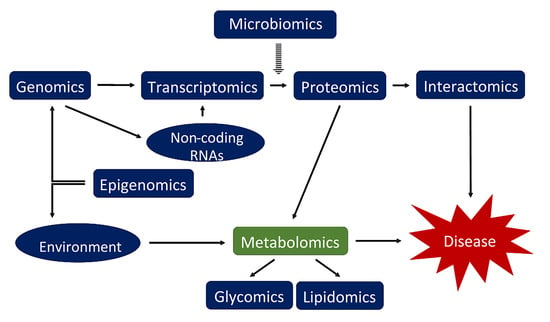
Metabolomics and Age-Related Macular Degeneration
Abstract
1. Introduction
2. Metabolic Processes in the Posterior Eye
2.1. Energy Sources in the Retina
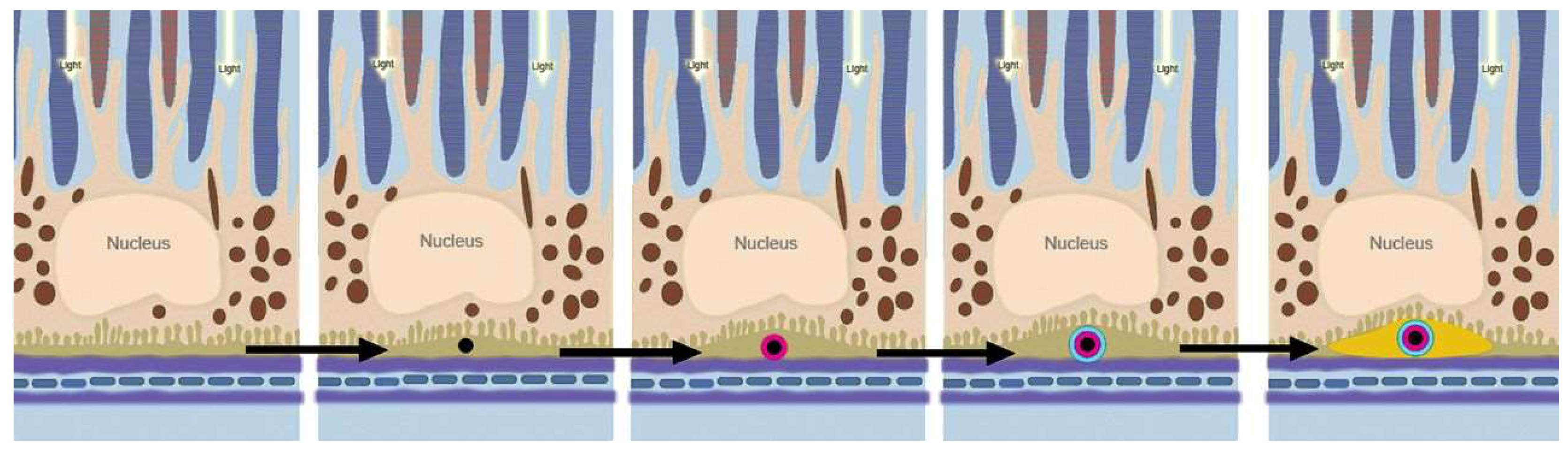
2.2. Lipofuscin Accumulation in the RPE
2.3. Sub-RPE Accumulations
2.3.1. Lipid Accumulation
2.3.2. Advanced Glycation End Product Accumulation
2.3.3. Drusen Accumulation and Development
2.3.4. Trace Metal Homeostasis
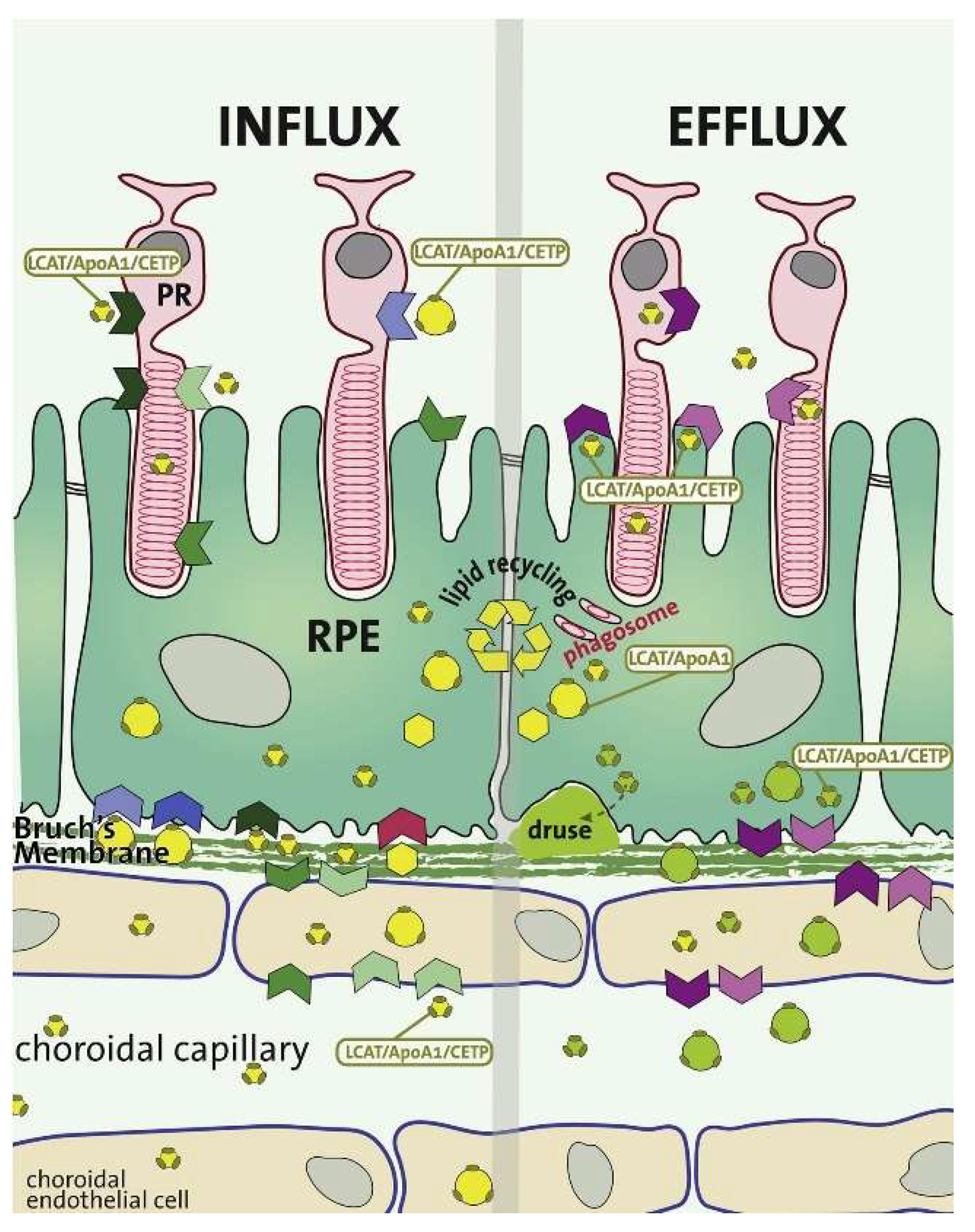
2.4. Choroid-BrM-RPE Interaction
3. Metabolomics in AMD
3.1. Introduction into Metabolomics
3.2. Retinal Tissues
3.3. RPE Cells
3.4. RPE Cells and the Retina
3.5. Biofluids
4. Alternative Approaches for AMD Metabolomics Studies
4.1. Tears
4.2. Vitreous and Aqueous Humor
4.3. In vivo Imaging
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Friedman, D.S.; O’Colmain, B.J.; Muñoz, B.; Tomany, S.C.; McCarty, C.; DeJong, P.T.V.M.; Nemesure, B.; Mitchell, P.; Kempen, J.; Congdon, N. Prevalence of age-related macular degeneration in the United States. Arch. Ophthalmol. 2004, 122, 564–572. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.G.; Klein, R.; Cheng, C.-Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Spraul, C.W.; Lang, G.E.; Grossniklaus, H.E.; Lang, G.K. Histologic and morphometric analysis of the choroid, Bruch’s membrane, and retinal pigment epithelium in postmortem eyes with age-related macular degeneration and histologic examination of surgically excised choroidal neovascular membranes. Surv. Ophthalmol. 1999, 44, S10–S32. [Google Scholar] [CrossRef]
- Chakravarthy, U.; Harding, S.P.; Rogers, C.A.; Downes, S.M.; Lotery, A.J.; Culliford, L.A.; Reeves, B.C. Alternative treatments to inhibit VEGF in age-related choroidal neovascularisation: 2-year findings of the IVAN randomised controlled trial. Lancet 2013, 382, 1258–1267. [Google Scholar] [CrossRef]
- Grunwald, J.E.; Daniel, E.; Huang, J.; Ying, G.-S.; Maguire, M.G.; Toth, C.A.; Jaffe, G.J.; Fine, S.L.; Blodi, B.; Klein, M.L.; et al. Risk of geographic atrophy in the comparison of age-related macular degeneration treatments trials. Ophthalmology 2014, 121, 150–161. [Google Scholar] [CrossRef]
- Colijn, J.M.; Buitendijk, G.H.S.S.; Prokofyeva, E.; Alves, D.; Cachulo, M.L.; Khawaja, A.P.; Cougnard-Gregoire, A.; Merle, B.M.J.J.; Korb, C.; Erke, M.G.; et al. Prevalence of age-related macular degeneration in Europe: The past and the future. Ophthalmology 2017, 124, 1753–1763. [Google Scholar] [CrossRef]
- Chakravarthy, U.; Augood, C.; Bentham, G.C.; de Jong, P.T.V.M.; Rahu, M.; Seland, J.; Soubrane, G.; Tomazzoli, L.; Topouzis, F.; Vingerling, J.R.; et al. Cigarette smoking and age-related macular degeneration in the EUREYE study. Ophthalmology 2007, 114, 1157–1163. [Google Scholar] [CrossRef]
- Christen, W.G.; Glynn, R.J.; Manson, J.E.; Ajani, U.A.; Buring, J.E. A prospective study of cigarette smoking and risk of age-related macular degeneration in men. JAMA 1996, 276, 1147–1151. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Hageman, G.S.; Anderson, D.H.; Johnson, L.V.; Hancox, L.S.; Taiber, A.J.; Hardisty, L.I.; Hageman, J.L.; Stockman, H.A.; Borchardt, J.D.; Gehrs, K.M.; et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2005, 102, 7227–7232. [Google Scholar] [CrossRef]
- Kanda, A.; Chen, W.; Othman, M.; Branham, K.E.H.; Brooks, M.; Khanna, R.; He, S.; Lyons, R.; Abecasis, G.R.; Swaroop, A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2007, 104, 16227–16232. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Loenhardt, T.; Janssen, A.; Fisher, S.A.; Rivera, A.; Keilhauer, C.N.; Weber, B.H.F. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat. Genet. 2008, 40, 892–896. [Google Scholar] [CrossRef] [PubMed]
- Micklisch, S.; Lin, Y.; Jacob, S.; Karlstetter, M.; Dannhausen, K.; Dasari, P.; von der Heide, M.; Dahse, H.-M.; Schmölz, L.; Grassmann, F.; et al. Age-related macular degeneration associated polymorphism rs10490924 in ARMS2 results in deficiency of a complement activator. J. Neuroinflamm. 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.W. Age-Related Macular Degeneration Revisited–Piecing the Puzzle: The LXIX Edward Jackson Memorial Lecture. Am. J. Ophthalmol. 2013, 155, 1–35.e13. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, U.; Wong, T.Y.; Fletcher, A.; Piault, E.; Evans, C.; Zlateva, G.; Buggage, R.; Pleil, A.; Mitchell, P. Clinical risk factors for age-related macular degeneration: A systematic review and meta-analysis. BMC Ophthalmol. 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Balashova, E.E.; Maslov, D.L.; Lokhov, P.G. A metabolomics approach to pharmacotherapy personalization. J. Pers. Med. 2018, 8, 28. [Google Scholar] [CrossRef] [PubMed]
- Kersten, E.; Paun, C.C.; Schellevis, R.L.; Hoyng, C.B.; Delcourt, C.; Lengyel, I.; Peto, T.; Ueffing, M.; Klaver, C.C.W.; Dammeier, S.; et al. Systemic and ocular fluid compounds as potential biomarkers in age-related macular degeneration. Surv. Ophthalmol. 2018, 63, 9–39. [Google Scholar] [CrossRef]
- Van Leeuwen, E.M.; Emri, E.; Merle, B.M.J.; Colijn, J.M.; Kersten, E.; Cougnard-Gregoire, A.; Dammeier, S.; Meester-Smoor, M.; Pool, F.M.; de Jong, E.K.; et al. A new perspective on lipid research in age-related macular degeneration. Prog. Retin. Eye Res. 2018. [Google Scholar] [CrossRef]
- Chen, L.; Gao, Y.; Wang, L.Z.; Cheung, N.; Tan, G.S.W.; Cheung, G.C.M.; Beuerman, R.W.; Wong, T.Y.; Chan, E.C.Y.; Zhou, L. Recent advances in the applications of metabolomics in eye research. Anal. Chim. Acta 2018, 1037, 28–40. [Google Scholar] [CrossRef]
- Tan, S.Z.; Begley, P.; Mullard, G.; Hollywood, K.A.; Bishop, P.N. Introduction to metabolomics and its applications in ophthalmology. Eye 2016, 30, 773–783. [Google Scholar] [CrossRef]
- Laíns, I.; Gantner, M.; Murinello, S.; Lasky-Su, J.A.; Miller, J.W.; Friedlander, M.; Husain, D. Metabolomics in the study of retinal health and disease. Prog. Retin. Eye Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dowling, J.E. Visual Adaptation. In The Retina: An Approachable Part of the Brain; Belknap Press of Harvard University Press: Cambridge, MA, USA, 2012; ISBN 9780674061545. [Google Scholar]
- Hurley, J.B.; Lindsay, K.J.; Du, J. Glucose, lactate, and shuttling of metabolites in vertebrate retinas. J. Neurosci. Res. 2015, 93, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, L.C.; Rosecan, L.R.; Laties, A.M.; Matschinsky, F.M. Altered retinal metabolism in diabetes. I. Microanalysis of lipid, glucose, sorbitol, and myo-inositol in the choroid and in the individual layers of the rabbit retina. J. Biol. Chem. 1986, 261, 4046–4051. [Google Scholar] [PubMed]
- Adler, A.J.; Southwick, R.E. Distribution of glucose and lactate in the interphotoreceptor matrix. Ophthalmic Res. 1992, 24, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Krebs, H.A. On the metabolism of the retina. Biochem. Z. 1927, 189, 57–59. [Google Scholar]
- Winkler, B.S. Glycolytic and oxidative metabolism in relation to retinal function. J. Gen. Physiol. 1981, 77, 667–692. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.; Van Driel, D.; Valter, K.; Rees, S.; Provis, J. The locations of mitochondria in mammalian photoreceptors: Relation to retinal vasculature. Brain Res. 2008, 1189, 58–69. [Google Scholar] [CrossRef]
- Molday, L.L.; Wu, W.W.H.; Molday, R.S. Retinoschisin (RS1), the protein encoded by the X-linked retinoschisis gene, is anchored to the surface of retinal photoreceptor and bipolar cells through its interactions with a Na/K ATPase-SARM1 complex. J. Biol. Chem. 2007, 282, 32792–32801. [Google Scholar] [CrossRef]
- Linton, J.D.; Holzhausen, L.C.; Babai, N.; Song, H.; Miyagishima, K.J.; Stearns, G.W.; Lindsay, K.; Wei, J.; Chertov, A.O.; Peters, T.A.; et al. Flow of energy in the outer retina in darkness and in light. Proc. Natl. Acad. Sci. USA 2010, 107, 8599–8604. [Google Scholar] [CrossRef]
- Du, J.; Rountree, A.; Cleghorn, W.M.; Contreras, L.; Lindsay, K.J.; Sadilek, M.; Gu, H.; Djukovic, D.; Raftery, D.; Satrústegui, J.; et al. Phototransduction influences metabolic flux and nucleotide metabolism in mouse retina. J. Biol. Chem. 2016, 291, 4698–4710. [Google Scholar] [CrossRef]
- Du, J.; Yanagida, A.; Knight, K.; Engel, A.L.; Vo, A.H.; Jankowski, C.; Sadilek, M.; Tran, V.T.B.; Manson, M.A.; Ramakrishnan, A.; et al. Reductive carboxylation is a major metabolic pathway in the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA 2016, 113, 14710–14715. [Google Scholar] [CrossRef] [PubMed]
- Pieczenik, S.R.; Neustadt, J. Mitochondrial dysfunction and molecular pathways of disease. Exp. Mol. Pathol. 2007, 83, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Zabka, T.S.; Singh, J.; Dhawan, P.; Liederer, B.M.; Oeh, J.; Kauss, M.A.; Xiao, Y.; Zak, M.; Lin, T.; McCray, B.; et al. Retinal toxicity, in vivo and in vitro, associated with inhibition of nicotinamide phosphoribosyltransferase. Toxicol. Sci. 2015, 144, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Sheline, C.T. NAD+ maintenance attenuates light induced photoreceptor degeneration. Exp. Eye Res. 2013, 108, 76–83. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, T.; Westenskow, P.D.; Gantner, M.L.; Usui, Y.; Schultz, A.; Bravo, S.; Aguilar, E.; Wittgrove, C.; Friedlander, M.S.; Paris, L.P.; et al. Hypoxia-induced metabolic stress in retinal pigment epithelial cells is sufficient to induce photoreceptor degeneration. Elife 2016, 5, e14319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Yasumura, D.; Li, X.; Matthes, M.; Lloyd, M.; Nielsen, G.; Ahern, K.; Snyder, M.; Bok, D.; Dunaief, J.L.; et al. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J. Clin. Investig. 2011, 121, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Kanow, M.A.; Giarmarco, M.M.; Jankowski, C.S.R.; Tsantilas, K.; Engel, A.L.; Du, J.; Linton, J.D.; Farnsworth, C.C.; Sloat, S.R.; Rountree, A.; et al. Biochemical adaptations of the retina and retinal pigment epithelium support a metabolic ecosystem in the vertebrate eye. Elife 2017, 6, e28899. [Google Scholar] [CrossRef]
- Feeney, L. Lipofuscin and melanin of human retinal pigment epithelium. Fluorescence, enzyme cytochemical, and ultrastructural studies. Investig. Ophthalmol. Vis. Sci. 1978, 17, 583–600. [Google Scholar]
- Dorey, C.K.; Wu, G.; Ebenstein, D.; Garsd, A.; Weiter, J.J. Cell loss in the aging retina. Relationship to lipofuscin accumulation and macular degeneration. Investig. Ophthalmol. Vis. Sci. 1989, 30, 1691–1699. [Google Scholar]
- Wolf, G. Lipofuscin and macular degeneration. Nutr. Rev. 2003, 61, 342–346. [Google Scholar] [CrossRef]
- Sparrow, J.R.; Fishkin, N.; Zhou, J.; Cai, B.; Jang, Y.P.; Krane, S.; Itagaki, Y.; Nakanishi, K. A2E, a byproduct of the visual cycle. Vis. Res. 2003, 43, 2983–2990. [Google Scholar] [CrossRef]
- Lakkaraju, A.; Finnemann, S.C.; Rodriguez-Boulan, E. The lipofuscin fluorophore A2E perturbs cholesterol metabolism in retinal pigment epithelial cells. Proc. Natl. Acad. Sci. USA 2007, 104, 11026–11031. [Google Scholar] [CrossRef] [PubMed]
- Shaban, H.; Gazzotti, P.; Richter, C.; Remé, C.E.; Grimm, C.; Wenzel, A.; Jä, M.; Esser, P.; Kociok, N.; Leist, M.; et al. Cytochrome c oxidase inhibition by N-retinyl-N-retinylidene ethanolamine, a compound suspected to cause age-related macula degeneration. Arch. Biochem. Biophys. 2001, 394, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Suter, M.; Remé, C.; Grimm, C.; Wenzel, A.; Jäättela, M.; Esser, P.; Kociok, N.; Leist, M.; Richter, C. Age-related macular degeneration. The lipofusion component N-retinyl-N-retinylidene ethanolamine detaches proapoptotic proteins from mitochondria and induces apoptosis in mammalian retinal pigment epithelial cells. J. Biol. Chem. 2000, 275, 39625–39630. [Google Scholar] [CrossRef] [PubMed]
- Leist, M.; Single, B.; Naumann, H.; Fava, E.; Simon, B.; Kühnle, S.; Nicotera, P. Inhibition of mitochondrial ATP generation by nitric oxide switches apoptosis to necrosis. Exp. Cell Res. 1999, 249, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Susin, S.A.; Lorenzo, H.K.; Zamzami, N.; Marzo, I.; Snow, B.E.; Brothers, G.M.; Mangion, J.; Jacotot, E.; Costantini, P.; Loeffler, M.; et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 1999, 397, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Tate, D.J.; Miceli, M.V.; Newsome, D.A.; Alcock, N.W.; Oliver, P.D. Influence of zinc on selected cellular functions of cultured human retinal pigment epithelium. Curr. Eye Res. 1995, 14, 897–903. [Google Scholar] [CrossRef]
- Tate, D.J.; Miceli, M.V.; Newsome, D.A. Zinc protects against oxidative damage in cultured human retinal pigment epithelial cells. Free Radic. Biol. Med. 1999, 26, 704–713. [Google Scholar] [CrossRef]
- Nicolas, M.G.; Fujiki, K.; Murayama, K.; Suzuki, M.T.; Shindo, N.; Hotta, Y.; Iwata, F.; Fujimura, T.; Yoshikawa, Y.; Cho, F.; et al. Studies on the mechanism of early onset macular degeneration in cynomolgus monkeys. II. Suppression of metallothionein synthesis in the retina in oxidative stress. Exp. Eye Res. 1996, 62, 399–408. [Google Scholar] [CrossRef]
- Newsome, D.A.; Swartz, M.; Leone, N.C.; Elston, R.C.; Miller, E. Oral zinc in macular degeneration. Arch. Ophthalmol. 1988, 106, 192–198. [Google Scholar] [CrossRef]
- Rajapakse, D.; Curtis, T.; Chen, M.; Xu, H. Zinc protects oxidative stress-induced RPE death by reducing mitochondrial damage and preventing lysosome rupture. Oxidative Med. Cell. Longev. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Age-Related Eye Disease Study Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch. Ophthalmol. 2001, 119, 1417–1436. [Google Scholar] [CrossRef]
- Julien, S.; Biesemeier, A.; Kokkinou, D.; Eibl, O.; Schraermeyer, U. Zinc deficiency leads to lipofuscin accumulation in the retinal pigment epithelium of pigmented rats. PLoS ONE 2011, 6, e29245. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, C.J.; Rakoczy, P.E.; Robertson, T.A.; Papadimitriou, J.M.; Constable, I.J. Kinetic studies on phagocytosis and lysosomal digestion of rod outer segments by human retinal pigment epithelial cells in vitro. Exp. Cell Res. 1994, 210, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Tokarz, P.; Kaarniranta, K.; Blasiak, J. Role of antioxidant enzymes and small molecular weight antioxidants in the pathogenesis of age-related macular degeneration (AMD). Biogerontology 2013, 14, 461–482. [Google Scholar] [CrossRef]
- Adler, L., IV; Boyer, N.P.; Anderson, D.M.; Spraggins, J.M.; Schey, K.L.; Hanneken, A.; Ablonczy, Z.; Crouch, R.K.; Koutalos, Y. Determination of N-retinylidene-N-retinylethanolamine (A2E) levels in central and peripheral areas of human retinal pigment epithelium. Photochem. Photobiol. Sci. 2015, 14, 1983–1990. [Google Scholar] [CrossRef] [PubMed]
- Gliem, M.; Müller, P.L.; Finger, R.P.; McGuinness, M.B.; Holz, F.G.; Charbel Issa, P. Quantitative fundus autofluorescence in early and intermediate age-related macular degeneration. JAMA Ophthalmol. 2016, 134, 817–824. [Google Scholar] [CrossRef]
- Orellana-Rios, J.; Yokoyama, S.; Agee, J.M.; Challa, N.; Freund, K.B.; Yannuzzi, L.A.; Smith, R.T. Quantitative fundus autofluorescence in non-neovascular age-related macular degeneration. Ophthalmic Surg. Lasers Imaging Retin. 2018, 49, S34–S42. [Google Scholar] [CrossRef]
- Wong, W.T.; Kam, W.; Cunningham, D.; Harrington, M.; Hammel, K.; Meyerle, C.B.; Cukras, C.; Chew, E.Y.; Sadda, S.R.; Ferris, F.L. Treatment of geographic atrophy by the topical administration of OT-551: Results of a phase II clinical trial. Investig. Ophthalmol. Vis. Sci. 2010, 51, 6131–6139. [Google Scholar] [CrossRef]
- Mata, N.L.; Lichter, J.B.; Vogel, R.; Han, Y.; Bui, T.V.; Singerman, L.J. Investigation of oral fenretinide for treatment of geographic atrophy in age-related macular degeneration. Retina 2013, 33, 498–507. [Google Scholar] [CrossRef]
- Pauleikhoff, D.; Harper, C.A.; Marshall, J.; Bird, A.C. Aging changes in Bruch’s membrane: A histochemical and morphologic study. Ophthalmology 1990, 97, 171–178. [Google Scholar] [CrossRef]
- Sheraidah, G.; Steinmetz, R.; Maguire, J.; Pauleikhoff, D.; Marshall, J.; Bird, A.C. Correlation between lipids extracted from Bruch’s membrane and age. Ophthalmology 1993, 100, 47–51. [Google Scholar] [CrossRef]
- Curcio, C.A.; Millican, C.L.; Bailey, T.; Kruth, H.S. Accumulation of cholesterol with age in human Bruch’s membrane. Investig. Ophthalmol. Vis. Sci. 2001, 42, 265–274. [Google Scholar]
- Li, C.-M.; Clark, M.E.; Rudolf, M.; Curcio, C.A. Distribution and composition of esterified and unesterified cholesterol in extra-macular drusen. Exp. Eye Res. 2007, 85, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Curcio, C.A.; Presley, J.B.; Malek, G.; Medeiros, N.E.; Avery, D.V.; Kruth, H.S. Esterified and unesterified cholesterol in drusen and basal deposits of eyes with age-related maculopathy. Exp. Eye Res. 2005, 81, 731–741. [Google Scholar] [CrossRef] [PubMed]
- Haimovici, R.; Gantz, D.L.; Rumelt, S.; Freddo, T.F.; Small, D.M. The lipid composition of drusen, Bruch’s membrane, and sclera by hot stage polarizing light microscopy. Investig. Ophthalmol. Vis. Sci. 2001, 42, 1592–1599. [Google Scholar]
- Rudolf, M.; Curcio, C.A. Esterified cholesterol is highly localized to Bruch’s membrane, as revealed by lipid histochemistry in wholemounts of human choroid. J. Histochem. Cytochem. 2009, 57, 731–739. [Google Scholar] [CrossRef]
- Pikuleva, I.A.; Curcio, C.A. Cholesterol in the retina: The best is yet to come. Prog. Retin. Eye Res. 2014, 41, 64–89. [Google Scholar] [CrossRef]
- Shen, J.; He, J.; Wang, F. Association of lipids with age-related macular degeneration. Discov. Med. 2016, 22, 129–145. [Google Scholar]
- Ishibashi, T.; Murata, T.; Hangai, M.; Nagai, R.; Horiuchi, S.; Lopez, P.F.; Hinton, D.R.; Ryan, S.J. Advanced glycation end products in age-related macular degeneration. Arch. Ophthalmol. 1998, 116, 1629–1632. [Google Scholar] [CrossRef]
- Glenn, J.V.; Mahaffy, H.; Wu, K.; Smith, G.; Nagai, R.; Simpson, D.A.C.; Boulton, M.E.; Stitt, A.W. Advanced glycation end product (AGE) accumulation on Bruch’s membrane: Links to age-related RPE dysfunction. Investig. Ophthalmol. Vis. Sci. 2009, 50, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Ishibashi, K.; Ishibashi, K.; Bhutto, I.A.; Tian, J.; Lutty, G.A.; Handa, J.T. The expression of advanced glycation endproduct receptors in rpe cells associated with basal deposits in human maculas. Exp. Eye Res. 2006, 82, 840–848. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Glenn, J.V.; Dasari, S.; McVicar, C.; Ward, M.; Colhoun, L.; Quinn, M.; Bierhaus, A.; Xu, H.; Stitt, A.W. RAGE regulates immune cell infiltration and angiogenesis in choroidal neovascularization. PLoS ONE 2014, 9, e89548. [Google Scholar] [CrossRef] [PubMed]
- Bird, A.C. Towards an understanding of age-related macular disease. Eye 2003, 17, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Balaratnasingam, C.; Yannuzzi, L.A.; Curcio, C.A.; Morgan, W.H.; Querques, G.; Capuano, V.; Souied, E.; Jung, J.; Freund, K.B. Associations between retinal pigment epithelium and drusen volume changes during the lifecycle of large drusenoid pigment epithelial detachments. Investig. Ophthalmol. Vis. Sci. 2016, 57, 5479–5489. [Google Scholar] [CrossRef] [PubMed]
- Sarks, S.; Cherepanoff, S.; Killingsworth, M.; Sarks, J. Relationship of basal laminar deposit and membranous debris to the clinical presentation of early age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2007, 48, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Clark, M.E.; Crossman, D.K.; Kojima, K.; Messinger, J.D.; Mobley, J.A.; Curcio, C.A. Abundant lipid and protein components of drusen. PLoS ONE 2010, 5, e10329. [Google Scholar] [CrossRef] [PubMed]
- Lotery, A.; Trump, D. Progress in defining the molecular biology of age related macular degeneration. Hum. Genet. 2007, 122, 219–236. [Google Scholar] [CrossRef]
- Khan, K.N.; Mahroo, O.A.; Khan, R.S.; Mohamed, M.D.; McKibbin, M.; Bird, A.; Michaelides, M.; Tufail, A.; Moore, A.T. Differentiating drusen: Drusen and drusen-like appearances associated with ageing, age-related macular degeneration, inherited eye disease and other pathological processes. Prog. Retin. Eye Res. 2016, 53, 70–106. [Google Scholar] [CrossRef]
- Tan, A.C.S.; Pilgrim, M.G.; Fearn, S.; Bertazzo, S.; Tsolaki, E.; Morrell, A.P.; Li, M.; Messinger, J.D.; Dolz-Marco, R.; Lei, J.; et al. Calcified nodules in retinal drusen are associated with disease progression in age-related macular degeneration. Sci. Transl. Med. 2018, 10, eaat4544. [Google Scholar] [CrossRef]
- Schlanitz, F.G.; Baumann, B.; Kundi, M.; Sacu, S.; Baratsits, M.; Scheschy, U.; Shahlaee, A.; Mittermüller, T.J.; Montuoro, A.; Roberts, P.; et al. Drusen volume development over time and its relevance to the course of age-related macular degeneration. Br. J. Ophthalmol. 2017, 101, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Nathoo, N.A.; Or, C.; Young, M.; Chui, L.; Fallah, N.; Kirker, A.W.; Albiani, D.A.; Merkur, A.B.; Forooghian, F. Optical coherence tomography-based measurement of drusen load predicts development of advanced age-related macular degeneration. Am. J. Ophthalmol. 2014, 158, 757–761. [Google Scholar] [CrossRef] [PubMed]
- Umeda, S.; Suzuki, M.T.; Okamoto, H.; Ono, F.; Mizota, A.; Terao, K.; Yoshikawa, Y.; Tanaka, Y.; Iwata, T. Molecular composition of drusen and possible involvement of anti-retinal autoimmunity in two different forms of macular degeneration in cynomolgus monkey (Macaca fascicularis). FASEB J. 2005, 19, 1683–1685. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.H.; Talaga, K.C.; Rivest, A.J.; Barron, E.; Hageman, G.S.; Johnson, L.V. Characterization of β amyloid assemblies in drusen: The deposits associated with aging and age-related macular degeneration. Exp. Eye Res. 2004, 78, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Ratnayaka, J.A.; Serpell, L.C.; Lotery, A.J. Dementia of the eye: The role of amyloid beta in retinal degeneration. Eye 2015, 29, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Hageman, G.S.; Mullins, R.F.; Russell, S.R.; Johnson, L.V.; Anderson, D.H. Vitronectin is a constituent of ocular drusen and the vitronectin gene is expressed in human retinal pigmented epithelial cells. FASEB J. 1999, 13, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.H.; Hageman, G.S.; Mullins, R.F.; Neitz, M.; Neitz, J.; Ozaki, S.; Preissner, K.T.; Johnson, L.V. Vitronectin gene expression in the adult human retina. Investig. Ophthalmol. Vis. Sci. 1999, 40, 3305–3315. [Google Scholar]
- Thompson, R.B.; Reffatto, V.; Bundy, J.G.; Kortvely, E.; Flinn, J.M.; Lanzirotti, A.; Jones, E.A.; McPhail, D.S.; Fearn, S.; Boldt, K.; et al. Identification of hydroxyapatite spherules provides new insight into subretinal pigment epithelial deposit formation in the aging eye. Proc. Natl. Acad. Sci. USA 2015, 112, 1565–1570. [Google Scholar] [CrossRef]
- Pilgrim, M.G.; Lengyel, I.; Lanzirotti, A.; Newville, M.; Fearn, S.; Emri, E.; Knowles, J.C.; Messinger, J.D.; Read, R.W.; Guidry, C.; et al. Subretinal pigment epithelial deposition of drusen components including hydroxyapatite in a primary cell culture model. Investig. Ophthalmol. Vis. Sci. 2017, 58, 708–719. [Google Scholar] [CrossRef]
- Brewer, G.J. Risks of copper and iron toxicity during aging in humans. Chem. Res. Toxicol. 2010, 23, 319–326. [Google Scholar] [CrossRef]
- Kurz, T.; Karlsson, M.; Brunk, U.T.; Nilsson, S.E. ARPE-19 retinal pigment epithelial cells are highly resistant to oxidative stress and exercise strict control over their lysosomal redox-active iron. Autophagy 2009, 5, 494–501. [Google Scholar] [CrossRef]
- Nan, R.; Gor, J.; Lengyel, I.; Perkins, S.J. Uncontrolled zinc- and copper-induced oligomerisation of the human complement regulator factor H and its possible implications for function and disease. J. Mol. Biol. 2008, 384, 1341–1352. [Google Scholar] [CrossRef] [PubMed]
- Flinn, J.M.; Kakalec, P.; Tappero, R.; Jones, B.; Lengyel, I. Correlations in distribution and concentration of calcium, copper and iron with zinc in isolated extracellular deposits associated with age-related macular degeneration. Metallomics 2014, 6, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Wood, J.P.M.; Osborne, N.N. Zinc and energy requirements in induction of oxidative stress to retinal pigmented epithelial cells. Neurochem. Res. 2003, 28, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Lengyel, I.; Flinn, J.M.; Peto, T.; Linkous, D.H.; Cano, K.; Bird, A.C.; Lanzirotti, A.; Frederickson, C.J.; van Kuijk, F.J.G.M. High concentration of zinc in sub-retinal pigment epithelial deposits. Exp. Eye Res. 2007, 84, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Friedman, E.; Kopald, H.H.; Smith, T.R.; Mimura, S. Retinal and choroidal blood flow determined with krypton-85 anesthetized animals. Investig. Ophthalmol. Vis. Sci. 1964, 3, 539–547. [Google Scholar]
- Alm, A.; Bill, A. Blood flow and oxygen extraction in the cat uvea at normal and high intraocular pressures. Acta Physiol. Scand. 1970, 80, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Roth, S.; Pietrzyk, Z. Blood flow after retinal ischemia in cats. Investig. Ophthalmol. Vis. Sci. 1994, 35, 3209–3217. [Google Scholar]
- Alm, A.; Bill, A. The oxygen supply to the retina, II. Effects of high intraocular pressure and of increased arterial carbon dioxide tension on uveal and retinal blood flow in cats. Acta Physiol. Scand. 1972, 84, 306–319. [Google Scholar] [CrossRef]
- Wang, L.; Kondo, M.; Bill, A. Glucose metabolism in cat outer retina. Effects of light and hyperoxia. Investig. Ophthalmol. Vis. Sci. 1997, 38, 48–55. [Google Scholar]
- Linsenmeier, R.A.; Padnick–Silver, L. Metabolic dependence of photoreceptors on the choroid in the normal and detached retina. Investig. Ophthalmol. Vis. Sci. 2000, 41, 3117–3123. [Google Scholar]
- Chirco, K.R.; Sohn, E.H.; Stone, E.M.; Tucker, B.A.; Mullins, R.F. Structural and molecular changes in the aging choroid: Implications for age-related macular degeneration. Eye 2017, 31, 10–25. [Google Scholar] [CrossRef] [PubMed]
- Wakatsuki, Y.; Shinojima, A.; Kawamura, A.; Yuzawa, M. Correlation of aging and segmental choroidal thickness measurement using swept source optical coherence tomography in healthy eyes. PLoS ONE 2015, 10, e0144156. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Gu, X.; Crabb, J.W.S.; Yue, X.; Shadrach, K.; Hollyfield, J.G.; Crabb, J.W.S. Quantitative proteomics: Comparison of the macular Bruch membrane/choroid complex from age-related macular degeneration and normal eyes. Mol. Cell. Proteom. 2010, 9, 1031–1046. [Google Scholar] [CrossRef] [PubMed]
- Boccard, J.; Veuthey, J.-L.; Rudaz, S. Knowledge discovery in metabolomics: An overview of MS data handling. J. Sep. Sci. 2010, 33, 290–304. [Google Scholar] [CrossRef] [PubMed]
- Kristal, B.S.; Vigneau-Callahan, K.E.; Matson, W.R. Simultaneous analysis of the majority of low-molecular-weight, redox-active compounds from mitochondria. Anal. Biochem. 1998, 263, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef]
- Würtz, P.; Kangas, A.J.; Soininen, P.; Lawlor, D.A.; Davey Smith, G.; Ala-Korpela, M. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: A primer on -omic technologies. Am. J. Epidemiol. 2017, 186, 1084–1096. [Google Scholar] [CrossRef]
- Lauwen, S.; de Jong, E.K.; Lefeber, D.J.; den Hollander, A.I. Omics biomarkers in ophthalmology. Investig. Ophthalmol. Vis. Sci. 2017, 58, BIO88–BIO98. [Google Scholar] [CrossRef]
- Brindle, J.T.; Antti, H.; Holmes, E.; Tranter, G.; Nicholson, J.K.; Bethell, H.W.L.; Clarke, S.; Schofield, P.M.; McKilligin, E.; Mosedale, D.E.; et al. Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat. Med. 2002, 8, 1439–1445. [Google Scholar] [CrossRef]
- Lindon, J.C.; Holmes, E.; Nicholson, J.K. So what’s the deal with metabonomics? Anal. Chem. 2003, 75, 384 A–391 A. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Wilson, I.D. Understanding “global” systems biology: Metabonomics and the continuum of metabolism. Nat. Rev. Drug Discov. 2003, 2, 668–676. [Google Scholar] [CrossRef] [PubMed]
- Rustam, Y.H.; Reid, G.E. Analytical challenges and recent advances in mass spectrometry based lipidomics. Anaytical Chem. 2018, 90, 374–397. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Lu, H.; Lee, Y.H. Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases. Mass Spectrom. Rev. 2018, 37, 772–792. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Westenskow, P.D.; Fang, M.; Friedlander, M.; Siuzdak, G. Quantitative metabolomics of photoreceptor degeneration and the effects of stem cell-derived retinal pigment epithelium transplantation. Philos. Trans. A Math. Phys. Eng. Sci. 2016, 374, 20150376. [Google Scholar] [CrossRef]
- Chen, Y.; Houghton, L.A.; Brenna, J.T.; Noy, N. Docosahexaenoic acid modulates the interactions of the interphotoreceptor retinoid-binding protein with 11-cis-retinal. J. Biol. Chem. 1996, 271, 20507–20515. [Google Scholar] [CrossRef]
- Chen, Y.; Saari, J.C.; Noy, N. Interactions of all-trans-retinol and long-chain fatty acids with interphotoreceptor retinoid-binding protein1. Biochemistry 1993, 32, 11311–11318. [Google Scholar] [CrossRef]
- Chen, Y.; Okano, K.; Maeda, T.; Chauhan, V.; Golczak, M.; Maeda, A.; Palczewski, K. Mechanism of all-trans-retinal toxicity with implications for stargardt disease and age-related macular degeneration. J. Biol. Chem. 2012, 287, 5059–5069. [Google Scholar] [CrossRef]
- Sparrow, J.R.; Wu, Y.; Kim, C.Y.; Zhou, J. Phospholipid meets all-trans-retinal: The making of RPE bisretinoids. J. Lipid Res. 2010, 51, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.-J.F.; Smart, M.J.K.; Ramsden, C.M.; Powner, M.B.; da Cruz, L.; Coffey, P.J. Development of human embryonic stem cell therapies for age-related macular degeneration. Trends Neurosci. 2013, 36, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Orban, T.; Johnson, W.M.; Dong, Z.; Maeda, T.; Maeda, A.; Sakai, T.; Tsuneoka, H.; Mieyal, J.J.; Palczewski, K. Serum levels of lipid metabolites in age-related macular degeneration. FASEB J. 2015, 29, 4579–4588. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.R.; Knight, K.; Engel, A.L.; Jankowski, C.; Wang, Y.; Manson, M.A.; Gu, H.; Djukovic, D.; Raftery, D.; Hurley, J.B.; et al. Human retinal pigment epithelial cells prefer proline as a nutrient and transport metabolic intermediates to the retinal side. J. Biol. Chem. 2017, 292, 12895–12905. [Google Scholar] [CrossRef]
- Osborn, M.P.; Park, Y.; Parks, M.B.; Burgess, L.G.; Uppal, K.; Lee, K.; Jones, D.P.; Brantley, M.A. Metabolome-wide association study of neovascular age-related macular degeneration. PLoS ONE 2013, 8, e72737. [Google Scholar] [CrossRef]
- Luo, D.; Deng, T.; Yuan, W.; Deng, H.; Jin, M. Plasma metabolomic study in Chinese patients with wet age-related macular degeneration. BMC Ophthalmol. 2017, 17, 165. [Google Scholar] [CrossRef]
- Laíns, I.; Duarte, D.; Barros, A.S.; Martins, A.S.; Gil, J.; Miller, J.W.J.B.; Marques, M.; Mesquita, T.; Kim, I.K.; da Luz Cachulo, M.; et al. Human plasma metabolomics in age-related macular degeneration (AMD) using nuclear magnetic resonance spectroscopy. PLoS ONE 2017, 12, e0177749. [Google Scholar] [CrossRef]
- Laíns, I.; Kelly, R.S.; Miller, J.W.J.B.; Silva, R.; Vavvas, D.G.; Kim, I.K.; Murta, J.N.; Lasky-Su, J.; Miller, J.W.J.B.; Husain, D.; et al. Human plasma metabolomics study across all stages of age-related macular degeneration identifies potential lipid biomarkers. Ophthalmology 2017, 125, 245–254. [Google Scholar] [CrossRef]
- Rowan, S.; Jiang, S.; Korem, T.; Szymanski, J.; Chang, M.-L.; Szelog, J.; Cassalman, C.; Dasuri, K.; McGuire, C.; Nagai, R.; et al. Involvement of a gut-retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2017, 114, E4472–E4481. [Google Scholar] [CrossRef]
- Mitchell, S.L.; Uppal, K.; Williamson, S.M.; Liu, K.; Burgess, L.G.; Tran, V.; Umfress, A.C.; Jarrell, K.L.; Cooke Bailey, J.N.; Agarwal, A.; et al. The carnitine shuttle pathway is altered in patients with neovascular age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4978–4985. [Google Scholar] [CrossRef]
- Overmyer, K.A.; Thonusin, C.; Qi, N.R.; Burant, C.F.; Evans, C.R. Impact of anesthesia and euthanasia on metabolomics of mammalian tissues: Studies in a C57BL/6J mouse model. PLoS ONE 2015, 10, e0117232. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.Z.; Mullard, G.; Hollywood, K.A.; Dunn, W.B.; Bishop, P.N. Characterisation of the metabolome of ocular tissues and post-mortem changes in the rat retina. Exp. Eye Res. 2016, 149, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Bok, D. A cell culture medium that supports the differentiation of human retinal pigment epithelium into functionally polarized monolayers. Mol. Vis. 2001, 7, 14–19. [Google Scholar] [PubMed]
- Maminishkis, A.; Chen, S.; Jalickee, S.; Banzon, T.; Shi, G.; Wang, F.E.; Ehalt, T.; Hammer, J.A.; Miller, S.S. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3612–3624. [Google Scholar] [CrossRef] [PubMed]
- Maminishkis, A.; Miller, S.S. Experimental models for study of retinal pigment epithelial physiology and pathophysiology. J. Vis. Exp. 2010, e2032. [Google Scholar] [CrossRef] [PubMed]
- Adijanto, J.; Philp, N.J. Cultured primary human fetal retinal pigment epithelium (hfRPE) as a model for evaluating RPE metabolism. Exp. Eye Res. 2014, 126, 77–84. [Google Scholar] [CrossRef]
- Du, J.; Linton, J.D.; Hurley, J.B. Probing metabolism in the intact retina using stable isotope tracers. Methods Enzymol. 2015, 561, 149–170. [Google Scholar] [CrossRef]
- Johnson, L.V.; Forest, D.L.; Banna, C.D.; Radeke, C.M.; Maloney, M.A.; Hu, J.; Spencer, C.N.; Walker, A.M.; Tsie, M.S.; Bok, D.; et al. Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proc. Natl. Acad. Sci. USA 2011, 108, 18277–18282. [Google Scholar] [CrossRef]
- Anderson, B.; Saltzman, H.A. Retinal oxygen utilization measured by hyperbaric blackout. Arch. Ophthalmol. 1964, 72, 792–795. [Google Scholar] [CrossRef]
- Ames, A.; Li, Y.Y.; Heher, E.C.; Kimble, C.R. Energy metabolism of rabbit retina as related to function: High cost of Na+ transport. J. Neurosci. 1992, 12, 840–853. [Google Scholar] [CrossRef]
- Yu, D.-Y.; Cringle, S.J. Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Prog. Retin. Eye Res. 2001, 20, 175–208. [Google Scholar] [CrossRef]
- Hess, H.H. The high calcium content of retinal pigmented epithelium. Exp. Eye Res. 1975, 21, 471–479. [Google Scholar] [CrossRef]
- Chen, H.; Tran, J.-T.A.; Eckerd, A.; Huynh, T.-P.; Elliott, M.H.; Brush, R.S.; Mandal, N.A. Inhibition of de novo ceramide biosynthesis by FTY720 protects rat retina from light-induced degeneration. J. Lipid Res. 2013, 54, 1616–1629. [Google Scholar] [CrossRef] [PubMed]
- Abrahan, C.E.; Miranda, G.E.; Agnolazza, D.L.; Politi, L.E.; Rotstein, N.P. Synthesis of sphingosine is essential for oxidative stress-induced apoptosis of photoreceptors. Investig. Ophthalmol. Vis. Sci. 2010, 51, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Rotstein, N.P.; Miranda, G.E.; Abrahan, C.E.; German, O.L. Regulating survival and development in the retina: Key roles for simple sphingolipids. J. Lipid Res. 2010, 51, 1247–1262. [Google Scholar] [CrossRef] [PubMed]
- Xie, B.; Shen, J.; Dong, A.; Rashid, A.; Stoller, G.; Campochiaro, P.A. Blockade of sphingosine-1-phosphate reduces macrophage influx and retinal and choroidal neovascularization. J. Cell. Physiol. 2009, 218, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Caballero, S.; Swaney, J.; Moreno, K.; Afzal, A.; Kielczewski, J.; Stoller, G.; Cavalli, A.; Garland, W.; Hansen, G.; Sabbadini, R.; et al. Anti-sphingosine-1-phosphate monoclonal antibodies inhibit angiogenesis and sub-retinal fibrosis in a murine model of laser-induced choroidal neovascularization. Exp. Eye Res. 2009, 88, 367–377. [Google Scholar] [CrossRef] [PubMed]
- Yonamine, I.; Bamba, T.; Nirala, N.K.; Jesmin, N.; Kosakowska-Cholody, T.; Nagashima, K.; Fukusaki, E.; Acharya, J.K.; Acharya, U. Sphingosine kinases and their metabolites modulate endolysosomal trafficking in photoreceptors. J. Cell Biol. 2011, 192, 557–567. [Google Scholar] [CrossRef]
- Kennedy, B.G.; Torabi, A.J.; Kurzawa, R.; Echtenkamp, S.F.; Mangini, N.J. Expression of transient receptor potential vanilloid channels TRPV5 and TRPV6 in retinal pigment epithelium. Mol. Vis. 2010, 16, 665–675. [Google Scholar]
- Wimmers, S.; Strauss, O. Basal calcium entry in retinal pigment epithelial cells is mediated by TRPC channels. Investig. Ophthalmol. Vis. Sci. 2007, 48, 5767–5772. [Google Scholar] [CrossRef]
- Bhutto, I.; Lutty, G. Understanding age-related macular degeneration (AMD): Relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol. Asp. Med. 2012, 33, 295–317. [Google Scholar] [CrossRef] [PubMed]
- Coscas, G.; Yamashiro, K.; Coscas, F.; De Benedetto, U.; Tsujikawa, A.; Miyake, M.; Gemmy Cheung, C.M.; Wong, T.Y.; Yoshimura, N. Comparison of exudative age-related macular degeneration subtypes in Japanese and French patients: Multicenter diagnosis with multimodal imaging. Am. J. Ophthalmol. 2014, 158, 309–318.e2. [Google Scholar] [CrossRef] [PubMed]
- Sohrab, M.; Wu, K.; Fawzi, A.A. A pilot study of morphometric analysis of choroidal vasculature in vivo, using en face optical coherence tomography. PLoS ONE 2012, 7, e48631. [Google Scholar] [CrossRef] [PubMed]
- McLeod, D.S.; Grebe, R.; Bhutto, I.; Merges, C.; Baba, T.; Lutty, G.A. Relationship between RPE and choriocapillaris in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2009, 50, 4982–4991. [Google Scholar] [CrossRef] [PubMed]
- Metelo, A.M.; Noonan, H.; Iliopoulos, O. HIF2α inhibitors for the treatment of VHL disease. Oncotarget 2015, 6, 23036–23037. [Google Scholar] [CrossRef] [PubMed]
- Metelo, A.M.; Noonan, H.R.; Li, X.; Jin, Y.; Baker, R.; Kamentsky, L.; Zhang, Y.; van Rooijen, E.; Shin, J.; Carpenter, A.E.; et al. Pharmacological HIF2α inhibition improves VHL disease-associated phenotypes in zebrafish model. J. Clin. Investig. 2015, 125, 1987–1997. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Cano, M.; Ebrahimi, K.; Wang, L.; Handa, J.T. The impact of oxidative stress and inflammation on RPE degeneration in non-neovascular AMD. Prog. Retin. Eye Res. 2017, 60, 201–218. [Google Scholar] [CrossRef]
- Brantley, M.A.; Osborn, M.P.; Sanders, B.J.; Rezaei, K.A.; Lu, P.; Li, C.; Milne, G.L.; Cai, J.; Sternberg, P.; Sternberg, P.; et al. Plasma biomarkers of oxidative stress and genetic variants in age-related macular degeneration. Am. J. Ophthalmol. 2012, 153, 460–467.e1. [Google Scholar] [CrossRef]
- Morrow, J.D.; Frei, B.; Longmire, A.W.; Gaziano, M.; Lynch, S.M.; Shyr, Y.; Strauss, W.E.; Oates, J.A.; Roberts, L.J., II. Increase in circulating products of lipid peroxidation (F2-isoprostanes) in smokers. N. Engl. J. Med. 1995, 332, 1198–1203. [Google Scholar] [CrossRef]
- Jones, D.P.; Mody, V.C., Jr.; Carlson, J.L.; Lynn, M.J.; Sternberg, P. Redox analysis of human plasma allows separation of pro-oxidant events of aging from decline in antioxidant defenses. Free Radic. Biol. Med. 2002, 33, 1290–1300. [Google Scholar] [CrossRef]
- Moriarty, S.E.; Shah, J.H.; Lynn, M.; Jiang, S.; Openo, K.; Jones, D.P.; Sternberg, P. Oxidation of glutathione and cysteine in human plasma associated with smoking. Free Radic. Biol. Med. 2003, 35, 1582–1588. [Google Scholar] [CrossRef] [PubMed]
- Merle, B.M.J.; Benlian, P.; Puche, N.; Bassols, A.; Delcourt, C.; Souied, E.H. Circulating omega-3 fatty acids and neovascular age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2010–2019. [Google Scholar] [CrossRef]
- Merle, B.M.J.; Delyfer, M.-N.; Korobelnik, J.-F.; Rougier, M.-B.; Malet, F.; Féart, C.; Le Goff, M.; Peuchant, E.; Letenneur, L.; Dartigues, J.-F.; et al. High concentrations of plasma n3 fatty acids are associated with decreased risk for late age-related macular degeneration. J. Nutr. 2013, 143, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.R.; Lange, B.M. Open-access metabolomics databases for natural product research: Present capabilities and future potential. Front. Bioeng. Biotechnol. 2015, 3, 22. [Google Scholar] [CrossRef] [PubMed]
- Tolleson, W.H. Human melanocyte biology, toxicology, and pathology. J. Environ. Sci. Health Part C Environ. Carcinog. Ecotoxicol. Rev. 2005, 23, 105–161. [Google Scholar] [CrossRef]
- Chen, G.; Walmsley, S.; Cheung, G.C.; Chen, L.; Cheng, C.-Y.; Beuerman, R.W.; Wong, T.Y.; Zhou, L.; Choi, H. Customized consensus spectral library building for untargeted quantitative metabolomics analysis with data independent acquisition mass spectrometry and MetaboDIA workflow. Anal. Chem. 2017, 89, 4897–4906. [Google Scholar] [CrossRef]
- Pushpoth, S.; Fitzpatrick, M.; Young, S.; Yang, Y.; Talks, J.; Wallace, G. Metabolomic analysis in patients with age related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3662. [Google Scholar]
- Pushpoth, S.; Fitzpatrick, M.; Talks, J.S.; Young, S.; Yang, Y.C.; Wallace, G.R. Metabolomic analysis of urine in patients with age related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2015, 56, 368. [Google Scholar]
- Wong, C.W.; Yanagi, Y.; Lee, W.-K.; Ogura, Y.; Yeo, I.; Yin Wong, T.; Ming Gemmy Cheung, C. Age-related macular degeneration and polypoidal choroidal vasculopathy in Asians. Prog. Retin. Eye Res. 2016, 53, 107–139. [Google Scholar] [CrossRef]
- Li, M.; Zhang, X.; Liao, N.; Ye, B.; Peng, Y.; Ji, Y.; Wen, F. Analysis of the serum lipid profile in polypoidal choroidal vasculopathy. Sci. Rep. 2016, 6, 38342. [Google Scholar] [CrossRef]
- Schoumacher, M.; Lambert, V.; Leenders, J.; Roblain, Q.; Govaerts, B.; Rakic, J.-M.; Noël, A.; De Tullio, P. NMR-based metabolomics for new target discovery and personalized medicine: Application to age-related macular degeneration (AMD). In EUROMAR (European Magnetic Resonance Meeting); Université de Nantes: Nantes, France, 2018. [Google Scholar]
- Schoumacher, M.; De Tullio, P.; Lambert, V.; Hansen, S.; Leenders, J.; Govaerts, B.; Pirotte, B.; Rakic, J.-M.; Noël, A. From metabolomics to identification of a new therapeutic approach for age-related macular degeneration (AMD). In 30ièmes Journées Franco-Belges de Pharmacochimie; Université d’Orléans: Amboise, France, 2016. [Google Scholar]
- Kowalczuk, L.; Matet, A.; Dor, M.; Bararpour, N.; Daruich, A.; Dirani, A.; Behar-Cohen, F.; Thomas, A.; Turck, N. Proteome and metabolome of subretinal fluid in central serous chorioretinopathy and rhegmatogenous retinal detachment: A pilot case study. Transl. Vis. Sci. Technol. 2018, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, M.; Zhang, X.; Zhang, Q.; Nie, J.; Zhang, M.; Liu, X.; Ma, L. The association between the lipids levels in blood and risk of age-related macular degeneration. Nutrients 2016, 8, 663. [Google Scholar] [CrossRef] [PubMed]
- Mishima, S.; Gasset, A.; Klyce, S.D.; Baum, J.L. Determination of tear volume and tear flow. Investig. Ophthalmol. Vis. Sci. 1966, 5, 264–276. [Google Scholar]
- Zhou, L.; Beuerman, R.W.; Foo, Y.; Liu, S.; Ang, L.P. Characterisation of human tear proteins using high-resolution mass spectrometry. Ann. Acad. Med. Singap. 2006, 35, 400–407. [Google Scholar] [PubMed]
- Zhou, L.; Zhao, S.Z.; Koh, S.K.; Chen, L.; Vaz, C.; Tanavde, V.; Li, X.R.; Beuerman, R.W. In-depth analysis of the human tear proteome. J. Proteom. 2012, 75, 3877–3885. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.M.; Tong, L.; Duan, X.; Petznick, A.; Wenk, M.R.; Shui, G. Extensive characterization of human tear fluid collected using different techniques unravels the presence of novel lipid amphiphiles. J. Lipid Res. 2014, 55, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhou, L.; Chan, E.C.Y.; Neo, J.; Beuerman, R.W. Characterization of the human tear metabolome by LC-MS/MS. J. Proteome Res. 2011, 10, 4876–4882. [Google Scholar] [CrossRef]
- Hagan, S.; Martin, E.; Enríquez-de-Salamanca, A. Tear fluid biomarkers in ocular and systemic disease: Potential use for predictive, preventive and personalised medicine. EPMA J. 2016, 7, 15. [Google Scholar] [CrossRef]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Di Ilio, C.; Sacchetta, P.; Del Boccio, P. Unraveling the molecular repertoire of tears as a source of biomarkers: Beyond ocular diseases. Proteom. Clin. Appl. 2015, 9, 169–186. [Google Scholar] [CrossRef]
- Borchman, D.; Foulks, G.N.; Yappert, M.C.; Tang, D.; Ho, D. V Spectroscopic evaluation of human tear lipids. Chem. Phys. Lipids 2007, 147, 87–102. [Google Scholar] [CrossRef]
- Saville, J.T.; Zhao, Z.; Willcox, M.D.P.; Blanksby, S.J.; Mitchell, T.W. Detection and quantification of tear phospholipids and cholesterol in contact lens deposits: The effect of contact lens material and lens care solution. Investig. Ophthalmol. Vis. Sci. 2010, 51, 2843–2851. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.M.; Tong, L.; Reux, B.; Lear, M.J.; Wenk, M.R.; Shui, G. Rapid and sensitive profiling of tear wax ester species using high performance liquid chromatography coupled with tandem mass spectrometry. J. Chromatogr. A 2013, 1308, 166–171. [Google Scholar] [CrossRef] [PubMed]
- Rantamäki, A.H.; Seppänen-Laakso, T.; Oresic, M.; Jauhiainen, M.; Holopainen, J.M. Human tear fluid lipidome: From composition to function. PLoS ONE 2011, 6, e19553. [Google Scholar] [CrossRef] [PubMed]
- Dean, A.W.; Glasgow, B.J. Mass spectrometric identification of phospholipids in human tears and tear lipocalin. Investig. Ophthalmol. Vis. Sci. 2012, 53, 1773–1782. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A. On the lipid composition of human meibum and tears: Comparative analysis of nonpolar lipids. Investig. Ophthalmol. Vis. Sci. 2008, 49, 3779–3789. [Google Scholar] [CrossRef] [PubMed]
- Butovich, I.A. Tear film lipids. Exp. Eye Res. 2013, 117, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Bleau, G.; Bodley, F.H.; Longpré, J.; Chapdelaine, A.; Roberts, K.D. Cholesterol sulfate. I. Occurrence and possible biological functions as an amphipathic lipid in the membrane of the human erythrocyte. Biochim. Biophys. Acta 1974, 352, 1–9. [Google Scholar] [CrossRef]
- Romano, A.; Rolant, F. A non-invasive method of blood glucose evaluation by tear glucose measurement, for the detection and control of diabetic states. Metab. Pediatr. Syst. Ophthalmol. 1988, 11, 78–80. [Google Scholar]
- Baca, J.T.; Finegold, D.N.; Asher, S.A. Tear glucose analysis for the noninvasive detection and monitoring of diabetes mellitus. Ocul. Surf. 2007, 5, 280–293. [Google Scholar] [CrossRef]
- Daum, K.M.; Hill, R.M. Human tear glucose. Investig. Ophthalmol. Vis. Sci. 1982, 22, 509–514. [Google Scholar]
- Yan, Q.; Peng, B.; Su, G.; Cohan, B.E.; Major, T.C.; Meyerhoff, M.E. Measurement of tear glucose levels with amperometric glucose biosensor/capillary tube configuration. Anal. Chem. 2011, 83, 8341–8346. [Google Scholar] [CrossRef]
- Peng, B.; Lu, J.; Balijepalli, A.S.; Major, T.C.; Cohan, B.E.; Meyerhoff, M.E. Evaluation of enzyme-based tear glucose electrochemical sensors over a wide range of blood glucose concentrations. Biosens. Bioelectron. 2013, 49, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Baca, J.T.; Taormina, C.R.; Feingold, E.; Finegold, D.N.; Grabowski, J.J.; Asher, S.A. Mass spectral determination of fasting tear glucose concentrations in nondiabetic volunteers. Clin. Chem. 2007, 53, 1370–1372. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.N. Measuring Glucose Levels in Tears as an Alternative to Blood Glucose Levels, Bard College, 2016, Vol. Paper 262. Available online: https://digitalcommons.bard.edu/senproj_s2016/262 (accessed on 21 December 2018).
- Park, M.; Jung, H.; Jeong, Y.; Jeong, K.-H. Plasmonic Schirmer strip for human tear-based gouty arthritis diagnosis using surface-enhanced raman scattering. ACS Nano 2017, 11, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Dame, Z.T.; Aziat, F.; Mandal, R.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.C.; Sajed, T.; Deng, L.; Lin, H.; et al. The human saliva metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Tsuruoka, M.; Hara, J.; Hirayama, A.; Sugimoto, M.; Soga, T.; Shankle, W.R.; Tomita, M. Capillary electrophoresis-mass spectrometry-based metabolome analysis of serum and saliva from neurodegenerative dementia patients. Electrophoresis 2013, 34, 2865–2872. [Google Scholar] [CrossRef] [PubMed]
- Figueira, J.; Jonsson, P.; Nordin Adolfsson, A.; Adolfsson, R.; Nyberg, L. NMR analysis of the human saliva metabolome distinguishes dementia patients from matched controls. Mol. Biosyst. 2016, 12, 2562–2571. [Google Scholar] [CrossRef] [PubMed]
- Haines, N.R.; Manoharan, N.; Olson, J.L.; D’alessandro, A.; Reisz, J.A. Metabolomics analysis of human vitreous in diabetic retinopathy and rhegmatogenous retinal detachment. J. Proteome Res. 2018, 17, 2421–2427. [Google Scholar] [CrossRef]
- Paris, L.P.; Johnson, C.H.; Aguilar, E.; Usui, Y.; Cho, K.; Hoang, L.T.; Feitelberg, D.; Benton, H.P.; Westenskow, P.D.; Kurihara, T.; et al. Global metabolomics reveals metabolic dysregulation in ischemic retinopathy. Metabolomics 2016, 12, 15. [Google Scholar] [CrossRef]
- Schwartzman, M.L.; Iserovich, P.; Gotlinger, K.; Bellner, L.; Dunn, M.W.; Sartore, M.; Pertile, M.G.; Leonardi, A.; Sathe, S.; Beaton, A.; et al. Profile of lipid and protein autacoids in diabetic vitreous correlates with the progression of diabetic retinopathy. Diabetes 2010, 59, 1780–1788. [Google Scholar] [CrossRef]
- Al-Shabrawey, M.; Mussell, R.; Kahook, K.; Tawfik, A.; Eladl, M.; Sarthy, V.; Nussbaum, J.; El-Marakby, A.; Park, S.Y.; Gurel, Z.; et al. Increased expression and activity of 12-lipoxygenase in oxygen-induced ischemic retinopathy and proliferative diabetic retinopathy: Implications in retinal neovascularization. Diabetes 2011, 60, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Barba, I.; Garcia-Ramírez, M.; Hernández, C.; Alonso, M.A.; Masmiquel, L.; García-Dorado, D.; Simó, R. Metabolic fingerprints of proliferative diabetic retinopathy: An 1H-NMR–based metabonomic approach using vitreous humor. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4416–4421. [Google Scholar] [CrossRef] [PubMed]
- Young, S.P.; Nessim, M.; Falciani, F.; Trevino, V.; Banerjee, S.P.; Scott, R.A.H.; Murray, P.I.; Wallace, G.R. Metabolomic analysis of human vitreous humor differentiates ocular inflammatory disease. Mol. Vis. 2009, 15, 1210–1217. [Google Scholar] [PubMed]
- Li, M.; Li, H.; Jiang, P.; Liu, X.; Xu, D.; Wang, F. Investigating the pathological processes of rhegmatogenous retinal detachment and proliferative vitreoretinopathy with metabolomics analysis. Mol. Biosyst. 2014, 10, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Wu, Z.; Zhang, Z.; Huang, X.; Zhang, Q. Metabolomic analysis of human vitreous in rhegmatogenous retinal detachment associated with choroidal detachment. Investig. Ophthalmol. Vis. Sci. 2015, 56, 5706–5713. [Google Scholar] [CrossRef]
- Mains, J.; Tan, L.E.; Zhang, T.; Young, L.; Shi, R.; Wilson, C. Species variation in small molecule components of animal vitreous. Investig. Ophthalmol. Vis. Sci. 2012, 53, 4778–4786. [Google Scholar] [CrossRef] [PubMed]
- Agudo-Barriuso, M.; Lahoz, A.; Nadal-Nicolás, F.M.; Sobrado-Calvo, P.; Piquer-Gil, M.; Díaz-Llopis, M.; Vidal-Sanz, M.; Mullor, J.L. Metabolomic changes in the rat retina after optic nerve crush. Investig. Ophthalmol. Vis. Sci. 2013, 54, 4249–4259. [Google Scholar] [CrossRef]
- Edwards, G.; Aribindi, K.; Guerra, Y.; Bhattacharya, S.K. Sphingolipids and ceramides of mouse aqueous humor: Comparative profiles from normotensive and hypertensive DBA/2J mice. Biochimie 2014, 105, 99–109. [Google Scholar] [CrossRef]
- Mayordomo-Febrer, A.; Lopez-Murcia, M.; Morales-Tatay, J.; Monleon-Salvado, D.; Pinazo-Durán, M.D. Metabolomics of the aqueous humor in the rat glaucoma model induced by a series of intracamerular sodium hyaluronate injection. Exp. Eye Res. 2015, 131, 84–92. [Google Scholar] [CrossRef]
- Edwards, G.; Aribindi, K.; Guerra, Y.; Lee, R.K.; Bhattacharya, S.K. Phospholipid profiles of control and glaucomatous human aqueous humor. Biochimie 2014, 101, 232–247. [Google Scholar] [CrossRef]
- Aribindi, K.; Guerra, Y.; Piqueras, M.D.C.; Banta, J.T.; Lee, R.K.; Bhattacharya, S.K. Cholesterol and glycosphingolipids of human trabecular meshwork and aqueous humor: Comparative profiles from control and glaucomatous donors. Curr. Eye Res. 2013, 38, 1017–1026. [Google Scholar] [CrossRef] [PubMed]
- Aljohani, A.J.; Edwards, G.; Guerra, Y.; Dubovy, S.; Miller, D.; Lee, R.K.; Bhattacharya, S.K. Human trabecular meshwork sphingolipid and ceramide profiles and potential latent fungal commensalism. Investig. Ophthalmol. Vis. Sci. 2014, 55, 3413–3422. [Google Scholar] [CrossRef] [PubMed]
- Barbas-Bernardos, C.; Armitage, E.G.; García, A.; Mérida, S.; Navea, A.; Bosch-Morell, F.; Barbas, C. Looking into aqueous humor through metabolomics spectacles—Exploring its metabolic characteristics in relation to myopia. J. Pharm. Biomed. Anal. 2016, 127, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Rao, J.; Rong, X.; Lou, S.; Zheng, Z.; Lu, Y. Metabolic characterization of human aqueous humor in relation to high myopia. Exp. Eye Res. 2017, 159, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Snytnikova, O.A.; Khlichkina, A.A.; Yanshole, L.V.; Yanshole, V.V.; Iskakov, I.A.; Egorova, E.V.; Stepakov, D.A.; Novoselov, V.P.; Tsentalovich, Y.P. Metabolomics of the human aqueous humor. Metabolomics 2017, 13. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Fluorophores. In Principles of Fluorescence Spectroscopy; Springer: Boston, MA, USA, 2006; pp. 63–95. [Google Scholar]
- Chance, B. Pyridine nucleotide as an indicator of the oxygen requirements for energy-linked functions of mitochondria. Circ. Res. 1976, 38, I31–I38. [Google Scholar]
- Lakowicz, J.R.; Szmacinski, H.; Nowaczyk, K.; Johnson, M.L. Fluorescence lifetime imaging of free and protein-bound NADH. Proc. Natl. Acad. Sci. USA 1992, 89, 1271–1275. [Google Scholar] [CrossRef]
- Sharick, J.T.; Favreau, P.F.; Gillette, A.A.; Sdao, S.M.; Merrins, M.J.; Skala, M.C. Protein-bound NAD(P)H lifetime is sensitive to multiple fates of glucose carbon. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Fluorescence-Lifetime Imaging Microscopy. In Principles of Fluorescence Spectroscopy; Springer: Boston, MA, USA, 2006; pp. 741–755. ISBN 9780387463124. [Google Scholar]
- Becker, W. Fluorescence lifetime imaging—Techniques and applications. J. Microsc. 2012, 247, 119–136. [Google Scholar] [CrossRef]
- Schweitzer, D.; Klemm, M.; Quick, S.; Deutsch, L.; Jentsch, S.; Hammer, M.; Dawczynski, J.; Kloos, C.H.; Mueller, U.A. Detection of early metabolic alterations in the ocular fundus of diabetic patients by time-resolved autofluorescence of endogenous fluorophores. In Clinical and Biomedical Spectroscopy and Imaging II; Ramanujam, N., Popp, J., Eds.; International Society for Optics and Photonics: Bellingham WA, USA, 2011; Volume SPIE 8087, p. 80871G. [Google Scholar]
- Blacker, T.S.; Mann, Z.F.; Gale, J.E.; Ziegler, M.; Bain, A.J.; Szabadkai, G.; Duchen, M.R. Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat. Commun. 2014, 5, 3936. [Google Scholar] [CrossRef]
- Patterson, G.H.; Piston, D.W.; Barisas, B.G. Förster distances between green fluorescent protein pairs. Anal. Biochem. 2000, 284, 438–440. [Google Scholar] [CrossRef] [PubMed]
- Richards-Kortum, R.; Drezek, R.; Sokolov, K.; Pavlova, I.; Pollen, M. Survey of Endogenous Biological Fluorophores. In Handbook of Biomedical Fluorescence; Mycek, M.A., Pogue, B.W., Eds.; Marcel Dekker, Inc.: New York, NY, USA; Basel, Switzerland, 2003; pp. 237–264. [Google Scholar]
- Urayama, P.; Mycek, M.-A. Fluorescence Lifetime Imaging Microscopy of Endogenous Biological Fluorescence. In Handbook of Biomedical Fluorescence; Mycek, M.-A., Pogue, B.W., Eds.; Macel Dekker, Inc.: New York, NY, USA; Basel, Switzerland, 2003; pp. 211–236. ISBN 0-8247-0955-1. [Google Scholar]
- Schweitzer, D.; Schenke, S.; Hammer, M.; Schweitzer, F.; Jentsch, S.; Birckner, E.; Becker, W.; Bergmann, A.A. Towards metabolic mapping of the human retina. Microsc. Res. Tech. 2007, 70, 410–419. [Google Scholar] [CrossRef] [PubMed]
- Marcu, L. Fluorescence lifetime techniques in medical applications. Ann. Biomed. Eng. 2012, 40, 304–331. [Google Scholar] [CrossRef] [PubMed]
- Miura, Y.; Huettmann, G.; Orzekowsky-Schroeder, R.; Steven, P.; Szaszák, M.; Koop, N.; Brinkmann, R. Two-photon microscopy and fluorescence lifetime imaging of retinal pigment epithelial cells under oxidative stress. Investig. Ophthalmol. Vis. Sci. 2013, 54, 3366–3377. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, D.; Gaillard, E.R.; Dillon, J.; Mullins, R.F.; Russell, S.; Hoffmann, B.; Peters, S.; Hammer, M.; Biskup, C. Time-resolved autofluorescence imaging of human donor retina tissue from donors with significant extramacular drusen. Investig. Ophthalmol. Vis. Sci. 2012, 53, 3376–3386. [Google Scholar] [CrossRef]
- Schweitzer, D.; Hammer, M.; Schweitzer, F.; Anders, R.; Doebbecke, T.; Schenke, S.; Gaillard, E.R.; Gaillard, E.R. In vivo measurement of time-resolved autofluorescence at the human fundus. J. Biomed. Opt. 2004, 9, 1214–1222. [Google Scholar] [CrossRef] [PubMed]
- Delori, F.C.; Dorey, C.K.; Staurenghi, G.; Arend, O.; Goger, D.G.; Weiter, J.J. In vivo fluorescence of the ocular fundus exhibits retinal pigment epithelium lipofuscin characteristics. Investig. Ophthalmol. Vis. Sci. 1995, 36, 718–729. [Google Scholar]
- Pau, H.; Degen, J.; Schmidtke, H.-H. Different regional changes of fluorescence spectra of clear human lenses and nuclear cataracts. Graefe’s Arch. Clin. Exp. Ophthalmol. 1993, 231, 656–661. [Google Scholar] [CrossRef]
- Klemm, M.; Schweitzer, D.; Peters, S.; Sauer, L.; Hammer, M.; Haueisen, J. FLIMX: A software package to determine and analyze the fluorescence lifetime in time-resolved fluorescence data from the human eye. PLoS ONE 2015, 10, e0131640. [Google Scholar] [CrossRef]
- Delori, F.C.; Webb, R.H.; Sliney, D.H. Maximum permissible exposures for ocular safety (ANSI 2000), with emphasis on ophthalmic devices. J. Opt. Soc. Am. A 2007, 24, 1250–1265. [Google Scholar] [CrossRef]
- Dysli, C.; Quellec, G.; Abegg, M.; Menke, M.N.; Wolf-Schnurrbusch, U.; Kowal, J.; Blatz, J.; La Schiazza, O.; Leichtle, A.B.; Wolf, S.; et al. Quantitative analysis of fluorescence lifetime measurements of the macula using the fluorescence lifetime imaging ophthalmoscope in healthy subjects. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2106–2113. [Google Scholar] [CrossRef] [PubMed]
- Dysli, C.; Dysli, M.; Zinkernagel, M.S.; Enzmann, V. Effect of pharmacologically induced retinal degeneration on retinal autofluorescence lifetimes in mice. Exp. Eye Res. 2016, 153, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Dysli, C.; Fink, R.; Wolf, S.; Zinkernagel, M.S. Fluorescence lifetimes of drusen in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2017, 58, 4856–4862. [Google Scholar] [CrossRef]
- Sauer, L.; Gensure, R.H.; Andersen, K.M.; Kreilkamp, L.; Hageman, G.S.; Hammer, M.; Bernstein, P.S. Patterns of fundus autofluorescence lifetimes in eyes of individuals with nonexudative age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD65–AMD77. [Google Scholar] [CrossRef] [PubMed]
- Spaide, R.F.; Curcio, C.A. Drusen characterization with multimodal imaging. Retina 2010, 30, 1441–1454. [Google Scholar] [CrossRef] [PubMed]
- Dysli, C.; Wolf, S.; Berezin, M.Y.; Sauer, L.; Hammer, M.; Zinkernagel, M.S. Fluorescence lifetime imaging ophthalmoscopy. Prog. Retin. Eye Res. 2017, 60, 120–143. [Google Scholar] [CrossRef] [PubMed]
- Sauer, L.; Andersen, K.M.; Dysli, C.; Zinkernagel, M.S.; Bernstein, P.S.; Hammer, M. Review of clinical approaches in fluorescence lifetime imaging ophthalmoscopy. J. Biomed. Opt. 2018, 23, 1–20. [Google Scholar] [CrossRef]
- Gray, N.; Lewis, M.R.; Plumb, R.S.; Wilson, I.D.; Nicholson, J.K. High-throughput microbore UPLC−MS metabolic phenotyping of urine for large-Scale epidemiology studies. J. Proteome Res. 2015, 14, 2714–2721. [Google Scholar] [CrossRef]
- Marko, C.K.; Laíns, I.; Husain, D.; Miller, J.W. AMD biomarkers identified by metabolomics. Retin. Phys. 2018, 15, 22–24. [Google Scholar]
- Rieckmann, J.C.; Geiger, R.; Hornburg, D.; Wolf, T.; Kveler, K.; Jarrossay, D.; Sallusto, F.; Shen-Orr, S.S.; Lanzavecchia, A.; Mann, M.; et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 2017, 18, 583–593. [Google Scholar] [CrossRef]
- Szmacinski, H.; Hegde, K.; Zeng, H.-H.; Eslami, K.; Puche, A.; Lakowicz, J.R.; Lengyel, I.; Thompson, R.B. Towards early detection of age-related macular degeneration with tetracyclines and FLIM. In Proceedings of SPIE; Vo-Dinh, T., Mahadevan-Jansen, A., Grundfest, W.S., Eds.; SPIE: Bellingham, WA, USA, 2018; Volume 10484. [Google Scholar]
- Merle, B.M.J.; Colijn, J.M.; Cougnard-Grégoire, A.; de Koning-Backus, A.P.M.; Delyfer, M.-N.; Kiefte-de Jong, J.C.; Meester-Smoor, M.; Féart, C.; Verzijden, T.; Samieri, C.; et al. Mediterranean diet and incidence of advanced age-related macular degeneration: The EYE-RISK Consortium. Ophthalmology 2018. [Google Scholar] [CrossRef] [PubMed]
- Lien, E.L.; Hammond, B.R. Nutritional influences on visual development and function. Prog. Retin. Eye Res. 2011, 30, 188–203. [Google Scholar] [CrossRef] [PubMed]
- Hollyfield, J.G.; Bonilha, V.L.; Rayborn, M.E.; Yang, X.; Shadrach, K.G.; Lu, L.; Ufret, R.L.; Salomon, R.G.; Perez, V.L. Oxidative damage-induced inflammation initiates age-related macular degeneration. Nat. Med. 2008, 14, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Meer, S.G.; Miyagi, M.; Rayborn, M.E.; Hollyfield, J.G.; Crabb, J.W.; Salomon, R.G. Carboxyethylpyrrole protein adducts and autoantibodies, biomarkers for age-related macular degeneration. J. Biol. Chem. 2003, 278, 42027–42035. [Google Scholar] [CrossRef]
- Rinninella, E.; Mele, M.; Merendino, N.; Cintoni, M.; Anselmi, G.; Caporossi, A.; Gasbarrini, A.; Minnella, A.; Rinninella, E.; Mele, M.C.; et al. The role of diet, micronutrients and the gut microbiota in age-related macular degeneration: New perspectives from the gut–retina axis. Nutrients 2018, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Biarnés, M.; Vassiliev, V.; Nogoceke, E.; Emri, E.; Rodríguez-Bocanegra, E.; Ferraro, L.; Garcia, M.; Fauser, S.; Monés, J.; Lengyel, I.; et al. Precision medicine for age-related macular degeneration: Current developments and prospects. Expert Rev. Precis. Med. Drug Dev. 2018, 3, 249–263. [Google Scholar] [CrossRef]
- Lorés-Motta, L.; de Jong, E.K.; den Hollander, A.I. Exploring the use of molecular biomarkers for precision medicine in age-related macular degeneration. Mol. Diagn. Ther. 2018, 22, 315–343. [Google Scholar] [CrossRef]
- Park, K.S.; Xu, C.L.; Cui, X.; Tsang, S.H. Reprogramming the metabolome rescues retinal degeneration. Cell. Mol. Life Sci. 2018, 75, 1559–1566. [Google Scholar] [CrossRef]
| Subjects and Biofluid Used | Number of Identified Metabolites | Metabolite Separation Method (Chromatography) | Detection Instrument Used | Reference |
|---|---|---|---|---|
| Mouse eye lysates | Not reported | Imtakt Scherzo SM-18 150 × 2 mm column Agilent 1200 capillary LC | Agilent 6538 UHD-QTOF MS (ESI+/ESI-) | [36] |
| Mouse eye lysates | 203 | XBridge C18 column (3.5 µm, 135 Å, 150 mm × 1.0 mm) Agilent 1260 HPLC | Agilent 6538 UHD Accurate Mass Q-TOF (ESI+) | [118] |
| Human serum (NVAMD); Mouse eye lysates | Not reported | Aeris Peptide XB-C18 column (3.6 µm, 100 × 2.10 mm) Phenomenex HPLC | Untargeted: Thermo Scientific LTQ Velos Orbitrap (ESI+/ESI-) Targeted: Thermo Scientific LXQ (ESI-) | [124] |
| hfRPE cells; Mouse retina; Apical/basal secretomes | 202 (101 in medium; 53 changed substantially) | Ethylene bridged hybrid Amid column (1.7 µm, 2.1 mm × 150 mm) Agilent 1260 HPLC | AB Sciex QTrap 5500 (MRM) | [125] |
| Human plasma (NVAMD) | 1168 (94 differed significantly) | Hamilton PRPX-110S (2.1 cm × 10 cm) anion exchange column | Thermo LTQ-FT spectrometer (ESI+) | [126] |
| Human plasma (NVAMD) | 864 (10 differed significantly) | Acquity HSS T3 UPLC column (1.8 µm, 2.1 × 100 mm) Agilent 1290 Infinity UHPLC | AB SCIEX Triple 6600 TOF (ESI+/ESI-) | [127] |
| Human plasma | 1188 spectra (30 low-Mw metabolites) | N/A | NMR: Bruker Avance DRX 500 spectrometer (300 K) operating at 500.13 MHz for protein, with a 5 mm TXI probe | [128] |
| Human plasma (nonexudative) | 698 endogenous (87 associated with AMD) | C18 (acidic positive and basic negative ionization); HILIC (negative ionization) Waters ACQUITY ultra-UPLC (Metabolon, Inc.) | Thermo Scientific Q-Exactive (HESI-II), Orbitrap mass analyzer | [129] |
| WT mouse plasma (MS) and urine (NMR) | MS: 309 NMR: 47 | Polar and nonpolar lipids: ACQUITY BEH C8 column (1.7 µm, 100 × 2.1 mm) Simadzu Nexera X2 UHPLC Hydrophilic metabolites: Atlantis HILIC column (3 µm, 150 × 2 mm) Shimadzu Nexera X2 UHPLC Additional polar metabolites: Phenomenex Luna NH2 column (150 × 2.0 mm) ACQUITY UHPLC | Polar and nonpolar lipids: Thermo Scientific Exactive Plus Orbitrap MS (ESI+) Hydrophilic metabolites: Thermo Fisher Scientific Q Exactive hybrid quadrupole Orbitrap MS (ESI+) Additional polar metabolites: AB SCIEX 5500 QTRAP MS (ESI- and MRM) NMR: Bruker Avance 600 spectrometer | [130] |
| Human plasma (NVAMD) | 159 features differed (39 with medium to high confidence) | Hamilton PRP-X110S, 2.1 × 10 cm (anion exhchange) a Higgins Analytical C18 column, 2.1 × 10 cm (reverse phase) a | Thermo Scientific LTQ Velos Orbitrap MS (ESI+) | [131] |
| Reference | Biofluid | Comparison | Metabolomic Technique Employed | Definitively Identified Metabolites | Level in AMD Cohort Compared to Controls |
|---|---|---|---|---|---|
| [124] | Blood serum | NVAMD patients (n = 22) and age-matched control patients (n = 22) | LC-MS | Docosahexaenoic acid | Lower |
| Amino acids | Higher | ||||
| Prostaglandin G2 | Higher NS | ||||
| [159] | Blood plasma | Intermediate AMD (drusen), late AMD (GA and CNV) (n = 77) and non-AMD control patients (n = 75) | HPLC and GC-negative-ion chemical ionization (NICI)-MS | Cystine Isofurans | Higher NS |
| [126] | Blood plasma | NVAMD patients (n = 26) and age-matched control patients (n = 19) | LC- Fourier transform MS (FTMS) | Acetylphenylalanine | Higher |
| Dipeptide; Tripeptides (modified cysteine and alanine a) | |||||
| Sethoxydim | |||||
| Tripeptides b | |||||
| Tripeptides (acetyltryptophana) | |||||
| Flavones; halofenozide | |||||
| Glycocholic acid | Lower | ||||
| Vitamin D-related metabolites; phytochemicals b | |||||
| Glycodeoxycholic acid+H+; Glycoursodeoxycholic acid+H+ | |||||
| Glycodeoxycholic acid+Na+; Glycoursodeoxycholic acid+Na+ | |||||
| Sencrassidol | |||||
| Didemethylsimmondsin | |||||
| Dipeptides b | |||||
| [127] | Blood plasma | NVAMD patients (n = 20) and age-matched control patients (n = 20) | UPLC-TOF-MS | N-Acetyl-l-alanine | Higher |
| l-Tyrosine | |||||
| l-Phenylalanine | |||||
| l-Methionine | |||||
| l-Arginine | |||||
| Isomaltose | |||||
| Ν1-Methyl-2-pyridone-5-carboxamide | Lower | ||||
| l-Palmitoylcarnitine | |||||
| Hydrocortisone | |||||
| Biliverdin | |||||
| [167] | Blood serum | NVAMD patients (n = 20), PCV patients (n = 20), and age-matched controls (n = 20) | UPLC-QTOF-MS | Glycerophospholipids c | Higher |
| Phosphatidylcholine | |||||
| Covalently modified amino acids c | |||||
| Di/tri-peptides c | |||||
| Tripeptides c | |||||
| ω-3 and ω-6 PUFAs c | |||||
| Pinolenic acid | |||||
| Docoxahexaenoic acid | |||||
| Eicosatetraenoic acid | |||||
| Carnitine sp. c | |||||
| [129] | Blood plasma | AMD patients (n = 314) and age-matched controls (n = 82), both across two locations | HILIC- and UPLC-MS | Creatine d | Higher |
| Oleic acid d | Higher f | ||||
| N(CH3)3 choline HDL d | |||||
| Acetate d | |||||
| Dimethylsulfone d | |||||
| Pyruvate d | Higher g | ||||
| Glutamine e | |||||
| Unsaturated F.A. e | |||||
| Unsaturated F.A. LDL + VLDL e | |||||
| Unsaturated F.A. d | Lower f | ||||
| Unsaturated F.A. LDL + VLDL d | |||||
| Histidine d | Lower h | ||||
| Acetoacetate d | |||||
| β-hydroxybutyrate d | |||||
| Unsaturated F.A. LDL + VLDL e | Lower f | ||||
| Glutamine e | Lower g | ||||
| Histidine e | |||||
| CH2CH2COOR F.A. e | |||||
| CH2CH2C=C F.A. e | |||||
| Albumin lysil e | |||||
| Alanine e | Lower h | ||||
| Histidine e | |||||
| Glyceryl C1,3H’ e | |||||
| [128] | Blood plasma | AMD patients (n = 89) and age-matched control patients (n = 30) | NMR | N2-methylguanosine | Higher |
| 1-Stearoyl-2-oleoyl GPC | Lower NS | ||||
| 1-Linoleoyl-2-arachidonoyl GPC | Lower | ||||
| Stearoyl-arachidonoyl glycerol | |||||
| Oleoyl-olyeol-glycerol | |||||
| Dihomo-linolenoyl carnitine | |||||
| 1-Stearoyl-2-arachidonoyl GPC | |||||
| Linoleoyl-linolenoyl glycerol | |||||
| 1-Stearoyl-2-linoleoyl-GPI f | |||||
| Oleoyl-linoleoyl-glycerol | |||||
| Oleoylcarnitine | |||||
| Ximenoylcarnitine | |||||
| 1-Stearoyl-2-arachidonoyl GPI i | |||||
| [131] | Blood plasma | NVAMD patients (n = 100) and control patients (n = 192) | LC-MS and LC-MS/MS | l-Oxalylalbizziine j | Higher |
| Isopentyl beta-d-glucoside j | |||||
| LysoPC(P-18:0) j | |||||
| LysoPC(P-18:1(9Z)) j | |||||
| LysoPC(16:1(9Z)) j | |||||
| Darunavir j | |||||
| Bepridil j | |||||
| 912-Hexadecadienoylcarnitine j | |||||
| 456-Trimethylscutellarein 7-glucoside j | |||||
| 1-Lyso-2-arachidonoyl-phosphidate j | |||||
| Americanin B j | |||||
| Corchoroside A j | |||||
| N-Ornithyl-l-taurine j | |||||
| Lyciumoside III j | Lower | ||||
| Phosphatidylethanolamine f,j | |||||
| Phytosphingosine j | |||||
| Lenticin j | |||||
| 9-Hexadecenoylcarnitine k | Higher | ||||
| Heptadecanoyl carnitine k | |||||
| 11Z-Octadecenylcarnitine k | |||||
| l-Palmitoylcarnitine k | |||||
| Stearoylcarnitine k | |||||
| [168,169] | Blood serum and urine | Neovascular and nonexudative AMD patients (n = 104) l | NMR | Arginine | Higher m,NS |
| Glucose | Lower m,NS | ||||
| Lactate | |||||
| Glutamine | |||||
| Reduced glutathione |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, C.N.; Green, B.D.; Thompson, R.B.; Den Hollander, A.I.; Lengyel, I.; On behalf of the EYE-RISK consortium. Metabolomics and Age-Related Macular Degeneration. Metabolites 2019, 9, 4. https://doi.org/10.3390/metabo9010004
Brown CN, Green BD, Thompson RB, Den Hollander AI, Lengyel I, On behalf of the EYE-RISK consortium. Metabolomics and Age-Related Macular Degeneration. Metabolites. 2019; 9(1):4. https://doi.org/10.3390/metabo9010004
Chicago/Turabian StyleBrown, Connor N., Brian D. Green, Richard B. Thompson, Anneke I. Den Hollander, Imre Lengyel, and On behalf of the EYE-RISK consortium. 2019. "Metabolomics and Age-Related Macular Degeneration" Metabolites 9, no. 1: 4. https://doi.org/10.3390/metabo9010004
APA StyleBrown, C. N., Green, B. D., Thompson, R. B., Den Hollander, A. I., Lengyel, I., & On behalf of the EYE-RISK consortium. (2019). Metabolomics and Age-Related Macular Degeneration. Metabolites, 9(1), 4. https://doi.org/10.3390/metabo9010004