Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics
Abstract
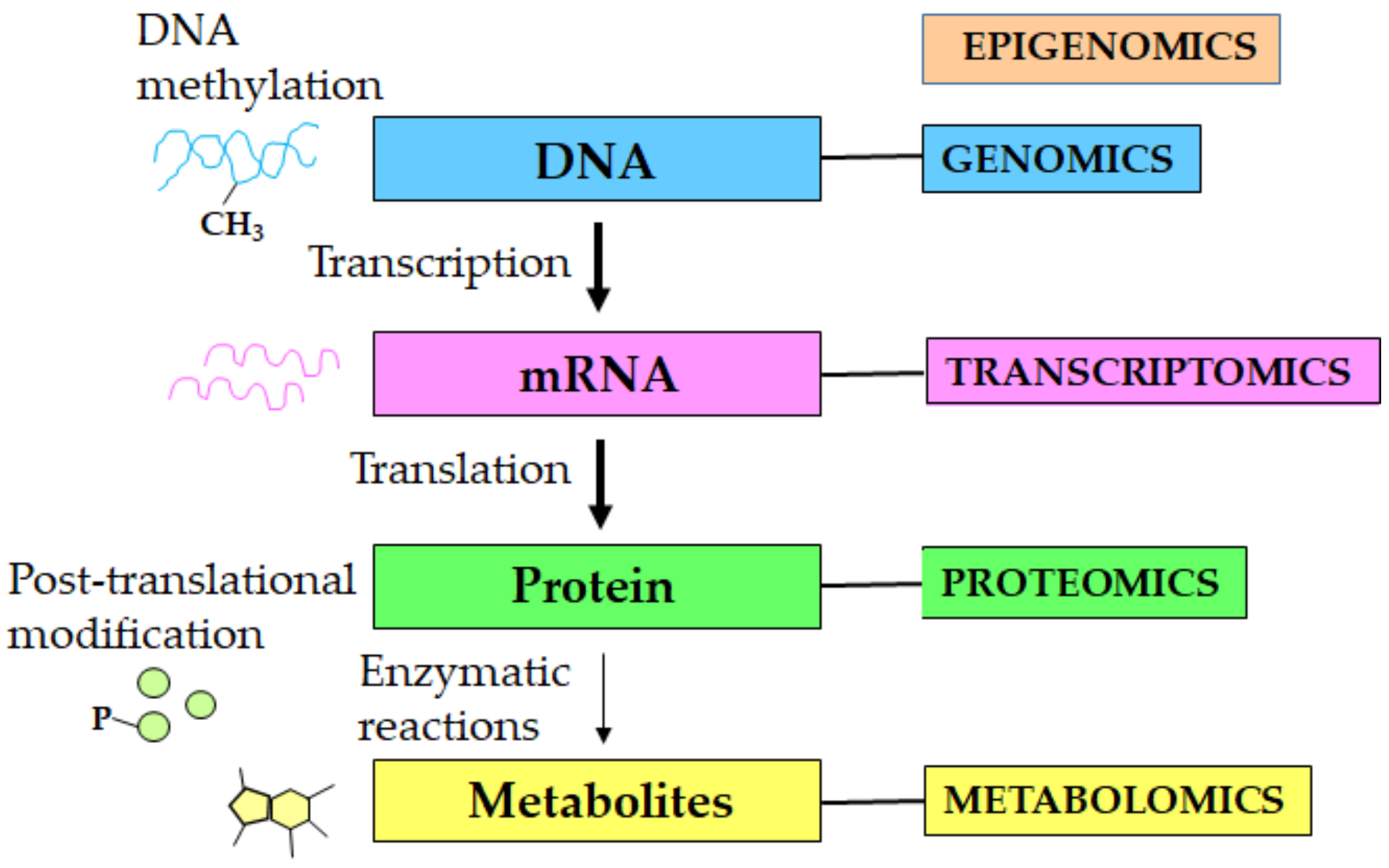
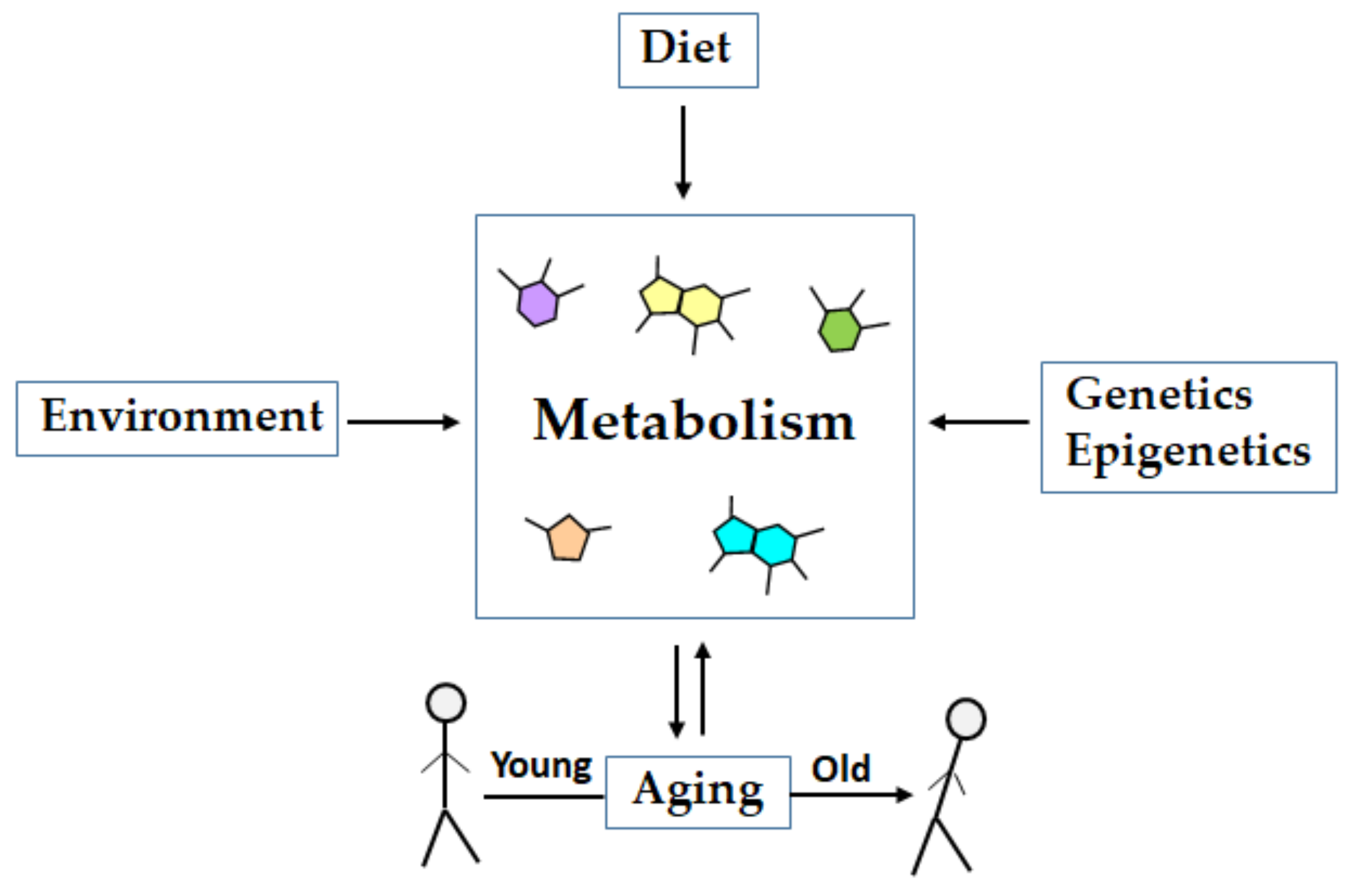
1. Introduction
2. Metabolomics of Aging
2.1. Biomarkers of Aging
2.2. Aging Studies in Model Organisms
2.3. Aging Studies in Humans
3. Conclusions and Perspective
Funding
Acknowledgments
Conflicts of Interest
References
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed]
- Forster, J.; Famili, I.; Fu, P.; Palsson, B.O.; Nielsen, J. Genome-scale reconstruction of the saccharomyces cerevisiae metabolic network. Genome Res. 2003, 13, 244–253. [Google Scholar] [CrossRef]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vazquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The human metabolome database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef]
- Fiehn, O. Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp. Funct. Genomics 2001, 2, 155–168. [Google Scholar] [CrossRef]
- Saito, K.; Matsuda, F. Metabolomics for functional genomics, systems biology, and biotechnology. Annu. Rev. Plant Biol. 2010, 61, 463–489. [Google Scholar] [CrossRef]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Kell, D.B.; Oliver, S.G. The metabolome 18 years on: A concept comes of age. Metabolomics 2016, 12, 148. [Google Scholar] [CrossRef] [PubMed]
- Raamsdonk, L.M.; Teusink, B.; Broadhurst, D.; Zhang, N.; Hayes, A.; Walsh, M.C.; Berden, J.A.; Brindle, K.M.; Kell, D.B.; Rowland, J.J.; et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 2001, 19, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Siuzdak, G. The role of metabolomics in brain metabolism research. J. Neuroimmune Pharmcol. 2015, 10, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Esterhuizen, K.; van der Westhuizen, F.H.; Louw, R. Metabolomics of mitochondrial disease. Mitochondrion 2017, 35, 97–110. [Google Scholar] [CrossRef]
- Sakaguchi, C.A.; Nieman, D.C.; Signini, E.F.; Abreu, R.M.; Catai, A.M. Metabolomics-based studies assessing exercise-induced alterations of the human metabolome: A systematic review. Metabolites 2019, 9, 164. [Google Scholar] [CrossRef]
- Gomase, V.S.; Changbhale, S.S.; Patil, S.A.; Kale, K.V. Metabolomics. Curr. Drug Metab. 2008, 9, 89–98. [Google Scholar] [CrossRef]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef]
- Aretz, I.; Meierhofer, D. Advantages and pitfalls of mass spectrometry based metabolome profiling in systems biology. Int. J. Mol. Sci. 2016, 17, 632. [Google Scholar] [CrossRef]
- Nagana Gowda, G.A.; Raftery, D. Recent advances in NMR-based metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef]
- Markley, J.L.; Bruschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef]
- Emwas, A.H.; Roy, R.; McKay, R.T.; Tenori, L.; Saccenti, E.; Gowda, G.A.N.; Raftery, D.; Alahmari, F.; Jaremko, L.; Jaremko, M.; et al. NMR spectroscopy for metabolomics research. Metabolites 2019, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Bailey, N.J.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Bouatra, S.; Aziat, F.; Mandal, R.; Guo, A.C.; Wilson, M.R.; Knox, C.; Bjorndahl, T.C.; Krishnamurthy, R.; Saleem, F.; Liu, P.; et al. The human urine metabolome. PLoS ONE 2013, 8, e73076. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Li, G.; Seymour, A.B. High-throughput and multiplexed lc/ms/mrm method for targeted metabolomics. Anal. Chem. 2010, 82, 5527–5533. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted metabolomics. Curr. Protoc. Mol. Biol. 2012, 30, 30.2.1–30.2.24. [Google Scholar] [CrossRef]
- Cajka, T.; Fiehn, O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef]
- Vinayavekhin, N.; Saghatelian, A. Untargeted metabolomics. Curr. Protoc. Mol. Biol. 2010, 90, 30.1.1–30.1.24. [Google Scholar] [CrossRef]
- Zhu, Z.J.; Schultz, A.W.; Wang, J.; Johnson, C.H.; Yannone, S.M.; Patti, G.J.; Siuzdak, G. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the metlin database. Nat. Protoc. 2013, 8, 451–460. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, X.; Wang, C.; Zhang, H.; Cai, Z. Non-targeted and targeted metabolomics approaches to diagnosing lung cancer and predicting patient prognosis. Oncotarget 2016, 7, 63437–63448. [Google Scholar] [CrossRef] [PubMed]
- Dias, D.A.; Jones, O.A.; Beale, D.J.; Boughton, B.A.; Benheim, D.; Kouremenos, K.A.; Wolfender, J.L.; Wishart, D.S. Current and future perspectives on the structural identification of small molecules in biological systems. Metabolites 2016, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Patti, G.J.; Yanes, O.; Siuzdak, G. Innovation: Metabolomics: The apogee of the omics trilogy. Nat. Rev. Mol. Cell Biol. 2012, 13, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via uplc-ms. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Members, M.S.I.B.; Sansone, S.A.; Fan, T.; Goodacre, R.; Griffin, J.L.; Hardy, N.W.; Kaddurah-Daouk, R.; Kristal, B.S.; Lindon, J.; Mendes, P.; et al. The metabolomics standards initiative. Nat. Biotechnol. 2007, 25, 846–848. [Google Scholar] [CrossRef]
- Zierer, J.; Menni, C.; Kastenmuller, G.; Spector, T.D. Integration of ‘omics’ data in aging research: From biomarkers to systems biology. Aging Cell 2015, 14, 933–944. [Google Scholar] [CrossRef]
- Valdes, A.M.; Glass, D.; Spector, T.D. Omics technologies and the study of human ageing. Nat. Rev. Genet. 2013, 14, 601–607. [Google Scholar] [CrossRef]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdak, G. Metlin: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. Massbank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef]
- Cui, Q.; Lewis, I.A.; Hegeman, A.D.; Anderson, M.E.; Li, J.; Schulte, C.F.; Westler, W.M.; Eghbalnia, H.R.; Sussman, M.R.; Markley, J.L. Metabolite identification via the madison metabolomics consortium database. Nat. Biotechnol. 2008, 26, 162–164. [Google Scholar] [CrossRef] [PubMed]
- Sud, M.; Fahy, E.; Cotter, D.; Brown, A.; Dennis, E.A.; Glass, C.K.; Merrill, A.H., Jr.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; et al. Lmsd: Lipid maps structure database. Nucleic Acids Res. 2007, 35, D527–D532. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Sud, M.; Cotter, D.; Subramaniam, S. Lipid maps online tools for lipid research. Nucleic Acids Res. 2007, 35, W606–W612. [Google Scholar] [CrossRef] [PubMed]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom. Rev. 2011, 30, 884–906. [Google Scholar] [CrossRef]
- Kaddurah-Daouk, R.; Kristal, B.S.; Weinshilboum, R.M. Metabolomics: A global biochemical approach to drug response and disease. Annu. Rev. Pharm. Toxicol. 2008, 48, 653–683. [Google Scholar] [CrossRef]
- Jones, D.P.; Park, Y.; Ziegler, T.R. Nutritional metabolomics: Progress in addressing complexity in diet and health. Annu. Rev. Nutr. 2012, 32, 183–202. [Google Scholar] [CrossRef]
- Alonso, A.; Marsal, S.; Julia, A. Analytical methods in untargeted metabolomics: State of the art in 2015. Front. Bioeng. Biotechnol. 2015, 3, 23. [Google Scholar] [CrossRef]
- Houten, S.M. Metabolomics: Unraveling the chemical individuality of common human diseases. Ann. Med. 2009, 41, 402–407. [Google Scholar] [CrossRef]
- Wild, C.P.; Scalbert, A.; Herceg, Z. Measuring the exposome: A powerful basis for evaluating environmental exposures and cancer risk. Environ. Mol. Mutagen. 2013, 54, 480–499. [Google Scholar] [CrossRef]
- Jove, M.; Portero-Otin, M.; Naudi, A.; Ferrer, I.; Pamplona, R. Metabolomics of human brain aging and age-related neurodegenerative diseases. J. Neuropathol. Exp. Neurol. 2014, 73, 640–657. [Google Scholar] [CrossRef]
- Wishart, D.S. Applications of metabolomics in drug discovery and development. Drugs R D 2008, 9, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Kaddurah-Daouk, R.; Krishnan, K.R. Metabolomics: A global biochemical approach to the study of central nervous system diseases. Neuropsychopharmacology 2009, 34, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Everett, J.R. Pharmacometabonomics in humans: A new tool for personalized medicine. Pharmacogenomics 2015, 16, 737–754. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Otin, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Srivastava, S. The mitochondrial basis of aging and age-related disorders. Genes 2017, 8, 398. [Google Scholar] [CrossRef]
- Yu, Z.; Zhai, G.; Singmann, P.; He, Y.; Xu, T.; Prehn, C.; Romisch-Margl, W.; Lattka, E.; Gieger, C.; Soranzo, N.; et al. Human serum metabolic profiles are age dependent. Aging Cell 2012, 11, 960–967. [Google Scholar] [CrossRef]
- Vijg, J.; Suh, Y. Genome instability and aging. Annu. Rev. Physiol. 2013, 75, 645–668. [Google Scholar] [CrossRef]
- Vermulst, M.; Denney, A.S.; Lang, M.J.; Hung, C.W.; Moore, S.; Moseley, M.A.; Thompson, J.W.; Madden, V.; Gauer, J.; Wolfe, K.J.; et al. Transcription errors induce proteotoxic stress and shorten cellular lifespan. Nat. Commun. 2015, 6, 8065. [Google Scholar] [CrossRef]
- Chaleckis, R.; Murakami, I.; Takada, J.; Kondoh, H.; Yanagida, M. Individual variability in human blood metabolites identifies age-related differences. Proc. Natl. Acad. Sci. USA 2016, 113, 4252–4259. [Google Scholar] [CrossRef]
- Ke, Z.; Mallik, P.; Johnson, A.B.; Luna, F.; Nevo, E.; Zhang, Z.D.; Gladyshev, V.N.; Seluanov, A.; Gorbunova, V. Translation fidelity coevolves with longevity. Aging Cell 2017, 16, 988–993. [Google Scholar] [CrossRef]
- Kirkwood, T.B.; Austad, S.N. Why do we age? Nature 2000, 408, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Chen, K. The essential mechanisms of aging: Irreparable damage accumulation of biochemical side-reactions. Exp. Gerontol. 2005, 40, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Jin, K. Modern biological theories of aging. Aging Dis 2010, 1, 72–74. [Google Scholar] [PubMed]
- Golubev, A.; Hanson, A.D.; Gladyshev, V.N. Non-enzymatic molecular damage as a prototypic driver of aging. J. Biol. Chem. 2017, 292, 6029–6038. [Google Scholar] [CrossRef]
- Brandhorst, S.; Choi, I.Y.; Wei, M.; Cheng, C.W.; Sedrakyan, S.; Navarrete, G.; Dubeau, L.; Yap, L.P.; Park, R.; Vinciguerra, M.; et al. A periodic diet that mimics fasting promotes multi-system regeneration, enhanced cognitive performance, and healthspan. Cell Metab. 2015, 22, 86–99. [Google Scholar] [CrossRef]
- Cartee, G.D.; Hepple, R.T.; Bamman, M.M.; Zierath, J.R. Exercise promotes healthy aging of skeletal muscle. Cell Metab. 2016, 23, 1034–1047. [Google Scholar] [CrossRef]
- Wei, M.; Brandhorst, S.; Shelehchi, M.; Mirzaei, H.; Cheng, C.W.; Budniak, J.; Groshen, S.; Mack, W.J.; Guen, E.; Di Biase, S.; et al. Fasting-mimicking diet and markers/risk factors for aging, diabetes, cancer, and cardiovascular disease. Sci. Transl. Med. 2017, 9, eaai8700. [Google Scholar] [CrossRef]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut microbiota and extreme longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef]
- Biagi, E.; Rampelli, S.; Turroni, S.; Quercia, S.; Candela, M.; Brigidi, P. The gut microbiota of centenarians: Signatures of longevity in the gut microbiota profile. Mech. Ageing Dev. 2017, 165, 180–184. [Google Scholar] [CrossRef]
- Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clin. Pharmcol. Ther. 2001, 69, 89–95. [Google Scholar] [CrossRef]
- WHO. Who International Programme on Chemical Safety Biomarkers in Risk Assessment: Validity and Validation; World Health Organization: Geneva, Switzerland, 2001. [Google Scholar]
- Strimbu, K.; Tavel, J.A. What are biomarkers? Curr. Opin. HIV AIDS 2010, 5, 463–466. [Google Scholar] [CrossRef] [PubMed]
- Sebastiani, P.; Thyagarajan, B.; Sun, F.; Schupf, N.; Newman, A.B.; Montano, M.; Perls, T.T. Biomarker signatures of aging. Aging Cell 2017, 16, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Chen, W.; McDermott, J.; Han, J.J. Molecular and phenotypic biomarkers of aging. F1000Research 2017, 6, 860. [Google Scholar] [CrossRef] [PubMed]
- Burkle, A.; Moreno-Villanueva, M.; Bernhard, J.; Blasco, M.; Zondag, G.; Hoeijmakers, J.H.; Toussaint, O.; Grubeck-Loebenstein, B.; Mocchegiani, E.; Collino, S.; et al. Mark-age biomarkers of ageing. Mech. Ageing Dev. 2015, 151, 2–12. [Google Scholar] [CrossRef] [PubMed]
- Crimmins, E.; Vasunilashorn, S.; Kim, J.K.; Alley, D. Biomarkers related to aging in human populations. Adv. Clin. Chem. 2008, 46, 161–216. [Google Scholar]
- Fuchs, S.; Bundy, J.G.; Davies, S.K.; Viney, J.M.; Swire, J.S.; Leroi, A.M. A metabolic signature of long life in caenorhabditis elegans. BMC Biol. 2010, 8, 14. [Google Scholar] [CrossRef]
- Avanesov, A.S.; Ma, S.; Pierce, K.A.; Yim, S.H.; Lee, B.C.; Clish, C.B.; Gladyshev, V.N. Age- and diet-associated metabolome remodeling characterizes the aging process driven by damage accumulation. Elife 2014, 3, e02077. [Google Scholar] [CrossRef]
- Hoffman, J.M.; Soltow, Q.A.; Li, S.; Sidik, A.; Jones, D.P.; Promislow, D.E. Effects of age, sex, and genotype on high-sensitivity metabolomic profiles in the fruit fly, drosophila melanogaster. Aging Cell 2014, 13, 596–604. [Google Scholar] [CrossRef]
- Tomas-Loba, A.; Bernardes de Jesus, B.; Mato, J.M.; Blasco, M.A. A metabolic signature predicts biological age in mice. Aging Cell 2013, 12, 93–101. [Google Scholar] [CrossRef]
- Houtkooper, R.H.; Argmann, C.; Houten, S.M.; Canto, C.; Jeninga, E.H.; Andreux, P.A.; Thomas, C.; Doenlen, R.; Schoonjans, K.; Auwerx, J. The metabolic footprint of aging in mice. Sci. Rep. 2011, 1, 134. [Google Scholar] [CrossRef]
- Shahmirzadi, A.A.; Edgar, D.; Liao, C.-Y.; Hsu, Y.-M.; Lucanic, M.; Shahmirzadi, A.A.; Wiley, C.; Riley, R.; Kaplowitz, B.; Gan, G.; et al. Alpha-ketoglutarate, an endogenous metabolite, extends lifespan and compresses morbidity in aging mice. bioRxiv 2019, 779157. [Google Scholar] [CrossRef]
- Su, Y.; Wang, T.; Wu, N.; Li, D.; Fan, X.; Xu, Z.; Mishra, S.K.; Yang, M. Alpha-ketoglutarate extends drosophila lifespan by inhibiting mtor and activating ampk. Aging 2019, 11, 4183–4197. [Google Scholar] [CrossRef] [PubMed]
- Mishur, R.J.; Khan, M.; Munkacsy, E.; Sharma, L.; Bokov, A.; Beam, H.; Radetskaya, O.; Borror, M.; Lane, R.; Bai, Y.; et al. Mitochondrial metabolites extend lifespan. Aging Cell 2016, 15, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Chin, R.M.; Fu, X.; Pai, M.Y.; Vergnes, L.; Hwang, H.; Deng, G.; Diep, S.; Lomenick, B.; Meli, V.S.; Monsalve, G.C.; et al. The metabolite alpha-ketoglutarate extends lifespan by inhibiting atp synthase and tor. Nature 2014, 510, 397–401. [Google Scholar] [CrossRef]
- Collino, S.; Montoliu, I.; Martin, F.P.; Scherer, M.; Mari, D.; Salvioli, S.; Bucci, L.; Ostan, R.; Monti, D.; Biagi, E.; et al. Metabolic signatures of extreme longevity in northern Italian centenarians reveal a complex remodeling of lipids, amino acids, and gut microbiota metabolism. PLoS ONE 2013, 8, e56564. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Jouandin, P.; Mohr, S.E.; Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 2019, e13034. [Google Scholar] [CrossRef]
- Parkhitko, A.A.; Binari, R.; Zhang, N.; Asara, J.M.; Demontis, F.; Perrimon, N. Tissue-specific down-regulation of s-adenosyl-homocysteine via suppression of dahcyl1/dahcyl2 extends health span and life span in drosophila. Genes Dev. 2016, 30, 1409–1422. [Google Scholar] [CrossRef]
- Laye, M.J.; Tran, V.; Jones, D.P.; Kapahi, P.; Promislow, D.E. The effects of age and dietary restriction on the tissue-specific metabolome of drosophila. Aging Cell 2015, 14, 797–808. [Google Scholar] [CrossRef]
- Ball, H.C.; Levari-Shariati, S.; Cooper, L.N.; Aliani, M. Comparative metabolomics of aging in a long-lived bat: Insights into the physiology of extreme longevity. PLoS ONE 2018, 13, e0196154. [Google Scholar] [CrossRef]
- Wijeyesekera, A.; Selman, C.; Barton, R.H.; Holmes, E.; Nicholson, J.K.; Withers, D.J. Metabotyping of long-lived mice using 1h NMR spectroscopy. J. Proteome Res. 2012, 11, 2224–2235. [Google Scholar] [CrossRef]
- De Guzman, J.M.; Ku, G.; Fahey, R.; Youm, Y.H.; Kass, I.; Ingram, D.K.; Dixit, V.D.; Kheterpal, I. Chronic caloric restriction partially protects against age-related alteration in serum metabolome. Age 2013, 35, 1091–1104. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Yim, S.H.; Lee, S.G.; Kim, E.B.; Lee, S.R.; Chang, K.T.; Buffenstein, R.; Lewis, K.N.; Park, T.J.; Miller, R.A.; et al. Organization of the mammalian metabolome according to organ function, lineage specialization, and longevity. Cell Metab. 2015, 22, 332–343. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Stauch, K.L.; Petrascheck, M.; Benton, H.P.; Epstein, A.A.; Fang, M.; Gorantla, S.; Tran, M.; Hoang, L.; Kurczy, M.E.; et al. Metabolic drift in the aging brain. Aging 2016, 8, 1000–1020. [Google Scholar] [CrossRef] [PubMed]
- Jewison, T.; Knox, C.; Neveu, V.; Djoumbou, Y.; Guo, A.C.; Lee, J.; Liu, P.; Mandal, R.; Krishnamurthy, R.; Sinelnikov, I.; et al. Ymdb: The yeast metabolome database. Nucleic Acids Res. 2012, 40, D815–D820. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Ikeda, S.; Niigata, K.; Tomita, M.; Sato, H.; Soga, T. Mmmdb: Mouse multiple tissue metabolome database. Nucleic Acids Res. 2012, 40, D809–D814. [Google Scholar] [CrossRef] [PubMed]
- Cevenini, E.; Bellavista, E.; Tieri, P.; Castellani, G.; Lescai, F.; Francesconi, M.; Mishto, M.; Santoro, A.; Valensin, S.; Salvioli, S.; et al. Systems biology and longevity: An emerging approach to identify innovative anti-aging targets and strategies. Curr. Pharm. Des. 2010, 16, 802–813. [Google Scholar] [CrossRef]
- Lu, T.; Pan, Y.; Kao, S.Y.; Li, C.; Kohane, I.; Chan, J.; Yankner, B.A. Gene regulation and DNA damage in the ageing human brain. Nature 2004, 429, 883–891. [Google Scholar] [CrossRef]
- Yeoman, M.; Scutt, G.; Faragher, R. Insights into cns ageing from animal models of senescence. Nat. Rev. Neurosci. 2012, 13, 435–445. [Google Scholar] [CrossRef]
- Horvath, S.; Zhang, Y.; Langfelder, P.; Kahn, R.S.; Boks, M.P.; van Eijk, K.; van den Berg, L.H.; Ophoff, R.A. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012, 13, R97. [Google Scholar] [CrossRef]
- Hannum, G.; Guinney, J.; Zhao, L.; Zhang, L.; Hughes, G.; Sadda, S.; Klotzle, B.; Bibikova, M.; Fan, J.B.; Gao, Y.; et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 2013, 49, 359–367. [Google Scholar] [CrossRef]
- Glass, D.; Vinuela, A.; Davies, M.N.; Ramasamy, A.; Parts, L.; Knowles, D.; Brown, A.A.; Hedman, A.K.; Small, K.S.; Buil, A.; et al. Gene expression changes with age in skin, adipose tissue, blood and brain. Genome Biol. 2013, 14, R75. [Google Scholar] [CrossRef]
- Stauch, K.L.; Purnell, P.R.; Villeneuve, L.M.; Fox, H.S. Proteomic analysis and functional characterization of mouse brain mitochondria during aging reveal alterations in energy metabolism. Proteomics 2015, 15, 1574–1586. [Google Scholar] [CrossRef] [PubMed]
- Anton, B.; Vitetta, L.; Cortizo, F.; Sali, A. Can we delay aging? The biology and science of aging. Ann. N. Y. Acad. Sci. 2005, 1057, 525–535. [Google Scholar] [CrossRef]
- Mootha, V.K.; Hirschhorn, J.N. Inborn variation in metabolism. Nat. Genet. 2010, 42, 97–98. [Google Scholar] [CrossRef]
- Illig, T.; Gieger, C.; Zhai, G.; Romisch-Margl, W.; Wang-Sattler, R.; Prehn, C.; Altmaier, E.; Kastenmuller, G.; Kato, B.S.; Mewes, H.W.; et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010, 42, 137–141. [Google Scholar] [CrossRef]
- Pimenta, L.P.; Kim, H.K.; Verpoorte, R.; Choi, Y.H. NMR-based metabolomics: A probe to utilize biodiversity. Methods Mol. Biol. 2013, 1055, 117–127. [Google Scholar]
- Baker, M. Metabolomics: From small molecules to big ideas. Nat. Methods 2011, 8, 117–121. [Google Scholar] [CrossRef]
- Jove, M.; Mate, I.; Naudi, A.; Mota-Martorell, N.; Portero-Otin, M.; De la Fuente, M.; Pamplona, R. Human aging is a metabolome-related matter of gender. J. Gerontol. A Biolmed. Sci. Med. Sci. 2016, 71, 578–585. [Google Scholar] [CrossRef]
- Cheng, S.; Larson, M.G.; McCabe, E.L.; Murabito, J.M.; Rhee, E.P.; Ho, J.E.; Jacques, P.F.; Ghorbani, A.; Magnusson, M.; Souza, A.L.; et al. Distinct metabolomic signatures are associated with longevity in humans. Nat. Commun. 2015, 6, 6791. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Kastenmuller, G.; Petersen, A.K.; Bell, J.T.; Psatha, M.; Tsai, P.C.; Gieger, C.; Schulz, H.; Erte, I.; John, S.; et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 2013, 42, 1111–1119. [Google Scholar] [CrossRef] [PubMed]
- Swann, J.R.; Spagou, K.; Lewis, M.; Nicholson, J.K.; Glei, D.A.; Seeman, T.E.; Coe, C.L.; Goldman, N.; Ryff, C.D.; Weinstein, M.; et al. Microbial-mammalian cometabolites dominate the age-associated urinary metabolic phenotype in Taiwanese and American populations. J. Proteome Res. 2013, 12, 3166–3180. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Covarrubias, V.; Beekman, M.; Uh, H.W.; Dane, A.; Troost, J.; Paliukhovich, I.; van der Kloet, F.M.; Houwing-Duistermaat, J.; Vreeken, R.J.; Hankemeier, T.; et al. Lipidomics of familial longevity. Aging Cell 2013, 12, 426–434. [Google Scholar] [CrossRef] [PubMed]
- Vaarhorst, A.A.; Beekman, M.; Suchiman, E.H.; van Heemst, D.; Houwing-Duistermaat, J.J.; Westendorp, R.G.; Slagboom, P.E.; Heijmans, B.T.; Leiden Longevity Study Group. Lipid metabolism in long-lived families: The leiden longevity study. Age 2011, 33, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Lawton, K.A.; Berger, A.; Mitchell, M.; Milgram, K.E.; Evans, A.M.; Guo, L.; Hanson, R.W.; Kalhan, S.C.; Ryals, J.A.; Milburn, M.V. Analysis of the adult human plasma metabolome. Pharmacogenomics 2008, 9, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.H.; Marioni, R.E.; Colicino, E.; Peters, M.J.; Ward-Caviness, C.K.; Tsai, P.C.; Roetker, N.S.; Just, A.C.; Demerath, E.W.; Guan, W.; et al. DNA methylation-based measures of biological age: Meta-analysis predicting time to death. Aging 2016, 8, 1844–1865. [Google Scholar] [CrossRef]
- Hertel, J.; Friedrich, N.; Wittfeld, K.; Pietzner, M.; Budde, K.; Van der Auwera, S.; Lohmann, T.; Teumer, A.; Volzke, H.; Nauck, M.; et al. Measuring biological age via metabonomics: The metabolic age score. J. Proteome Res. 2016, 15, 400–410. [Google Scholar] [CrossRef]
- Auro, K.; Joensuu, A.; Fischer, K.; Kettunen, J.; Salo, P.; Mattsson, H.; Niironen, M.; Kaprio, J.; Eriksson, J.G.; Lehtimaki, T.; et al. A metabolic view on menopause and ageing. Nat. Commun. 2014, 5, 4708. [Google Scholar] [CrossRef]
- Chak, C.M.; Lacruz, M.E.; Adam, J.; Brandmaier, S.; Covic, M.; Huang, J.; Meisinger, C.; Tiller, D.; Prehn, C.; Adamski, J.; et al. Ageing investigation using two-time-point metabolomics data from kora and carla studies. Metabolites 2019, 9, 44. [Google Scholar] [CrossRef]
- Rist, M.J.; Roth, A.; Frommherz, L.; Weinert, C.H.; Kruger, R.; Merz, B.; Bunzel, D.; Mack, C.; Egert, B.; Bub, A.; et al. Metabolite patterns predicting sex and age in participants of the karlsruhe metabolomics and nutrition (karmen) study. PLoS ONE 2017, 12, e0183228. [Google Scholar] [CrossRef]
- Makinen, V.P.; Ala-Korpela, M. Metabolomics of aging requires large-scale longitudinal studies with replication. Proc. Natl. Acad. Sci. USA 2016, 113, E3470. [Google Scholar] [CrossRef]
- Belsky, D.W.; Caspi, A.; Houts, R.; Cohen, H.J.; Corcoran, D.L.; Danese, A.; Harrington, H.; Israel, S.; Levine, M.E.; Schaefer, J.D.; et al. Quantification of biological aging in young adults. Proc. Natl. Acad. Sci. USA 2015, 112, E4104–E4110. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Jimenez, C.P.; Eling, N.; Chen, H.C.; Vallejos, C.A.; Kolodziejczyk, A.A.; Connor, F.; Stojic, L.; Rayner, T.F.; Stubbington, M.J.T.; Teichmann, S.A.; et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 2017, 355, 1433–1436. [Google Scholar] [CrossRef] [PubMed]
- Ori, A.; Toyama, B.H.; Harris, M.S.; Bock, T.; Iskar, M.; Bork, P.; Ingolia, N.T.; Hetzer, M.W.; Beck, M. Integrated transcriptome and proteome analyses reveal organ-specific proteome deterioration in old rats. Cell Syst. 2015, 1, 224–237. [Google Scholar] [CrossRef] [PubMed]
- Janssens, G.E.; Meinema, A.C.; Gonzalez, J.; Wolters, J.C.; Schmidt, A.; Guryev, V.; Bischoff, R.; Wit, E.C.; Veenhoff, L.M.; Heinemann, M. Protein biogenesis machinery is a driver of replicative aging in yeast. Elife 2015, 4, e08527. [Google Scholar] [CrossRef]
| Metabolites | Biofluids | Aging (↑↓) | Longevity (↑↓) | References |
|---|---|---|---|---|
| Arginine | Serum | ↓ | - | [119] |
| Ornithine, serine | Serum | ↑ | - | [119] |
| Creatinine, leucine, isoleucine, uric acid, sarcosine, phosphate, glycine, sphingomyelin (C18:1), phosphatidylcholines | Plasma | ↑ | - | [120] |
| Sedoheptulose | Urine | ↓ | - | [120] |
| Phosphoserine (40:5), monoacylglyceride (22:1), diacylglyceride (33:2), resolvin | Plasma | ↓ | - | [109] |
| 25-hydroxy-hexacosanoic acid, eicosapentaenoic acid, phosphocholine (42:9), phosphoserine (42:3), 15-keto-prostaglandin F2α | Plasma | ↓ | - | [109] |
| l-γ-glutamyl-l-leucine | Plasma | ↑ | - | [109] |
| 1,5-Anhydroglucitol, ophthalmic acid, carnosine, acetyl-carnosine, UDP-acetyl-glucosamine, NAD+, NADP+, leucine, isoleucine | Blood | ↓ | - | [59] |
| N6-acetyl-lysine, citrulline, pantothenate, dimethyl-guanosine, N-acetyl-arginine | Blood | ↑ | - | [59] |
| Lipoproteins | Serum | ↑ | - | [118] |
| Tryptophan | Serum | ↓ | - | [56,94] |
| C-glycosyl tryptophan, | Blood | ↓ | - | [111] |
| Creatine, β-hydroxy-β-methylbutyrate | Urine | ↓ | - | [112] |
| Acylcarnitines, diacyl phosphatidylcholines | Serum | ↑ | - | [56] |
| Amino acids | Serum | ↓ | - | [56] |
| Tricarboxylic acid intermediates | Plasma | ↑ | - | [115] |
| Creatine, urea, ornithine, polyamines | Plasma | ↑ | - | [115,120] |
| Essential, non-essential amino acids | Plasma | ↑ | - | [115] |
| Oxoproline, hippurate | Plasma | ↑ | - | [115] |
| Fatty acids, carnitine | Plasma | ↑ | - | [115] |
| Cholesterol, β-hydroxybutyrate | Plasma, serum | ↑ | - | [115,118] |
| Dehydroepiandrosterone-sulfate | Plasma | ↓ | - | [115] |
| Isocitrate, taurochlorate | Plasma | - | ↓ | [110] |
| Sphingomyelins | Serum | - | ↓↑ | [94,113] |
| Glycerophospholipids | Serum | - | ↓↑ | [94] |
| Phenylacetylglutamine, p-cresol sulfate | Urine | ↑ | ↑ | [94,112] |
| Ether phosphocholine, monounsaturated/polyunsaturated fatty acids ratio | Plasma | - | ↑ | [113] |
| Phosphoethanolamine | Plasma | - | ↓ | [113] |
| Low density lipoprotein size | Serum | - | ↑ | [114] |
| Triglycerides | Serum | ↑ | ↓ | [113,114,118] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srivastava, S. Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics. Metabolites 2019, 9, 301. https://doi.org/10.3390/metabo9120301
Srivastava S. Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics. Metabolites. 2019; 9(12):301. https://doi.org/10.3390/metabo9120301
Chicago/Turabian StyleSrivastava, Sarika. 2019. "Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics" Metabolites 9, no. 12: 301. https://doi.org/10.3390/metabo9120301
APA StyleSrivastava, S. (2019). Emerging Insights into the Metabolic Alterations in Aging Using Metabolomics. Metabolites, 9(12), 301. https://doi.org/10.3390/metabo9120301




