Defining Metabolic Rewiring in Lung Squamous Cell Carcinoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Samples
2.2. Metabolites Extraction
2.3. Untargeted Metabolite Fingerprinting by Flow Infusion Electrospray Ionization High Resolution Mass Spectrometry (FIE-HRMS)
2.4. Statistical Analysis
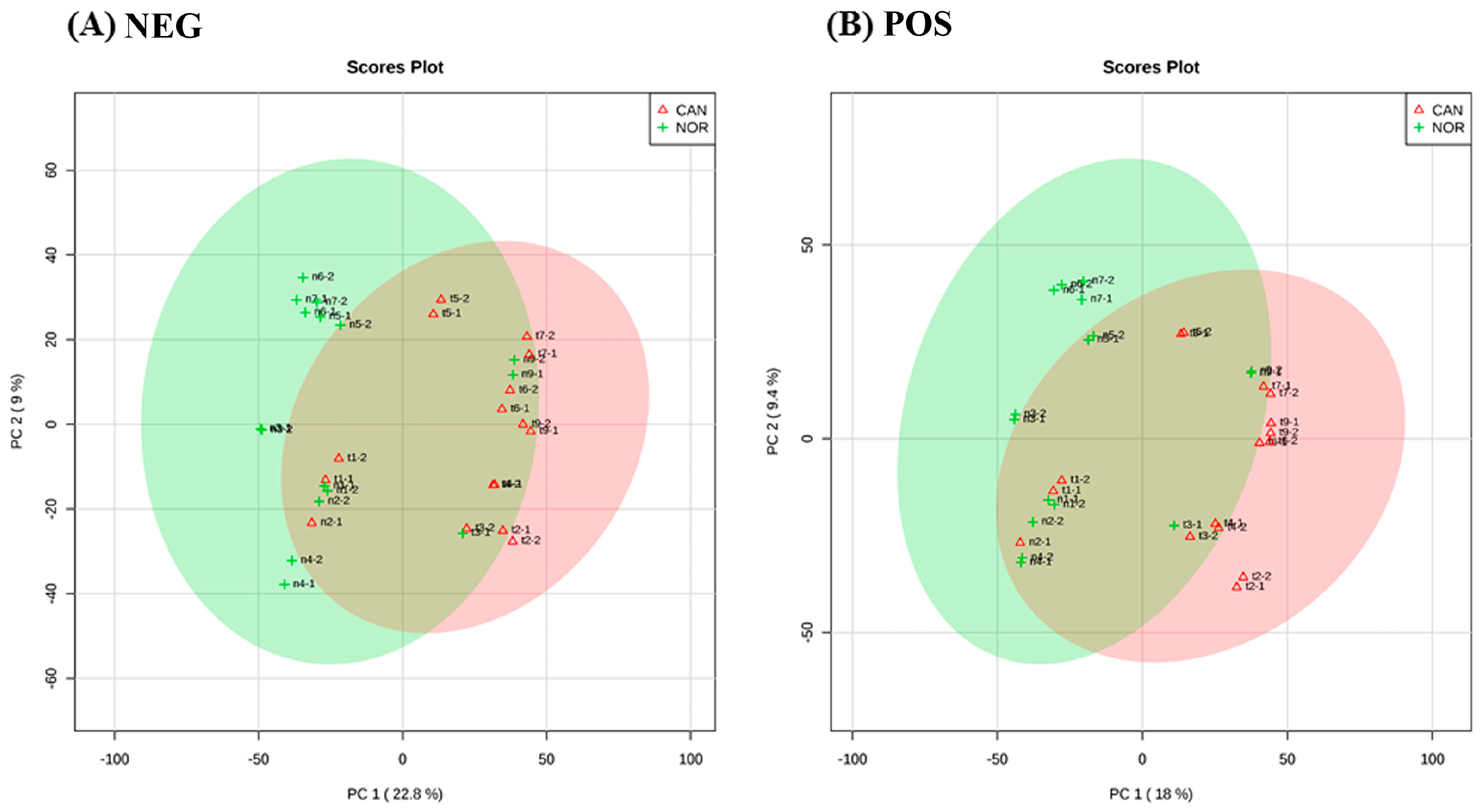
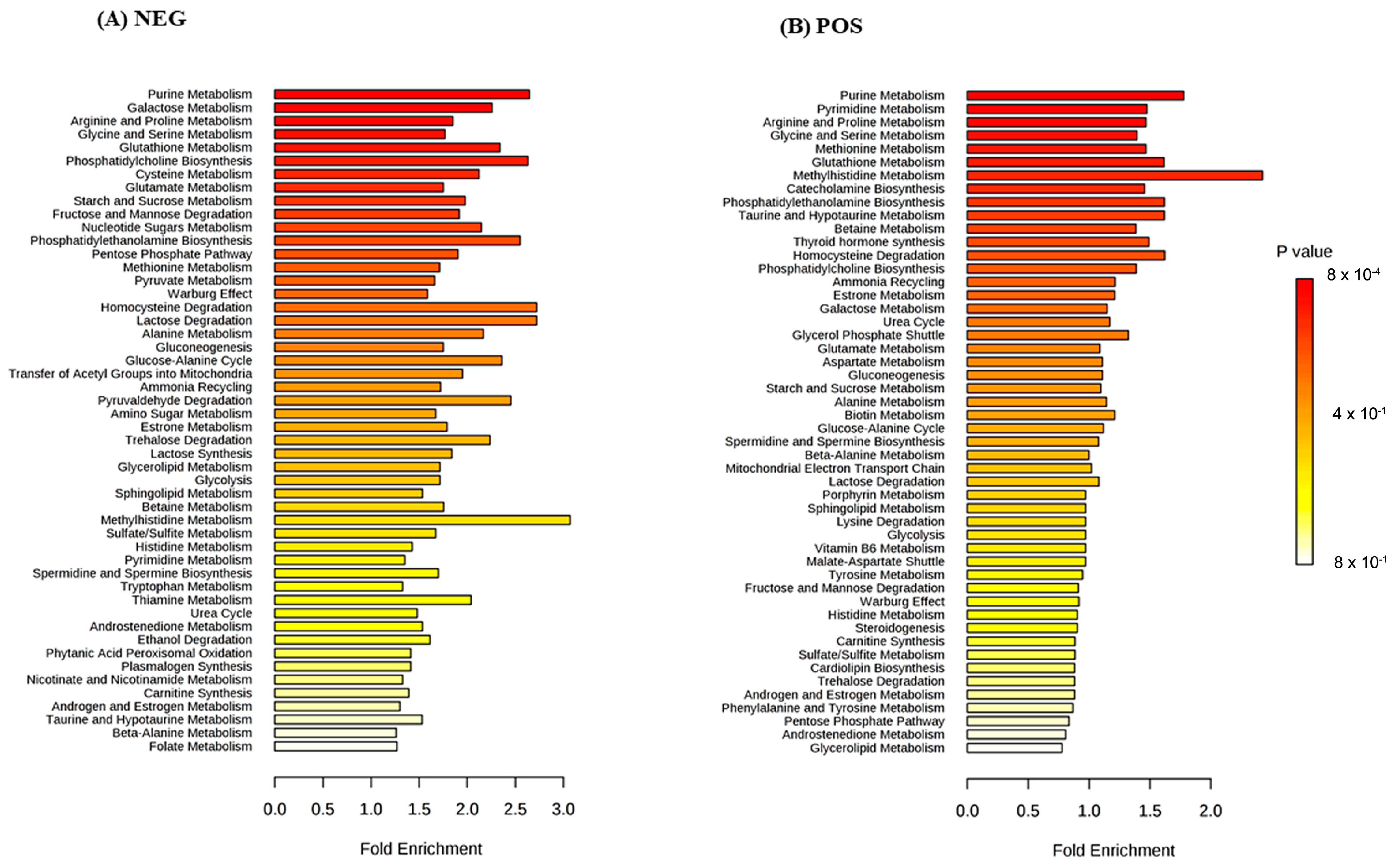
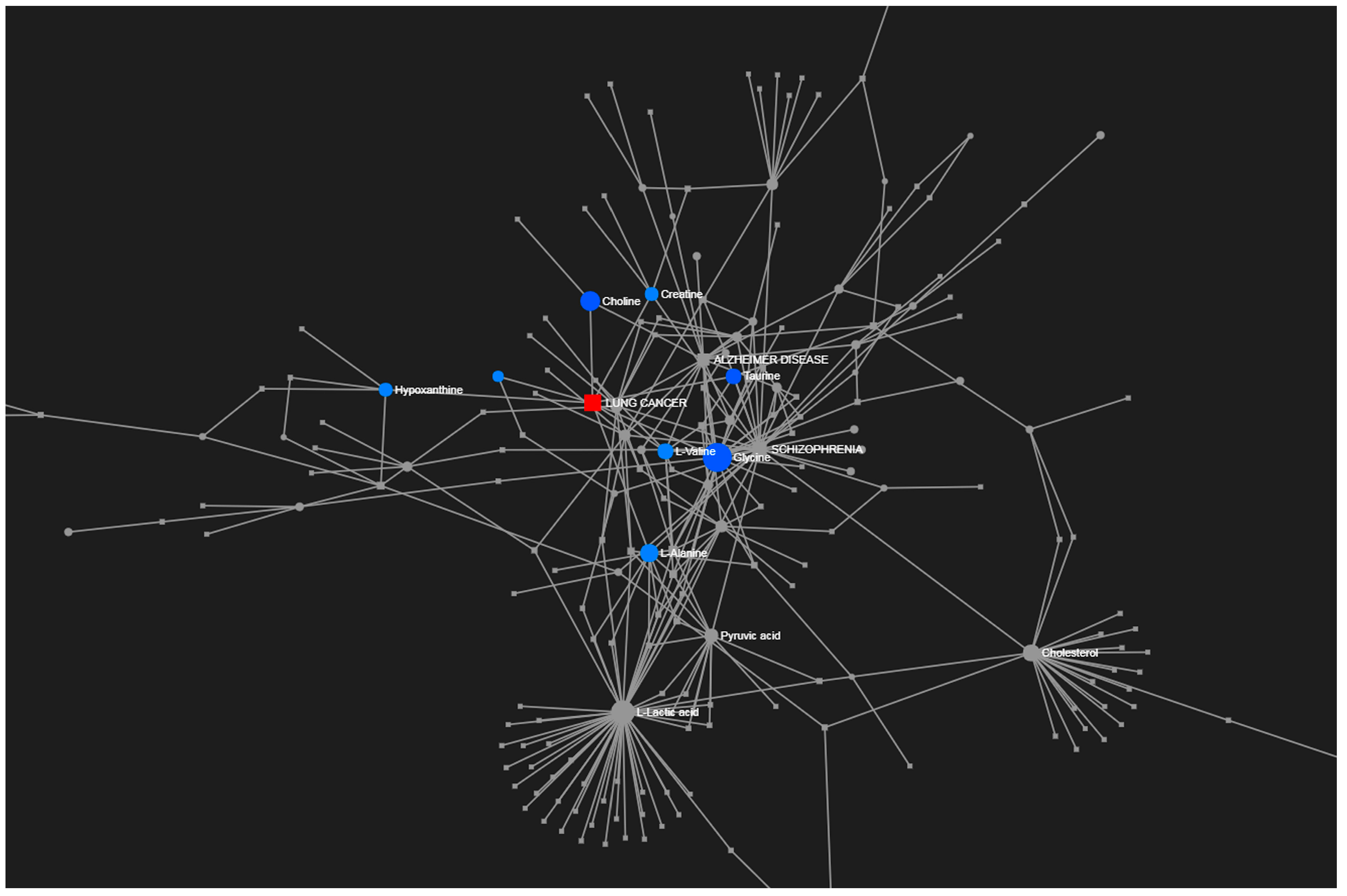
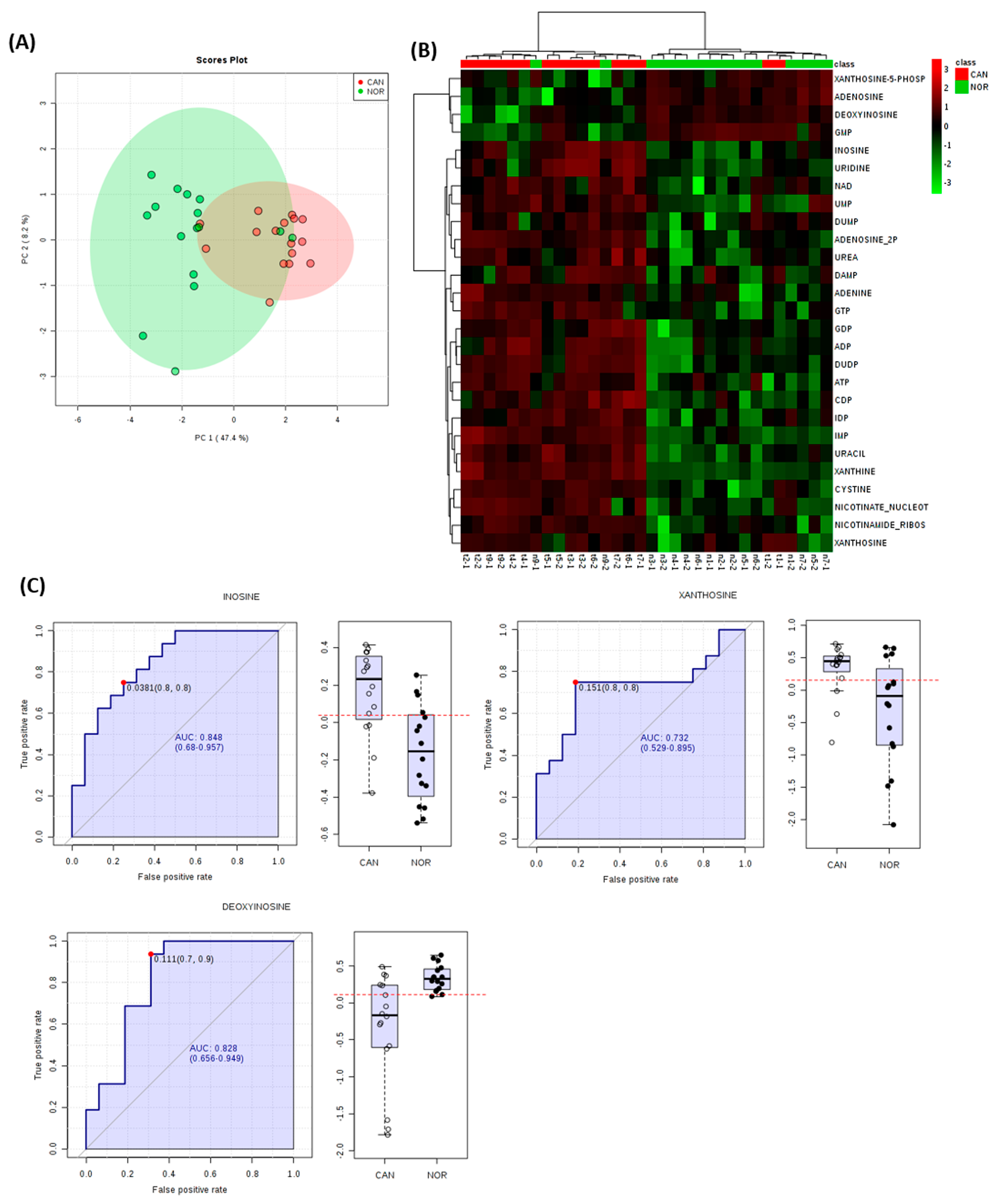
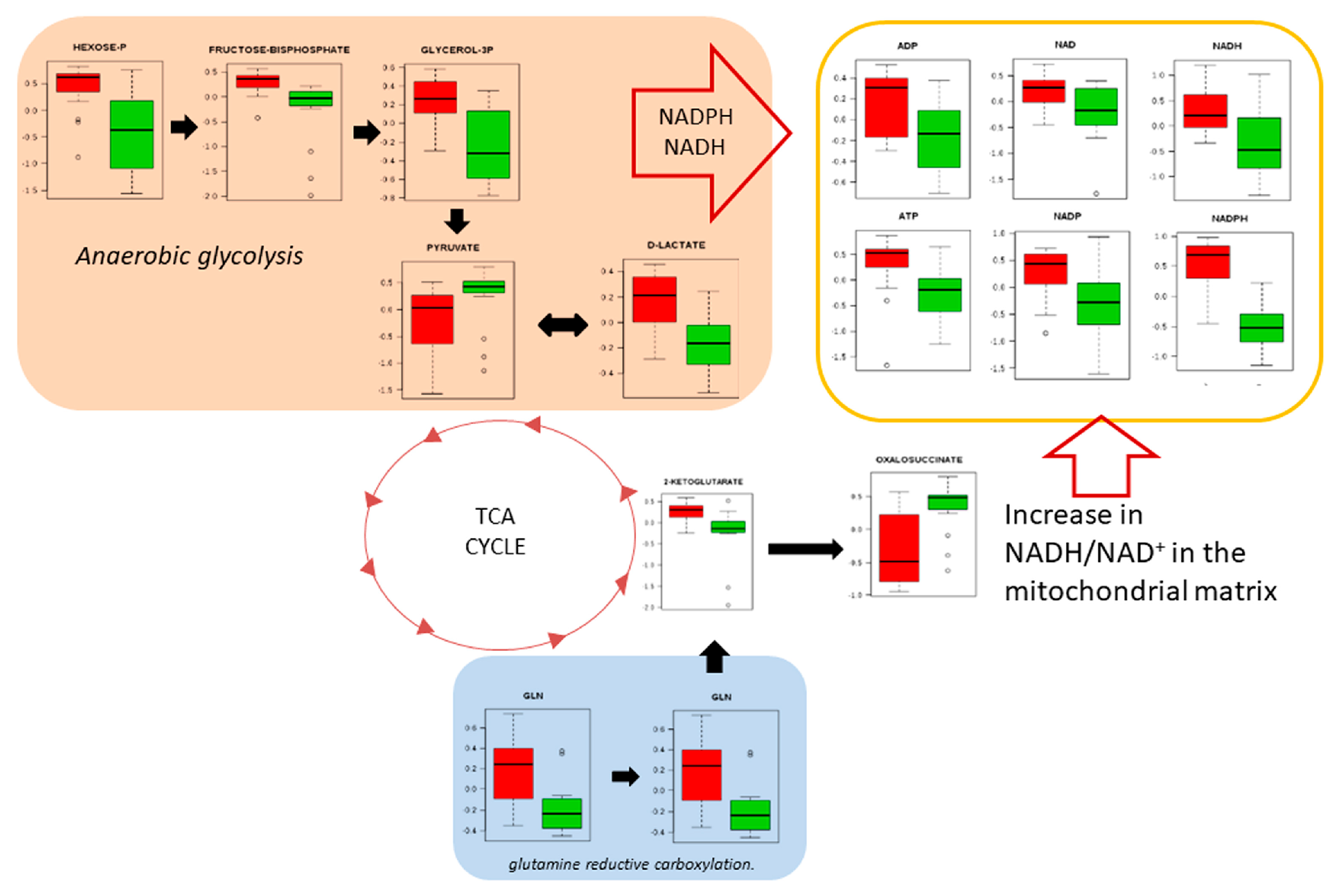
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Heymach, J.V.; Lippman, S.M. Lung cancer. N. Engl. J. Med. 2008, 359, 1367–1380. [Google Scholar] [CrossRef] [PubMed]
- The American Thoracic Society and The European Respiratory Society. Pretreatment evaluation of non-small-cell lung cancer. Am. J. Respir. Crit. Care Med. 1997, 156, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Langer, C.J.; Obasaju, C.; Bunn, P.; Bonomi, P.; Gandara, D.; Hirsch, F.R.; Kim, E.S.; Natale, R.B.; Novello, S.; Paz-Ares, L.; et al. Incremental Innovation and Progress in Advanced Squamous Cell Lung Cancer: Current Status and Future Impact of Treatment. J. Thorac. Oncol. 2016, 11, 2066–2081. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Overy, D.P.; Enot, D.P.; Tailliart, K.; Jenkins, H.; Parker, D.; Beckmann, M.; Draper, J. Explanatory signal interpretation and metabolite identification strategies for nominal mass FIE-MS metabolite fingerprints. Nat. Protoc. 2008, 3, 471–485. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Overy, S.; Quick, W.P. Evaluation of automated electrospray-TOF mass spectrometry for metabolic fingerprinting of the plant metabolome. Metabolomics 2005, 1, 137–148. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Robles, A.I.; Harris, C.C. Integration of multiple “OMIC” biomarkers: A precision medicine strategy for lung cancer. Lung Cancer 2017, 107, 50–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.; Agnihotri, S.; Micallef, J.; Mukherjee, J.; Sabha, N.; Cairns, R.; Hawkins, C.; Guha, A. Hexokinase 2 is a key mediator of aerobic glycolysis and promotes tumor growth in human glioblastoma multiforme. J. Exp. Med. 2011, 208, 313–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, T.W.; Lane, A.N.; Higashi, R.M.; Farag, M.A.; Gao, H.; Bousamra, M.; Miller, D.M. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol. Cancer. 2009, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S. Glucose-regulated proteins in cancer: Molecular mechanisms and therapeutic potential. Nat. Rev. Cancer 2014, 14, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Klupczynska, A.; Derezinski, P.; Garrett, T.J.; Rubio, V.Y.; Dyszkiewicz, W.; Kasprzyk, M.; Kokot, Z.J. Study of early stage non-small-cell lung cancer using Orbitrap-based global serum metabolomics. J. Cancer Res. Clin. 2017, 143, 649–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaitheesvaran, B.; Xu, J.; Yee, J.; Lu, Q.-Y.L.; Go, V.L.; Xiao, G.G.; Lee, W.N. The Warburg effect: A balance of flux analysis. Metabolomics 2015, 11, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Rocha, C.M.; Barros, A.S.; Gil, A.M.; Goodfellow, B.J.; Humpfer, E.; Spraul, M.; Carreira, I.M.; Melo, J.B.; Bernardo, J.; Gomes, A.; et al. Metabolic profiling of human lung cancer tissue by 1H high resolution magic angle spinning (HRMAS) NMR spectroscopy. J. Proteome Res. 2010, 9, 319–332. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, K.F.; Zhang, X.Y. Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: Mini review. Oncotarget 2017, 8, 115774–115786. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.J.; Lewis, K.E.; Beckmann, M.; Allison, G.G.; Ghosal, R.; Lewis, P.D.; Mur, L.A. The metabolomic detection of lung cancer biomarkers in sputum. Lung Cancer 2016, 94, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baptista, R.; Fazakerley, D.M.; Beckmann, M.; Baillie, L.; Mur, L.A.J. Untargeted metabolomics reveals a new mode of action of pretomanid (PA-824). Sci. Rep. 2018, 8, 5084. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Hackstadt, A.J.; Hess, A.M. Filtering for increased power for microarray data analysis. BMC Bioinform. 2009, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, F.; Ross, A.; Schlotterbeck, G.; Senn, H. Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in H-1 NMR metabonomics. Anal. Chem. 2006, 78, 4281–4290. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Z.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting Network Activity from High Throughput Metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, D.; Kim, S.Y. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008, 9, 189–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draper, J.; Enot, D.P.; Parker, D.; Beckmann, M.; Snowdon, S.; Lin, W.; Zubair, H. Metabolite signal identification in accurate mass metabolomics data with MZedDB, an interactive m/z annotation tool utilising predicted ionisation behaviour ‘rules’. BMC Bioinform. 2009, 10, 227. [Google Scholar] [CrossRef] [PubMed]
- Alberghina, L.; Gaglio, D. Redox control of glutamine utilization in cancer. Cell Death Dis. 2014, 5, e1561. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Jordan, K.W.; Adkins, C.B.; Su, L.; Halpern, E.F.; Mark, E.J.; Christiani, D.C.; Cheng, L.L. Comparison of squamous cell carcinoma and adenocarcinoma of the lung by metabolomic analysis of tissue-serum pairs. Lung Cancer 2010, 68, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Moreno, P.; Jimenez-Jimenez, C.; Garrido-Rodriguez, M.; Calderon-Santiago, M.; Molina, S.; Lara-Chica, M.; Priego-Capote, F.; Salvatierra, A.; Muñoz, E.; Calzado, M.A. Metabolomic profiling of human lung tumor tissues—Nucleotide metabolism as a candidate for therapeutic interventions and biomarkers. Mol Oncol. 2018, 12, 1778–1796. [Google Scholar] [CrossRef] [PubMed]
- Hori, S.; Nishiumi, S.; Kobayashi, K.; Shinohara, M.; Hatakeyama, Y.; Kotani, Y.; Hatano, N.; Maniwa, Y.; Nishio, W.; Bamba, T.; et al. A metabolomic approach to lung cancer. Lung Cancer 2011, 74, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Brunelli, L.; Caiola, E.; Marabese, M.; Broggini, M.; Pastorelli, R. Capturing the metabolomic diversity of KRAS mutants in non-small-cell lung cancer cells. Oncotarget 2014, 5, 4722–4731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amelio, I.; Cutruzzola, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, M.; Nilsson, R.; Sharma, S.; Madhusudhan, N.; Kitami, T.; Souza, A.L.; Kafri, R.; Kirschner, M.W.; Clish, C.B.; Mootha, V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012, 336, 1040–1044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E.; et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef] [PubMed]
- Keshet, R.; Szlosarek, P.; Carracedo, A.; Erez, A. Rewiring urea cycle metabolism in cancer to support anabolism. Nat. Rev. Cancer 2018, 18, 634–645. [Google Scholar] [CrossRef] [PubMed]
- Burke, A.J.; Sullivan, F.J.; Giles, F.J.; Glynn, S.A. The yin and yang of nitric oxide in cancer progression. Carcinogenesis 2013, 34, 503–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Y.B. Creatine kinase in cell cycle regulation and cancer. Amino Acids 2016, 48, 1775–1784. [Google Scholar] [CrossRef] [PubMed]
- Yokota, H.; Guo, J.; Matoba, M.; Higashi, K.; Tonami, H.; Nagao, Y. Lactate, choline, and creatine levels measured by vitro 1H-MRS as prognostic parameters in patients with non-small-cell lung cancer. J. Magn. Reson. Imaging. 2007, 25, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Gazdar, A.F.; Zweig, M.H.; Carney, D.N.; Van Steirteghen, A.C.; Baylin, S.B.; Minna, J.D. Levels of creatine kinase and its BB isoenzyme in lung cancer specimens and cultures. Cancer Res. 1981, 41, 2773–2777. [Google Scholar] [PubMed]
- Joseph, J.; Cardesa, A.; Carreras, J. Creatine kinase activity and isoenzymes in lung, colon and liver carcinomas. Brit. J. Cancer 1997, 76, 600–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; He, Y.; Ge, G.; Li, L.; Zhou, P.; Zhu, Y.; Tang, H.; Huang, Y.; Li, W.; Zhang, L. Lactate dehydrogenase and creatine kinase as poor prognostic factors in lung cancer: A retrospective observational study. PLoS ONE 2017, 12, e0182168. [Google Scholar] [Green Version]
- Fahrmann, J.F.; Grapov, D.; Wanichthanarak, K.; DeFelice, B.C.; Salemi, M.R.; Rom, W.N.; Gandara, D.R.; Phinney, B.S.; Fiehn, O.; Pass, H.; et al. Integrated metabolomics and proteomics highlight altered nicotinamide and polyamine pathways in lung adenocarcinoma. Carcinogenesis 2017, 38, 271–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, S.Z.; Bordey, A. GABA’s control of stem and cancer cell proliferation in adult neural and peripheral niches. Physiology 2009, 24, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Niu, X.; Liu, F.; Zhou, Y.; Zhou, Z.; Zhou, D.; Wang, T.; Li, Z.; Ye, X.; Yu, Y.; Weng, X.; et al. Genome-wide DNA Methylation Analysis Reveals GABBR2 as a Novel Epigenetic Target for EGFR 19 Deletion Lung Adenocarcinoma with Induction Erlotinib Treatment. Clin. Cancer Res. 2017, 23, 5003–5014. [Google Scholar] [CrossRef] [PubMed]
- Sarrouilhe, D.; Mesnil, M. Serotonin and human cancer: A critical view. Biochimie 2018. [Google Scholar] [CrossRef]
- Leboyer, M.; Philippe, A.; Bouvard, M.; Guilloud-Bataille, M.; Bondoux, D.; Tabuteau, F.; Feingold, J.; Mouren-Simeoni, M.C.; Launay, J.M. Whole blood serotonin and plasma beta-endorphin in autistic probands and their first-degree relatives. Biol. Psychiat. 1999, 45, 158–163. [Google Scholar] [CrossRef]
- Tu, S.; Zhang, X.L.; Wan, H.F.; Xia, Y.Q.; Liu, Z.Q.; Yang, X.H.; Wan, F.S. Effect of taurine on cell proliferation and apoptosis human lung cancer A549 cells. Oncol. Lett. 2018, 15, 5473–5480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Agouza, I.M.; Eissa, S.S.; El Houseini, M.M.; El-Nashar, D.E.; Abd El Hameed, O.M. Taurine: A novel tumor marker for enhanced detection of breast cancer among female patients. Angiogenesis 2011, 14, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Simon, M.C. Glutathione metabolism in cancer progression and treatment resistance. J. Cell Biol. 2018, 217, 2291–2298. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. Protecting the genome: Defence against nucleotide glycation and emerging role of glyoxalase I overexpression in multidrug resistance in cancer chemotherapy. Biochem. Soc. Trans. 2003, 31, 1372–1377. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J.; Waris, S.; Fleming, T.; Santarius, T.; Larkin, S.J.; Winklhofer-Roob, B.M.; Stratton, M.R.; Rabbani, N. Imidazopurinones are markers of physiological genomic damage linked to DNA instability and glyoxalase 1-associated tumour multidrug resistance. Nucleic Acids Res. 2010, 38, 5432–5442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutschenreuther, A.; Bigl, M.; Hemdan, N.Y.; Debebe, T.; Gaunitz, F.; Birkenmeier, G. Modulation of GLO1 Expression Affects Malignant Properties of Cells. Int. J. Mol. Sci. 2016, 17, 2133. [Google Scholar] [CrossRef] [PubMed]
- Rohrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Duvel, K.; Yecies, J.L.; Menon, S.; Raman, P.; Lipovsky, A.I.; Souza, A.L.; Triantafellow, E.; Ma, Q.; Gorski, R.; Cleaver, S.; et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell. 2010, 39, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Freeman, M.R.; Cinar, B.; Kim, J.; Mukhopadhyay, N.K.; Di Vizio, D.; Adam, R.M.; Solomon, K.R. Transit of hormonal and EGF receptor-dependent signals through cholesterol-rich membranes. Steroids 2007, 72, 210–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Irwin, M.E.; Mueller, K.L.; Bohin, N.; Ge, Y.B.; Boerner, J.L. Lipid Raft Localization of EGFR Alters the Response of Cancer Cells to the EGFR Tyrosine Kinase Inhibitor Gefitinib. J. Cell. Physiol. 2011, 226, 2316–2328. [Google Scholar] [CrossRef] [PubMed]
- Zastre, J.A.; Sweet, R.L.; Hanberry, B.S.; Ye, S. Linking vitamin B1 with cancer cell metabolism. Cancer Metab. 2013, 1, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buesa, J.M.; Garcia-Teijido, P.; Losa, R.; Fra, J. Treatment of ifosfamide encephalopathy with intravenous thiamin. Clin. Cancer Res. 2003, 9, 4636–4637. [Google Scholar] [PubMed]
- Ames, B.; Lewis, L.D.; Chaffee, S.; Kim, J.; Morse, R. Ifosfamide-induced encephalopathy and movement disorder. Pediatr. Blood Cancer 2010, 54, 624–626. [Google Scholar] [CrossRef] [PubMed]
- Hanberry, B.S.; Berger, R.; Zastre, J.A. High-dose vitamin B1 reduces proliferation in cancer cell lines analogous to dichloroacetate. Cancer Chemother Pharmacol. 2014, 73, 585–594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.T.; Chao, P.C.; Yin, M.C. Riboflavin at High Doses Enhances Lung Cancer Cell Proliferation, Invasion, and Migration. J. Food Sci. 2013, 78, H343–H349. [Google Scholar] [CrossRef] [PubMed]
- Merigliano, C.; Mascolo, E.; Burla, R.; Saggio, I.; Verni, F. The Relationship Between Vitamin B6, Diabetes and Cancer. Front. Genet. 2018, 9, 388. [Google Scholar] [CrossRef] [PubMed]
- Hartman, T.J.; Woodson, K.; Stolzenberg-Solomon, R.; Virtamo, J.; Selhub, J.; Barrett, M.J.; Albanes, D. Association of the B-vitamins pyridoxal 5’-phosphate (B-6), B-12, and folate with lung cancer risk in older men. Am. J. Epidemiol. 2001, 153, 688–694. [Google Scholar] [CrossRef] [PubMed]
- Gylling, B.; Myte, R.; Schneede, J.; Hallmans, G.; Haggstrom, J.; Johansson, I.; Ulvik, A.; Ueland, P.M.; Van Guelpen, B.; Palmqvist, R. Vitamin B-6 and colorectal cancer risk: A prospective population-based study using 3 distinct plasma markers of vitamin B-6 status. Am. J. Clin. Nutr. 2017, 105, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Routy, J.P.; Routy, B.; Graziani, G.M.; Mehraj, V. The Kynurenine Pathway Is a Double-Edged Sword in Immune-Privileged Sites and in Cancer: Implications for Immunotherapy. Int. J. Tryptophan Res. 2016, 9, 67–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Variables | N |
|---|---|
| Age at diagnosis (years) | |
| Median (range) | 55 (39–71) |
| Gender | |
| Male | 5 |
| Female | 3 |
| Tobacco use | |
| Yes | 7 |
| No | 1 |
| Alcohol use | |
| Yes | 5 |
| No | 3 |
| T category | |
| T1-T2 | 6 |
| T3-T4 | 2 |
| Nodal status (pathological) | |
| Negative (N0) | 6 |
| Positive (N1, N2) | 2 |
| Disease stage | |
| IA | 1 |
| IIA | 1 |
| IIIA | 2 |
| IB | 3 |
| IIB | 1 |
| Tumour grade | |
| Well differentiated | 1 |
| Moderately differentiated | 5 |
| Poorly differentiated | 2 |
| Perineural invasion | |
| Yes | 1 |
| No | 7 |
| Angiolymphatic invasion | |
| Yes | 6 |
| No | 2 |
| Outcome | |
| Alive with disease | 6 |
| Death cause by disease | 2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paes de Araújo, R.; Bertoni, N.; Seneda, A.L.; Felix, T.F.; Carvalho, M.; Lewis, K.E.; Hasimoto, É.N.; Beckmann, M.; Drigo, S.A.; Reis, P.P.; et al. Defining Metabolic Rewiring in Lung Squamous Cell Carcinoma. Metabolites 2019, 9, 47. https://doi.org/10.3390/metabo9030047
Paes de Araújo R, Bertoni N, Seneda AL, Felix TF, Carvalho M, Lewis KE, Hasimoto ÉN, Beckmann M, Drigo SA, Reis PP, et al. Defining Metabolic Rewiring in Lung Squamous Cell Carcinoma. Metabolites. 2019; 9(3):47. https://doi.org/10.3390/metabo9030047
Chicago/Turabian StylePaes de Araújo, Rachel, Natália Bertoni, Ana L. Seneda, Tainara F. Felix, Márcio Carvalho, Keir E. Lewis, Érica N. Hasimoto, Manfred Beckmann, Sandra A. Drigo, Patricia P. Reis, and et al. 2019. "Defining Metabolic Rewiring in Lung Squamous Cell Carcinoma" Metabolites 9, no. 3: 47. https://doi.org/10.3390/metabo9030047
APA StylePaes de Araújo, R., Bertoni, N., Seneda, A. L., Felix, T. F., Carvalho, M., Lewis, K. E., Hasimoto, É. N., Beckmann, M., Drigo, S. A., Reis, P. P., & Mur, L. A. J. (2019). Defining Metabolic Rewiring in Lung Squamous Cell Carcinoma. Metabolites, 9(3), 47. https://doi.org/10.3390/metabo9030047







