1. BRBs Modulate the Metabolic Profile of Oral Cancer Induced Rats
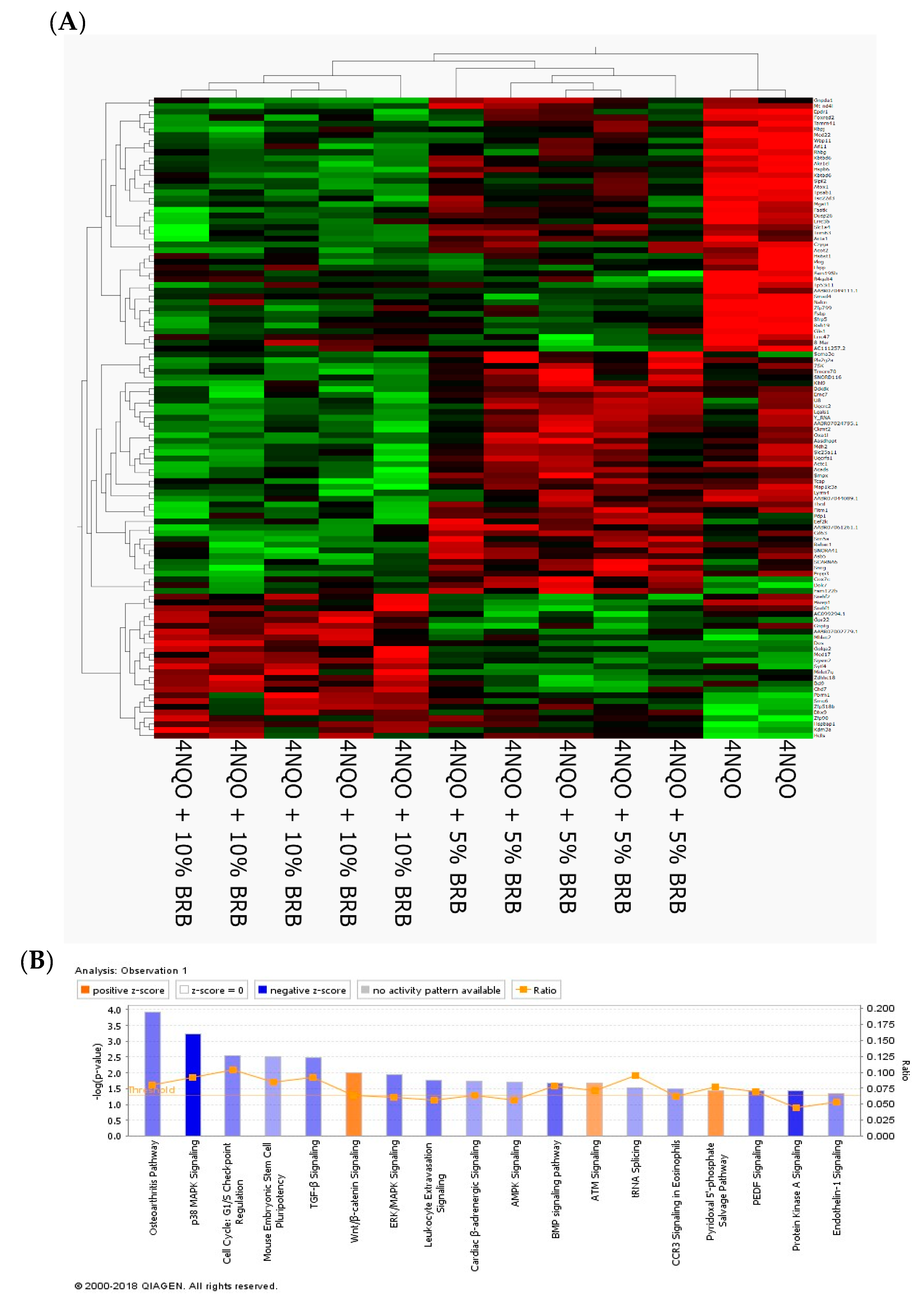
2.2. Modulation of Global Transcriptional Profiles during Oral Cancer Chemoprevention by BRBs
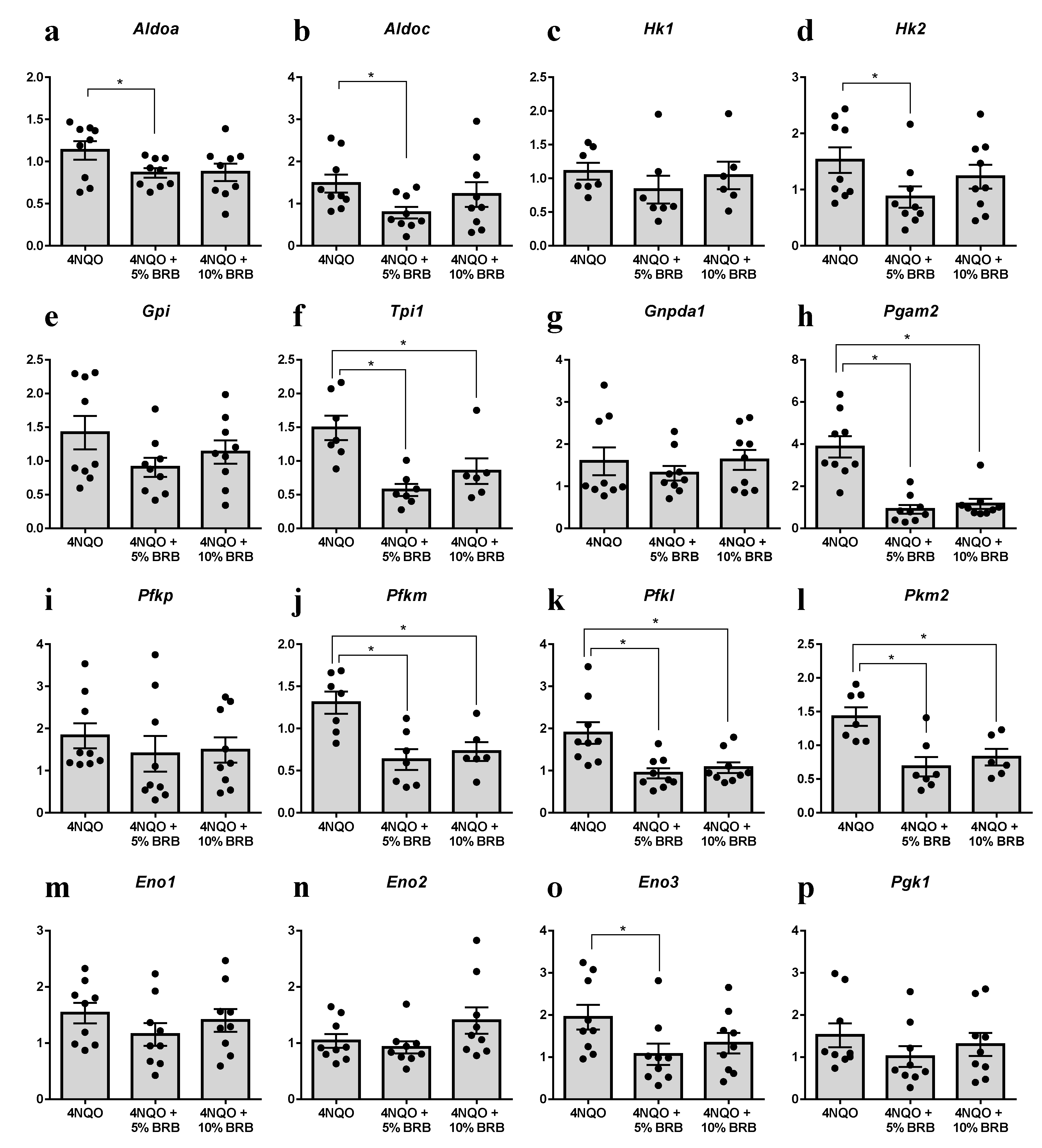
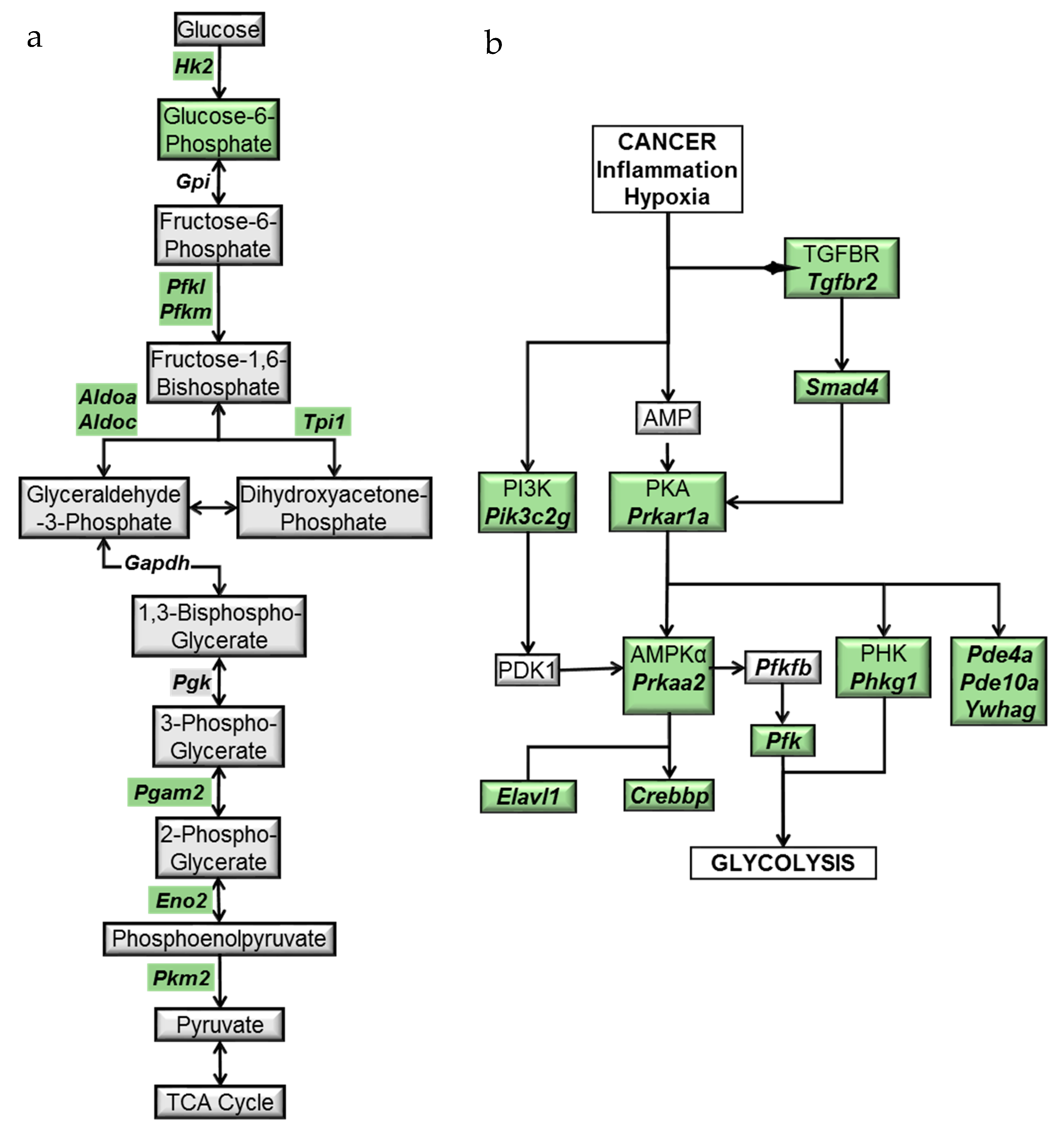
2.3. Modulation of the Glycolytic Pathway by BRB during Experimental Oral Carcinogenesis
2.4. BRB Downregulates Protein Kinase A and AMP-Activated Protein Kinase Signaling Pathways
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Chemicals
4.3. Experimental Oral Carcinogenesis
4.4. RNA Sequencing Analysis
4.5. Reverse Transcription Quantitative PCR
4.6. Liquid Chromatography-Mass Spectrometry
4.7. Nuclear Magnetic Resonance
4.8. Metabolite Data Extraction and Compound Identification
4.9. Metabolite Data Extraction, Statistical Analysis and Compound Identification
4.10. Metabolic Pathway Analysis
4.11. Ingenuity Pathway Analysis
4.12. RNA Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Rivera, C. Essentials of oral cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 11884–11894. [Google Scholar] [PubMed]
- Cohen, N.; Fedewa, S.; Chen, A.Y. Epidemiology and Demographics of the Head and Neck Cancer Population. Oral. Maxillofac. Surg. Clin. N. Am. 2018, 30, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Adjei Boakye, E.; Buchanan, P.; Hinyard, L.; Osazuwa-Peters, N.; Schootman, M.; Piccirillo, J.F. Incidence and Risk of Second Primary Malignant Neoplasm After a First Head and Neck Squamous Cell Carcinoma. JAMA Otolaryngol. Head Neck Surg. 2018, 144, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Han, S.; Yao, Z.; Li, X.; Huang, P.; Zhang, M.; Chen, Y.; He, J. A study of epidemiologic and recurrence factors of oral cancer. J. Oral. Maxillofac. Surg. 2012, 70, 2205–2210. [Google Scholar] [CrossRef] [PubMed]
- Knobloch, T.J.; Uhrig, L.K.; Pearl, D.K.; Casto, B.C.; Warner, B.M.; Clinton, S.K.; Sardo-Molmenti, C.L.; Ferguson, J.M.; Daly, B.T.; Riedl, K.; et al. Suppression of Proinflammatory and Prosurvival Biomarkers in Oral Cancer Patients Consuming a Black Raspberry Phytochemical-Rich Troche. Cancer Prev. Res. (Phila.) 2016, 9, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Warner, B.M.; Casto, B.C.; Knobloch, T.J.; Accurso, B.T.; Weghorst, C.M. Chemoprevention of oral cancer by topical application of black raspberries on high at-risk mucosa. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2014, 118, 674–683. [Google Scholar] [CrossRef] [PubMed]
- Bobinaite, R.; Viskelis, P.; Venskutonis, P.R. Variation of total phenolics, anthocyanins, ellagic acid and radical scavenging capacity in various raspberry (Rubus spp.) cultivars. Food Chem. 2012, 132, 1495–1501. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Subar, A.F.; Bosire, C.; Dawsey, S.M.; Kahle, L.L.; Zimmerman, T.P.; Abnet, C.C.; Heller, R.; Graubard, B.I.; Cook, M.B.; et al. Dietary Flavonoid Intake Reduces the Risk of Head and Neck but Not Esophageal or Gastric Cancer in US Men and Women. J. Nutr. 2017, 147, 1729–1738. [Google Scholar] [CrossRef]
- Teegarden, M.D.; Knobloch, T.J.; Weghorst, C.M.; Cooperstone, J.L.; Peterson, D.G. Storage conditions modulate the metabolomic profile of a black raspberry nectar with minimal impact on bioactivity. Food Funct. 2018, 9, 4593–4601. [Google Scholar] [CrossRef]
- Oghumu, S.; Casto, B.C.; Ahn-Jarvis, J.; Weghorst, L.C.; Maloney, J.; Geuy, P.; Horvath, K.Z.; Bollinger, C.E.; Warner, B.M.; Summersgill, K.F.; et al. Inhibition of Pro-inflammatory and Anti-apoptotic Biomarkers during Experimental Oral Cancer Chemoprevention by Dietary Black Raspberries. Front Immunol. 2017, 8, 1325. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, D.S.; Wang, L.S.; Zimmerman, N.P.; Ransom, B.W.; Carmella, S.G.; Kuo, C.T.; Chen, J.H.; Oshima, K.; Huang, Y.W.; Hecht, S.S.; et al. Dietary Consumption of Black Raspberries or Their Anthocyanin Constituents Alters Innate Immune Cell Trafficking in Esophageal Cancer. Cancer Immunol. Res. 2016, 4, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Mace, T.A.; King, S.A.; Ameen, Z.; Elnaggar, O.; Young, G.; Riedl, K.M.; Schwartz, S.J.; Clinton, S.K.; Knobloch, T.J.; Weghorst, C.M.; et al. Bioactive compounds or metabolites from black raspberries modulate T lymphocyte proliferation, myeloid cell differentiation and Jak/STAT signaling. Cancer Immunol. Immunother. 2014, 63, 889–900. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, J.; An, J.; Lee, S.; Lee, H.; Kong, H.; Song, Y.; Choi, H.R.; Kwon, J.W.; Shin, D.; et al. Restoration of Declined Immune Responses and Hyperlipidemia by Rubus occidenalis in Diet-Induced Obese Mice. Biomol. Ther. (Seoul) 2017, 25, 140–148. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mallery, S.R.; Tong, M.; Shumway, B.S.; Curran, A.E.; Larsen, P.E.; Ness, G.M.; Kennedy, K.S.; Blakey, G.H.; Kushner, G.M.; Vickers, A.M.; et al. Topical application of a mucoadhesive freeze-dried black raspberry gel induces clinical and histologic regression and reduces loss of heterozygosity events in premalignant oral intraepithelial lesions: Results from a multicentered, placebo-controlled clinical trial. Clin. Cancer Res. 2014, 20, 1910–1924. [Google Scholar] [CrossRef] [PubMed]
- Mallery, S.R.; Zwick, J.C.; Pei, P.; Tong, M.; Larsen, P.E.; Shumway, B.S.; Lu, B.; Fields, H.W.; Mumper, R.J.; Stoner, G.D. Topical application of a bioadhesive black raspberry gel modulates gene expression and reduces cyclooxygenase 2 protein in human premalignant oral lesions. Cancer Res. 2008, 68, 4945–4957. [Google Scholar] [CrossRef] [PubMed]
- Bazzano, M.; Laghi, L.; Zhu, C.; Magi, G.E.; Serri, E.; Spaterna, A.; Tesei, B.; Laus, F. Metabolomics of tracheal wash samples and exhaled breath condensates in healthy horses and horses affected by equine asthma. J. Breath Res. 2018, 12, 046015. [Google Scholar] [CrossRef]
- Xu, T.; Zhou, L.; Shi, Y.; Liu, L.; Zuo, L.; Jia, Q.; Du, S.; Kang, J.; Zhang, X.; Sun, Z. Metabolomics approach in lung tissue of septic rats and the interventional effects of Xuebijing injection using UHPLC-Q-Orbitrap-HRMS. J. Biochem. 2018. [Google Scholar] [CrossRef]
- Nwosu, Z.C.; Battello, N.; Rothley, M.; Pioronska, W.; Sitek, B.; Ebert, M.P.; Hofmann, U.; Sleeman, J.; Wolfl, S.; Meyer, C.; et al. Liver cancer cell lines distinctly mimic the metabolic gene expression pattern of the corresponding human tumours. J. Exp. Clin. Cancer Res. 2018, 37, 211. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, P.; Wang, X.; Wang, Y.; Zhou, Z.; Zhu, W. NMR- and LC-MS/MS-based urine metabolomic investigation of the subacute effects of hexabromocyclododecane in mice. Env. Sci. Pollut. Res. Int. 2016, 23, 8500–8507. [Google Scholar] [CrossRef]
- Jensen, B.C.; Parry, T.L.; Huang, W.; Ilaiwy, A.; Bain, J.R.; Muehlbauer, M.J.; O’Neal, S.K.; Patterson, C.; Johnson, G.L.; Willis, M.S. Non-Targeted Metabolomics Analysis of the Effects of Tyrosine Kinase Inhibitors Sunitinib and Erlotinib on Heart, Muscle, Liver and Serum Metabolism In Vivo. Metabolites 2017, 7, 31. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Solmonson, A.; Lodi, A.; Nowinski, S.M.; Sentandreu, E.; Riley, C.L.; Mills, E.M.; Tiziani, S. The early metabolomic response of adipose tissue during acute cold exposure in mice. Sci. Rep. 2017, 7, 3455. [Google Scholar] [CrossRef] [PubMed]
- Kanojia, D.; Vaidya, M.M. 4-nitroquinoline-1-oxide induced experimental oral carcinogenesis. Oral. Oncol. 2006, 42, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Kresty, L.A.; Mallery, S.R.; Stoner, G.D. Black raspberries in cancer clinical trials: Past, present and future. J. Berry Res. 2016, 6, 251–261. [Google Scholar] [CrossRef]
- Ngo, H.; Tortorella, S.M.; Ververis, K.; Karagiannis, T.C. The Warburg effect: Molecular aspects and therapeutic possibilities. Mol. Biol. Rep. 2015, 42, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Grimm, M.; Cetindis, M.; Lehmann, M.; Biegner, T.; Munz, A.; Teriete, P.; Kraut, W.; Reinert, S. Association of cancer metabolism-related proteins with oral carcinogenesis—Indications for chemoprevention and metabolic sensitizing of oral squamous cell carcinoma? J. Transl. Med. 2014, 12, 208. [Google Scholar] [CrossRef]
- Ha, J.; Guan, K.L.; Kim, J. AMPK and autophagy in glucose/glycogen metabolism. Mol. Asp. Med. 2015, 46, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Oghumu, S.; Knobloch, T.J.; Terrazas, C.; Varikuti, S.; Ahn-Jarvis, J.; Bollinger, C.E.; Iwenofu, H.; Weghorst, C.M.; Satoskar, A.R. Deletion of macrophage migration inhibitory factor inhibits murine oral carcinogenesis: Potential role for chronic pro-inflammatory immune mediators. Int. J. Cancer 2016, 139, 1379–1390. [Google Scholar] [CrossRef]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015, 356, 156–164. [Google Scholar] [CrossRef]
- Huang, Y.P.; Chang, N.W. Proteomic analysis of oral cancer reveals new potential therapeutic targets involved in the Warburg effect. Clin. Exp. Pharm. Physiol. 2017, 44, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Mims, J.; Bansal, N.; Bharadwaj, M.S.; Chen, X.; Molina, A.J.; Tsang, A.W.; Furdui, C.M. Energy metabolism in a matched model of radiation resistance for head and neck squamous cell cancer. Radiat. Res. 2015, 183, 291–304. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Hong, Y.; Lu, Y.; Qiu, S.; Chaganty, B.K.; Zhang, L.; Wang, X.; Li, Q.; Fan, Z. Acetyl-CoA carboxylase rewires cancer metabolism to allow cancer cells to survive inhibition of the Warburg effect by cetuximab. Cancer Lett. 2017, 384, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Du, S.; Guan, Z.; Hao, L.; Song, Y.; Wang, L.; Gong, L.; Liu, L.; Qi, X.; Hou, Z.; Shao, S. Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration. PLoS ONE 2014, 9, e85804. [Google Scholar] [CrossRef] [PubMed]
- Pfleiderer, G.; Thoner, M.; Wachsmuth, E.D. Histological examination of the aldolase monomer composition of cells from human kidney and hypernephroid carcinoma. Beitr. Pathol. 1975, 156, 266–279. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, B.; Liu, J.; Qin, Y.; Liang, C.; Shi, S.; Jin, K.; Liang, D.; Xu, W.; Xu, H.; et al. ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Lett. 2016, 374, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, X.; Wu, M.; Yang, J.; Liu, M.; Zhang, W.; Xiang, B.; Wang, X.; Li, X.; Li, G.; et al. New prognosis biomarkers identified by dynamic proteomic analysis of colorectal cancer. Mol. Biosyst. 2012, 8, 3077–3088. [Google Scholar] [CrossRef] [PubMed]
- Heron, P.; Abellan-Flos, M.; Salmon, L.; Sygusch, J. Bisphosphonate inhibitors of mammalian glycolytic aldolase. J. Med. Chem. 2018. [Google Scholar] [CrossRef]
- Lincet, H.; Icard, P. How do glycolytic enzymes favour cancer cell proliferation by nonmetabolic functions? Oncogene 2015, 34, 3751–3759. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, S.; Li, Y.; Tang, Z.; Kong, W. Hexokinase 2 overexpression promotes the proliferation and survival of laryngeal squamous cell carcinoma. Tumour. Biol. 2014, 35, 3743–3753. [Google Scholar] [CrossRef]
- Patra, K.C.; Wang, Q.; Bhaskar, P.T.; Miller, L.; Wang, Z.; Wheaton, W.; Chandel, N.; Laakso, M.; Muller, W.J.; Allen, E.L.; et al. Hexokinase 2 is required for tumor initiation and maintenance and its systemic deletion is therapeutic in mouse models of cancer. Cancer Cell 2013, 24, 213–228. [Google Scholar] [CrossRef] [PubMed]
- Min, J.W.; Kim, K.I.; Kim, H.A.; Kim, E.K.; Noh, W.C.; Jeon, H.B.; Cho, D.H.; Oh, J.S.; Park, I.C.; Hwang, S.G.; et al. INPP4B-mediated tumor resistance is associated with modulation of glucose metabolism via hexokinase 2 regulation in laryngeal cancer cells. Biochem. Biophys. Res. Commun. 2013, 440, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Kwee, S.A.; Hernandez, B.; Chan, O.; Wong, L. Choline kinase alpha and hexokinase-2 protein expression in hepatocellular carcinoma: Association with survival. PLoS ONE 2012, 7, e46591. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, D.; Fitzgerald, D.; Shreeve, S.M.; Hua, E.; Bronder, J.L.; Weil, R.J.; Davis, S.; Stark, A.M.; Merino, M.J.; Kurek, R.; et al. Analyses of resected human brain metastases of breast cancer reveal the association between up-regulation of hexokinase 2 and poor prognosis. Mol. Cancer. Res. 2009, 7, 1438–1445. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Ojo, D.; Yan, J.; Tang, D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015, 356, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.; Ding, M.; Wang, B.; Lu, Z.; Zhao, Q.; Shaw, K.; Yung, W.K.; Weinstein, J.N.; Tan, M.; Yao, J. Tissue-specific isoform switch and DNA hypomethylation of the pyruvate kinase PKM gene in human cancers. Oncotarget 2014, 5, 8202–8210. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yang, Z.; Zou, Q.; Yuan, Y.; Li, J.; Liang, L.; Zeng, G.; Chen, S. PKM2 and ACVR 1C are prognostic markers for poor prognosis of gallbladder cancer. Clin. Transl. Oncol. 2014, 16, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, J.; Phillips, J.J.; Zheng, S.; Wiencke, J.; Ronen, S.M.; Pieper, R.O. Pyruvate kinase M2 expression, but not pyruvate kinase activity, is up-regulated in a grade-specific manner in human glioma. PLoS ONE 2013, 8, e57610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; He, C.; He, C.; Chen, B.; Liu, Y.; Kong, M.; Wang, C.; Lin, L.; Dong, Y.; Sheng, H. Nuclear PKM2 expression predicts poor prognosis in patients with esophageal squamous cell carcinoma. Pathol. Res. Pr. 2013, 209, 510–515. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xiao, Z.; Chen, Z.; Zhang, X.; Li, J.; Wu, X.; Li, X.; Yi, H.; Li, M.; Zhu, G.; et al. Proteome analysis of human lung squamous carcinoma. Proteomics 2006, 6, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Gharib, T.G.; Huang, C.C.; Thomas, D.G.; Shedden, K.A.; Taylor, J.M.; Kardia, S.L.; Misek, D.E.; Giordano, T.J.; Iannettoni, M.D.; et al. Proteomic analysis of lung adenocarcinoma: Identification of a highly expressed set of proteins in tumors. Clin. Cancer Res. 2002, 8, 2298–2305. [Google Scholar] [PubMed]
- Unwin, R.D.; Craven, R.A.; Harnden, P.; Hanrahan, S.; Totty, N.; Knowles, M.; Eardley, I.; Selby, P.J.; Banks, R.E. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics 2003, 3, 1620–1632. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, L.; Li, W.; Chen, T.; Bin, Z.; Zhao, L.; Wang, H.; Wang, X.; Xu, L.; Liu, X.; et al. TAp73-induced phosphofructokinase-1 transcription promotes the Warburg effect and enhances cell proliferation. Nat. Commun. 2018, 9, 4683. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Wang, W.; Wu, J.; Feng, B.; Chen, W.; Wang, M.; Tang, J.; Wang, F.; Cheng, F.; Pu, L.; et al. Comparative proteomic profiling of human bile reveals SSP411 as a novel biomarker of cholangiocarcinoma. PLoS ONE 2012, 7, e47476. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Wu, H.; Lei, Y.; Zhang, H.; Liu, R.; Zhao, Y.; Chen, X.; Zeng, D.; Tong, A.; Chen, L.; et al. Quantitative proteomics identification of phosphoglycerate mutase 1 as a novel therapeutic target in hepatocellular carcinoma. Mol. Cancer 2010, 9, 81. [Google Scholar] [CrossRef] [PubMed]
- Viano, D.C.; Lau, I.V.; Asbury, C.; King, A.I.; Begeman, P. Biomechanics of the human chest, abdomen, and pelvis in lateral impact. Accid. Anal. Prev. 1989, 21, 553–574. [Google Scholar] [CrossRef]
- Tsai, S.C.; Tsai, M.H.; Chiu, C.F.; Lu, C.C.; Kuo, S.C.; Chang, N.W.; Yang, J.S. AMPK-dependent signaling modulates the suppression of invasion and migration by fenofibrate in CAL 27 oral cancer cells through NF-kappaB pathway. Environ. Toxicol. 2016, 31, 866–876. [Google Scholar] [CrossRef]
- Han, F.; Li, C.F.; Cai, Z.; Zhang, X.; Jin, G.; Zhang, W.N.; Xu, C.; Wang, C.Y.; Morrow, J.; Zhang, S.; et al. The critical role of AMPK in driving Akt activation under stress, tumorigenesis and drug resistance. Nat. Commun. 2018, 9, 4728. [Google Scholar] [CrossRef]
- Eichner, L.J.; Brun, S.N.; Herzig, S.; Young, N.P.; Curtis, S.D.; Shackelford, D.B.; Shokhirev, M.N.; Leblanc, M.; Vera, L.I.; Hutchins, A.; et al. Genetic Analysis Reveals AMPK Is Required to Support Tumor Growth in Murine Kras-Dependent Lung Cancer Models. Cell Metab. 2018. [Google Scholar] [CrossRef]
- Monteverde, T.; Muthalagu, N.; Port, J.; Murphy, D.J. Evidence of cancer-promoting roles for AMPK and related kinases. FEBS J. 2015, 282, 4658–4671. [Google Scholar] [CrossRef]
- Faubert, B.; Vincent, E.E.; Poffenberger, M.C.; Jones, R.G. The AMP-activated protein kinase (AMPK) and cancer: Many faces of a metabolic regulator. Cancer Lett. 2015, 356, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. Molecular Pathways: Is AMPK a Friend or a Foe in Cancer? Clin. Cancer Res. 2015, 21, 3836–3840. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lu, Y.; Lu, H.; Luo, J.; Hong, Y.; Fan, Z. AMPK-mediated energy homeostasis and associated metabolic effects on cancer cell response and resistance to cetuximab. Oncotarget 2015, 6, 11507–11518. [Google Scholar] [CrossRef]
- Wu, Y.; Sarkissyan, M.; McGhee, E.; Lee, S.; Vadgama, J.V. Combined inhibition of glycolysis and AMPK induces synergistic breast cancer cell killing. Breast Cancer Res. Treat. 2015, 151, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Shi, N.; Chen, F.; Zhang, X.; Clinton, S.K.; Tang, X.; Sun, Z.; Chen, T. Suppression of Oxidative Stress and NFkappaB/MAPK Signaling by Lyophilized Black Raspberries for Esophageal Cancer Prevention in Rats. Nutrients 2017, 9, 413. [Google Scholar] [CrossRef] [PubMed]
- Arun, P.; Brown, M.S.; Ehsanian, R.; Chen, Z.; Van Waes, C. Nuclear NF-kappaB p65 phosphorylation at serine 276 by protein kinase A contributes to the malignant phenotype of head and neck cancer. Clin. Cancer Res. 2009, 15, 5974–5984. [Google Scholar] [CrossRef] [PubMed]
- Qu, C.F.; Shang, D.H.; Qi, Z.Z.; Sun, C.F. The expression and activity of protein kinase A I in oral squamous cell carcinoma. Shanghai Kou Qiang Yi Xue 2006, 15, 58–62. [Google Scholar] [PubMed]
- Hussain, M.; Tang, F.; Liu, J.; Zhang, J.; Javeed, A. Dichotomous role of protein kinase A type I (PKAI) in the tumor microenvironment: A potential target for ‘two-in-one’ cancer chemoimmunotherapeutics. Cancer Lett. 2015, 369, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Li, J.; Zhang, D.; Stoner, G.D.; Huang, C. Molecular mechanisms involved in chemoprevention of black raspberry extracts: From transcription factors to their target genes. Nutr. Cancer 2006, 54, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Kartha, V.K.; Alamoud, K.A.; Sadykov, K.; Nguyen, B.C.; Laroche, F.; Feng, H.; Lee, J.; Pai, S.I.; Varelas, X.; Egloff, A.M.; et al. Functional and genomic analyses reveal therapeutic potential of targeting beta-catenin/CBP activity in head and neck cancer. Genome Med. 2018, 10, 54. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Sharpe, R.; Pearson, A.; Campbell, J.; Natrajan, R.; Ashworth, A.; Turner, N.C. An siRNA screen identifies the GNAS locus as a driver in 20q amplified breast cancer. Oncogene 2014, 33, 2478–2486. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Deng, Y.; Fu, X. MiR-509-5p suppresses the proliferation, migration, and invasion of non-small cell lung cancer by targeting YWHAG. Biochem. Biophys. Res. Commun. 2017, 482, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Bingol, K.; Li, D.W.; Zhang, B.; Bruschweiler, R. Comprehensive Metabolite Identification Strategy Using Multiple Two-Dimensional NMR Spectra of a Complex Mixture Implemented in the COLMARm Web Server. Anal. Chem. 2016, 88, 12411–12418. [Google Scholar] [CrossRef]
- Bingol, K.; Li, D.W.; Bruschweiler-Li, L.; Cabrera, O.A.; Megraw, T.; Zhang, F.; Bruschweiler, R. Unified and isomer-specific NMR metabolomics database for the accurate analysis of (13)C-(1)H HSQC spectra. ACS Chem. Biol. 2015, 10, 452–459. [Google Scholar] [CrossRef]
| Predicted Metabolic Pathways | Total Metabolite Hits | Significant Metabolite Hits |
|---|---|---|
| Steroid hormone biosynthesis | 15 | 11 |
| Glyoxylate and dicarboxylate metabolism | 6 | 5 |
| Vitamin B6 metabolism | 5 | 4 |
| Cysteine and methionine metabolism | 9 | 5 |
| Riboflavin metabolism | 2 | 2 |
| Tryptophan metabolism | 11 | 5 |
| One carbon pool by folate | 6 | 3 |
| Porphyrin and chlorophyll metabolism | 9 | 4 |
| Glycerophospholipid metabolism | 7 | 3 |
| Pyrimidine metabolism | 13 | 5 |
| Citrate cycle (TCA cycle) | 4 | 2 |
| Ascorbate and aldarate metabolism | 5 | 2 |
| Primary bile acid biosynthesis | 5 | 2 |
| Pantothenate and CoA biosynthesis | 5 | 2 |
| Purine metabolism | 24 | 8 |
| Folate biosynthesis | 6 | 2 |
| Inositol phosphate metabolism | 7 | 2 |
| Arginine and proline metabolism | 18 | 5 |
| Starch and sucrose metabolism | 12 | 3 |
| Pentose phosphate pathway | 8 | 2 |
| Tyrosine metabolism | 13 | 3 |
| Glutathione metabolism | 9 | 2 |
| Biosynthesis of unsaturated fatty acids | 9 | 2 |
| Glycolysis or Gluconeogenesis | 10 | 2 |
| Alanine, aspartate and glutamate metabolism | 10 | 2 |
| Fructose and mannose metabolism | 11 | 2 |
| beta-Alanine metabolism | 11 | 2 |
| Aminoacyl-tRNA biosynthesis | 17 | 3 |
| Galactose metabolism | 19 | 3 |
| Amino sugar and nucleotide sugar metabolism | 20 | 3 |
| Metabolite Groups | Metabolite | Treatment Groups | ||
|---|---|---|---|---|
| Normal Diet | 10% BRB Diet | 5% BRB Diet | ||
| Unique Metabolites | 4-Methyl-2-oxovaleric acid | *D | *ND | ND |
| Delta-Hexanolactone | ND | D | ND | |
| Quinone | ND | D | ND | |
| Hydroquinone | ND | D | ND | |
| 4-Aminohippuric acid | ND | ND | D | |
| Homovanillic acid | ND | D | D | |
| Hydroxyphenylacetylglycine | ND | D | D | |
| N-Acetylcysteine | ND | D | D | |
| Downregulated Metabolites | alpha-D-Glucose-6-phosphate | 1 | 0.7 | 0.48 |
| alpha-D-Glucose-1-phosphate | 1 | 0.68 | 0.57 | |
| N_methylnicotinamide | 1 | 0.67 | 0.64 | |
| Lactose | 1 | 0.63 | 0.67 | |
| Dihydrouracil | 1 | 0.62 | 0.68 | |
| Taurine | 1 | 0.61 | 0.67 | |
| D-Galactose | 1 | 0.57 | 0.56 | |
| D-Xylose | 1 | 0.49 | 0.52 | |
| Pyruvic acid | 1 | 0.47 | 0.42 | |
| Spermine | 1 | 0.23 | 0.34 | |
| Upregulated Metabolites | Alpha-Hydroxyhippuric acid | 1 | 5.25 | 1.86 |
| 3-Methyl-oxopentanoic acid | 1 | 3.73 | 1.54 | |
| Chitosan | 1 | 2.47 | 3.36 | |
| Pipecolic acid | 1 | 2.15 | 1.82 | |
| N-Acetyl-D-glucosamine-phosphate | 1 | 1.99 | 1.49 | |
| 3-Methoxy-4-Hydroxyphenylglycol-sulfate | 1 | 1.6 | 1.84 | |
| D-Glucuronic acid | 1 | 1.59 | 1.54 | |
| D-Sorbitol | 1 | 1.59 | 1.78 | |
| Myo-Inositol | 1 | 1.57 | 1.51 | |
| Stachyose | 1 | 1.53 | 1.83 | |
| DL-alpha-Glycerol-phosphate | 1 | 1.35 | 1.4 | |
| Gene | 5% BRB | 10% BRB | Expected Expression | ||
|---|---|---|---|---|---|
| Expression | p Value | Expression | p Value | ||
| Cdc25a | 5.5 | 0.451 | 17.9 | 0.0304 | Down |
| Crebbp | −1450.5 | 0.0114 | −401.9 | 0.403 | Up |
| Gnas | −1256.4 | 0.217 | −3132.4 | 0.00908 | Up |
| Mapk13 | −29.0 | 0.141 | −47.0 | 0.0279 | Up |
| Mppe1 | −1.3 | 0.281 | −2.9 | 0.0307 | Up |
| Pde10a | −11.0 | 0.0256 | −9.0 | 0.0566 | Up |
| Pde4a | −60.9 | 0.0748 | −75.7 | 0.0336 | Up |
| Phkg1 | −9.1 | 0.696 | −53.5 | 0.0417 | Up |
| Pik3c2g | −3.7 | 0.00638 | -3.7 | 0.00638 | Up |
| Ppp1r14c | 16.0 | 0.0712 | 20.6 | 0.0272 | Down |
| Prkaa2 | −136.8 | 0.0409 | −149.2 | 0.0287 | Up |
| Prkar1a | −79.5 | 0.469 | −270.1 | 0.0303 | Up |
| Ptpre | 7.7 | 0.00969 | 5.5 | 0.0443 | Down |
| Smad4 | −479.6 | 0.000238 | −367.4 | 0.0015 | Up |
| Tgfbr2 | −82.4 | 0.113 | −123.0 | 0.0278 | Up |
| Ywhag | −624.7 | 0.033 | −628.7 | 0.0321 | Up |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| Aldoa | CCCCAAGTTATCAAGTCCAAGG | GTCGGCTCCATCCTTCTTATAC |
| Aldob | TCAAGGAGAAGGGAATTGTGG | TTCTTGTACTGAGCACAGCG |
| Aldoc | CCTGTCCCATCAAATATAGCCC | TGGCATTGAGGTTGAGTGATG |
| Gnpda1 | TCACACCCTCTTCAATGGC | GATTCTTTGATGGTGATCTTGCC |
| Hk1 | ACACCCATTGTCACCGAAG | AAGAACCCACTTGCGAAATTG |
| Hk2 | GCTGTGAAAATGTTGCCTACC | CATTGTCCGTCACCCTTACTC |
| Hk3 | AGTGGAGCTGAATGTGGTTG | TCTTCCATATAGCAGGCGTTG |
| Gck | GCCACAATGATCTCCTGCTAC | TCCCTCATCCCCTTCCAC |
| Gpi | AGGTGCTGGACAAGATGAAG | GAGAGCTTCAGTCACCATGAG |
| Pfkm | TGGTACGAGTTGGCATCTTC | ACTTCCAATCACCGTGCC |
| Pfkl | GGAAAGCCTATCTCATCCAGC | CCATACCCATCTTGCTACTCAG |
| Pfkp | TCACTGAGAGGAAATCAAGCTG | TGTAGGTGTTCAGGTTGCC |
| Tpi1 | AGATAATGTGAAGGACTGGTGC | CAGAGACGTTGCATTTCAGC |
| Gapdh | TGGTTACAATACAGTTACAGACTAGG | GTGGTTGTAAATGGAAGAAACATCT |
| Pgk1 | TGACTTTGGACAAGCTGGAC | CAGAATTTGATGCTTGGGACAG |
| Pgam2 | CCTAAGATTAAGGCTGGCAAGAG | GGTTCAGCTCATAGACAATGGG |
| Eno1 | CCGAGACAATGATAAGACCCG | TCTCCTGCTCCACAACATTC |
| Eno2 | TCAAGGACAAGTACGGCAAG | CATACCAATCACCATCTTTTCCG |
| Eno3 | GAATCCCGACCTTGTACTTCC | GCCAACTTGTTTCCAGCATG |
| Pkm2 | GTGGAGATGCTGAAGGAGATG | AGGTCGGTAGAGAATGGGATC |
| Pklr | CACCTCTGCCTTCTGGATATC | TCGTGCAATGTTCATCCCTG |
| Bact | CACTTTCTACAATGAGCTGCG | CTGGATGGCTACGTACATGG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Knobloch, T.J.; Ryan, N.M.; Bruschweiler-Li, L.; Wang, C.; Bernier, M.C.; Somogyi, A.; Yan, P.S.; Cooperstone, J.L.; Mo, X.; Brüschweiler, R.P.; et al. Metabolic Regulation of Glycolysis and AMP Activated Protein Kinase Pathways during Black Raspberry-Mediated Oral Cancer Chemoprevention. Metabolites 2019, 9, 140. https://doi.org/10.3390/metabo9070140
Knobloch TJ, Ryan NM, Bruschweiler-Li L, Wang C, Bernier MC, Somogyi A, Yan PS, Cooperstone JL, Mo X, Brüschweiler RP, et al. Metabolic Regulation of Glycolysis and AMP Activated Protein Kinase Pathways during Black Raspberry-Mediated Oral Cancer Chemoprevention. Metabolites. 2019; 9(7):140. https://doi.org/10.3390/metabo9070140
Chicago/Turabian StyleKnobloch, Thomas J., Nathan M. Ryan, Lei Bruschweiler-Li, Cheng Wang, Matthew C. Bernier, Arpad Somogyi, Pearlly S. Yan, Jessica L. Cooperstone, Xiaokui Mo, Rafael P. Brüschweiler, and et al. 2019. "Metabolic Regulation of Glycolysis and AMP Activated Protein Kinase Pathways during Black Raspberry-Mediated Oral Cancer Chemoprevention" Metabolites 9, no. 7: 140. https://doi.org/10.3390/metabo9070140
APA StyleKnobloch, T. J., Ryan, N. M., Bruschweiler-Li, L., Wang, C., Bernier, M. C., Somogyi, A., Yan, P. S., Cooperstone, J. L., Mo, X., Brüschweiler, R. P., Weghorst, C. M., & Oghumu, S. (2019). Metabolic Regulation of Glycolysis and AMP Activated Protein Kinase Pathways during Black Raspberry-Mediated Oral Cancer Chemoprevention. Metabolites, 9(7), 140. https://doi.org/10.3390/metabo9070140








