Metabolomic Profiling of Bile Acids in an Experimental Model of Prodromal Parkinson’s Disease
Abstract
1. Introduction
2. Results
2.1. Univariate Analysis
2.2. Logistic Regression Analysis
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Purification of Recombinant α-syn, Assembly of Preformed Fibrils and Stereotactic Injections
4.3. Serum Collection
4.4. Bile Acid Quantification
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- De Lau, L.M.; Breteler, M.M. Epidemiology of Parkinson’s disease. Lancet Neurol. 2006, 5, 525–535. [Google Scholar] [CrossRef]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Tysnes, O.-B.; Storstein, A. Epidemiology of Parkinson’s disease. J. Neural Transm. 2017, 124, 901–905. [Google Scholar] [CrossRef] [PubMed]
- Schapira, A.H.V.; Chaudhuri, K.R.; Jenner, P. Non-motor features of Parkinson disease. Nat. Rev. Neurosci. 2017, 18, 435–450. [Google Scholar] [CrossRef] [PubMed]
- Postuma, R.B.; Berg, D. Advances in markers of prodromal Parkinson disease. Nat. Rev. Neurol. 2016, 12, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Havelund, J.F.; Heegaard, N.H.H.; Faergeman, N.J.K.; Gramsbergen, J.B. Biomarker Research in Parkinson’s Disease Using Metabolite Profiling. Metabolites 2017, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Rey, N.L.; Steiner, J.A.; Maroof, N.; Luk, K.C.; Madaj, Z.; Trojanowski, J.Q.; Lee, V.M.-Y.; Brundin, P. Widespread transneuronal propagation of α-synucleinopathy triggered in olfactory bulb mimics prodromal Parkinson’s disease. J. Exp. Med. 2016, 213, 1759–1778. [Google Scholar] [CrossRef] [PubMed]
- Espay, A.J.; Schwarzschild, M.A.; Tanner, C.M.; Fernandez, H.H.; Simon, D.K.; Leverenz, J.B.; Merola, A.; Chen-Plotkin, A.; Brundin, P.; Erro, R.; et al. Biomarker-driven phenotyping in Parkinson’s disease: A translational missing link in disease-modifying clinical trials. Mov. Disord. Off. J. Mov. Dis. Soc. 2017, 32, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Espay, A.J.; Brundin, P.; Lang, A.E. Precision medicine for disease modification in Parkinson disease. Nat. Rev. Neurol. 2017, 13, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.F.; Rey, N.L.; Yilmaz, A.; Kumar, P.; Madaj, Z.; Maddens, M.; Bahado-Singh, R.O.; Becker, K.; Schulz, E.; Meyerdirk, L.K.; et al. Biochemical Profiling of the Brain and Blood Metabolome in a Mouse Model of Prodromal Parkinson’s Disease Reveals Distinct Metabolic Profiles. J. Proteom Res. 2018, 17, 2460–2469. [Google Scholar] [CrossRef] [PubMed]
- Camilleri, M.; Gores, G.J. Therapeutic targeting of bile acids. Am. J. Phys. Gastrointest. Liver Phys. 2015, 309, G209–G215. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Dis. 2008, 7, 678–693. [Google Scholar] [CrossRef] [PubMed]
- Perino, A.; Schoonjans, K. TGR5 and Immunometabolism: Insights from Physiology and Pharmacology. Trends Pharmacol. Sci. 2015, 36, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Parry, G.J.; Rodrigues, C.M.; Aranha, M.M.; Hilbert, S.J.; Davey, C.; Kelkar, P.; Low, W.C.; Steer, C.J. Safety, tolerability, and cerebrospinal fluid penetration of ursodeoxycholic Acid in patients with amyotrophic lateral sclerosis. Clin. Neuropharmacol. 2010, 33, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Mano, N.; Goto, T.; Uchida, M.; Nishimura, K.; Ando, M.; Kobayashi, N.; Goto, J. Presence of protein-bound unconjugated bile acids in the cytoplasmic fraction of rat brain. J. Lipid Res. 2004, 45, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Bron, B.; Waldram, R.; Silk, D.B.; Williams, R. Serum, cerebrospinal fluid, and brain levels of bile acids in patients with fulminant hepatic failure. Gut 1977, 18, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Olazaran, J.; Gil-de-Gomez, L.; Rodriguez-Martin, A.; Valenti-Soler, M.; Frades-Payo, B.; Marin-Munoz, J.; Antunez, C.; Frank-Garcia, A.; Acedo-Jimenez, C.; Morlan-Gracia, L.; et al. A blood-based, 7-metabolite signature for the early diagnosis of Alzheimer’s disease. J. Alzheimer’s Dis. 2015, 45, 1157–1173. [Google Scholar] [CrossRef] [PubMed]
- Bathena, S.P.; Mukherjee, S.; Olivera, M.; Alnouti, Y. The profile of bile acids and their sulfate metabolites in human urine and serum. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2013, 942–943, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Abdelkader, N.F.; Safar, M.M.; Salem, H.A. Ursodeoxycholic Acid Ameliorates Apoptotic Cascade in the Rotenone Model of Parkinson’s Disease: Modulation of Mitochondrial Perturbations. Mol. Neurobiol. 2016, 53, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Theofilopoulos, S.; Wang, Y.; Kitambi, S.S.; Sacchetti, P.; Sousa, K.M.; Bodin, K.; Kirk, J.; Salto, C.; Gustafsson, M.; Toledo, E.M.; et al. Brain endogenous liver X receptor ligands selectively promote midbrain neurogenesis. Nat. Chem. Biol. 2013, 9, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Marksteiner, J.; Blasko, I.; Kemmler, G.; Koal, T.; Humpel, C. Bile acid quantification of 20 plasma metabolites identifies lithocholic acid as a putative biomarker in Alzheimer’s disease. Metabolomics 2018, 14, 1. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Elliott, C.T.; McGuinness, B.; Passmore, P.; Kehoe, P.G.; Holscher, C.; McClean, P.L.; Graham, S.F.; Green, B.D. Metabolomic Profiling of Bile Acids in Clinical and Experimental Samples of Alzheimer’s Disease. Metabolites 2017, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Ghebremedhin, E.; Rub, U.; Bratzke, H.; Del Tredici, K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004, 318, 121–134. [Google Scholar] [CrossRef] [PubMed]
- MahmoudianDehkordi, S.; Arnold, M.; Nho, K.; Ahmad, S.; Jia, W.; Xie, G.; Louie, G.; Kueider-Paisley, A.; Moseley, M.A.; Thompson, J.W.; et al. Altered bile acid profile associates with cognitive impairment in Alzheimer’s disease-An emerging role for gut microbiome. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2018. [Google Scholar] [CrossRef] [PubMed]
- Braak, H.; Del Tredici, K. Neuropathological Staging of Brain Pathology in Sporadic Parkinson’s disease: Separating the Wheat from the Chaff. J. Parkinson’s Dis. 2017, 7, S71–S85. [Google Scholar] [CrossRef] [PubMed]
- Beach, T.G.; White, C.L., 3rd; Hladik, C.L.; Sabbagh, M.N.; Connor, D.J.; Shill, H.A.; Sue, L.I.; Sasse, J.; Bachalakuri, J.; Henry-Watson, J.; et al. Olfactory bulb alpha-synucleinopathy has high specificity and sensitivity for Lewy body disorders. Acta Neuropathol. 2009, 117, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Beach, T.G.; Adler, C.H.; Lue, L.; Sue, L.I.; Bachalakuri, J.; Henry-Watson, J.; Sasse, J.; Boyer, S.; Shirohi, S.; Brooks, R.; et al. Unified staging system for Lewy body disorders: Correlation with nigrostriatal degeneration, cognitive impairment and motor dysfunction. Acta Neuropathol. 2009, 117, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rong, Z.; Xiang, D.; Zhang, C.; Liu, D. Detection technologies and metabolic profiling of bile acids: A comprehensive review. Lipid Health Dis. 2018, 17, 121. [Google Scholar] [CrossRef] [PubMed]
- Lieu, T.; Jayaweera, G.; Bunnett, N.W. GPBA: A GPCR for bile acids and an emerging therapeutic target for disorders of digestion and sensation. Br. J. Pharmacol. 2014, 171, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, A.F. The continuing importance of bile acids in liver and intestinal disease. Arch. Int. Med. 1999, 159, 2647–2658. [Google Scholar] [CrossRef]
- Benedetti, A.; Alvaro, D.; Bassotti, C.; Gigliozzi, A.; Ferretti, G.; La Rosa, T.; Di Sario, A.; Baiocchi, L.; Jezequel, A.M. Cytotoxicity of bile salts against biliary epithelium: A study in isolated bile ductule fragments and isolated perfused rat liver. Hepatology 1997, 26, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Mello-Vieira, J.; Sousa, T.; Coutinho, A.; Fedorov, A.; Lucas, S.D.; Moreira, R.; Castro, R.E.; Rodrigues, C.M.; Prieto, M.; Fernandes, F. Cytotoxic bile acids, but not cytoprotective species, inhibit the ordering effect of cholesterol in model membranes at physiologically active concentrations. Biochim. Biophys. Acta 2013, 1828, 2152–2163. [Google Scholar] [CrossRef] [PubMed]
- Geier, A.; Wagner, M.; Dietrich, C.G.; Trauner, M. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim. Biophys. Acta 2007, 1773, 283–308. [Google Scholar] [CrossRef] [PubMed]
- Cortez, L.M.; Campeau, J.; Norman, G.; Kalayil, M.; Van der Merwe, J.; McKenzie, D.; Sim, V.L. Bile Acids Reduce Prion Conversion, Reduce Neuronal Loss, and Prolong Male Survival in Models of Prion Disease. J. Virol. 2015, 89, 7660–7672. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.M.; Fan, G.; Wong, P.Y.; Kren, B.T.; Steer, C.J. Ursodeoxycholic acid may inhibit deoxycholic acid-induced apoptosis by modulating mitochondrial transmembrane potential and reactive oxygen species production. Mol. Med. 1998, 4, 165–178. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.M.; Sola, S.; Sharpe, J.C.; Moura, J.J.; Steer, C.J. Tauroursodeoxycholic acid prevents Bax-induced membrane perturbation and cytochrome C release in isolated mitochondria. Biochemistry 2003, 42, 3070–3080. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.M.; Fan, G.; Ma, X.; Kren, B.T.; Steer, C.J. A novel role for ursodeoxycholic acid in inhibiting apoptosis by modulating mitochondrial membrane perturbation. J. Clin. Investig. 1998, 101, 2790–2799. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’Mahony, S.M. The microbiome-gut-brain axis: From bowel to behavior. Neurogastroenterol. Motil. 2011, 23, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Rey, N.L.; George, S.; Steiner, J.A.; Madaj, Z.; Luk, K.C.; Trojanowski, J.Q.; Lee, V.M.-Y.; Brundin, P. Spread of aggregates after olfactory bulb injection of α-synuclein fibrils is associated with early neuronal loss and is reduced long term. Acta Neuropathol. 2018, 135, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Rey, N.L.; Petit, G.H.; Bousset, L.; Melki, R.; Brundin, P. Transfer of human alpha-synuclein from the olfactory bulb to interconnected brain regions in mice. Acta Neuropathol. 2013, 126, 555–573. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. MetaboAnalyst 3.0—Making metabolomics more meaningful. Nucleic Acid Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef] [PubMed]
- Tibshirani, R. Regression Shrinkage and Selection via the Lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acid Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
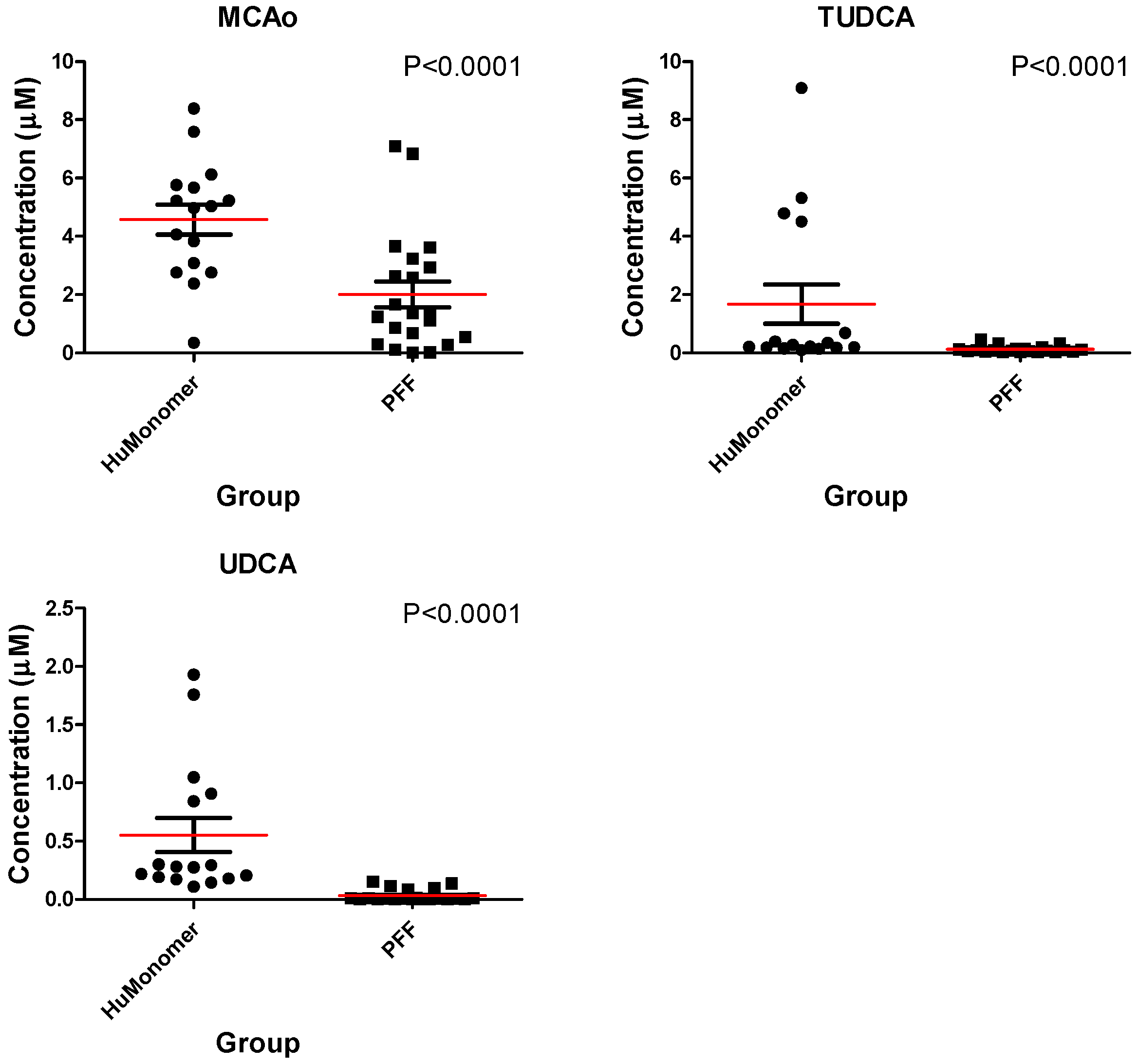
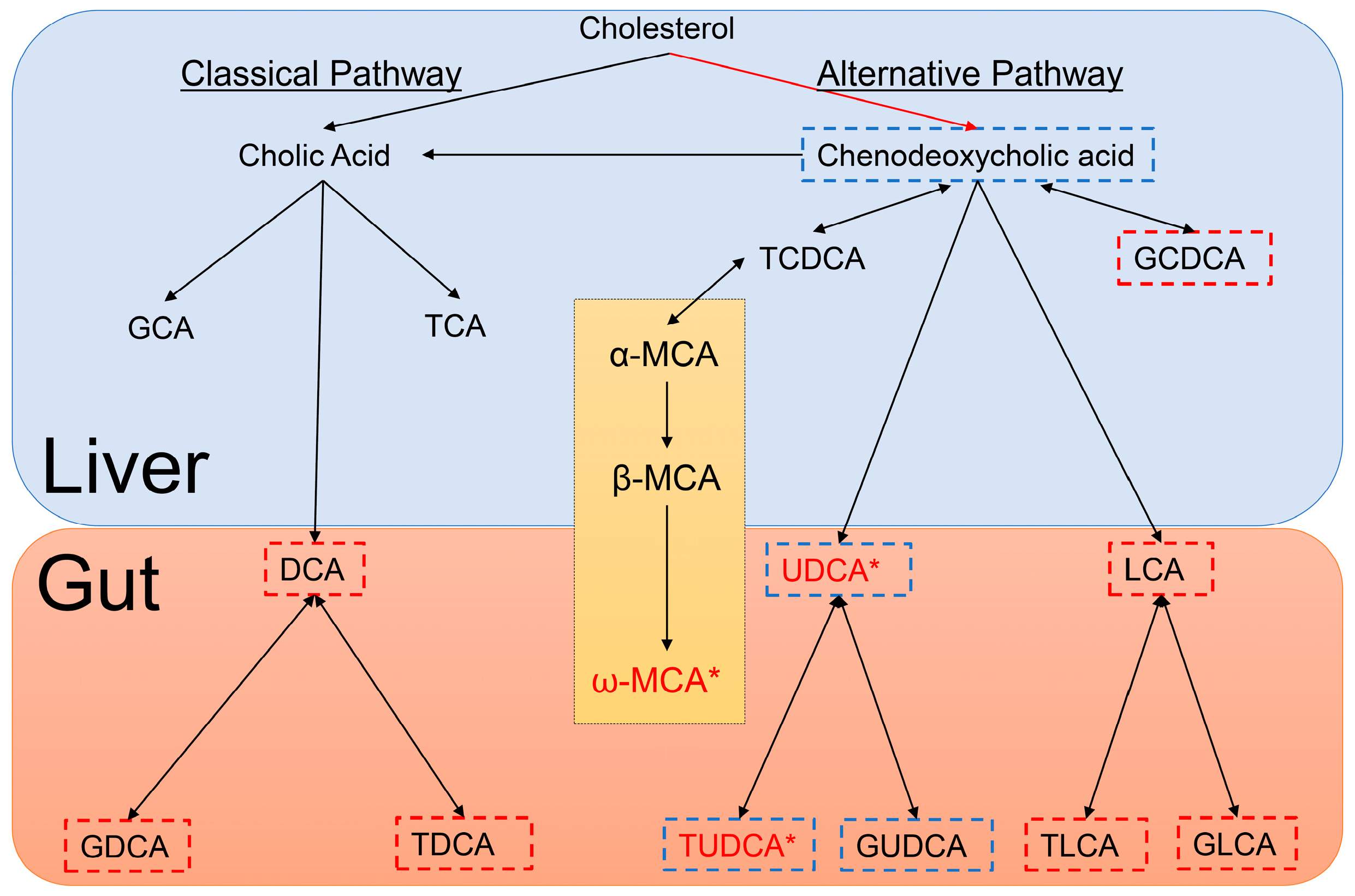
| HMDB# | Name | Mean (SD) of HuMonomer | Mean (SD) of PFF | p-Value | q-Value (FDR) | Fold Change | LOD | LLOQ |
|---|---|---|---|---|---|---|---|---|
| HMDB0000619 | Cholic Acid | 11.09 (20.89) | 10.12 (18.99) | 0.24 | 0.39 | 1.10 | 0.004 | 0.03 |
| HMDB0000518 | Chenodeoxycholic acid | 0.89 (1.22) | 0.77 (1.53) | 0.06 | 0.19 | 1.15 | 0.005 | 0.02 |
| HMDB0000626 | Deoxycholic acid | 1.63 (2.07) | 1.52 (2.61) | 0.20 | 0.39 | 1.08 | 0.005 | 0.02 |
| HMDB0000138 | Glycocholic acid | 0.07 (0.07) | 0.06 (0.06) | 0.67 | 0.85 | 1.14 | 0.003 | 0.03 |
| HMDB0000637 | Glycochenodeoxycholic acid | 0.06 (0.14) | 0.07 (0.14) | 0.19 | 0.39 | −1.07 | 0.01 | 0.02 |
| HMDB0000631 | Glycodeoxycholic acid | 0.66 (0.77) | 0.35 (0.46) | 0.37 | 0.55 | 1.90 | 0.01 | 0.01 |
| HMDB0000733 | Hyodeoxycholic acid | 0.65 (0.51) | 0.44 (0.52) | 0.04 | 0.16 | 1.47 | 0.005 | 0.02 |
| HMDB0000761 | Lithocholic acid | 0.10 (0.13) | 0.10 (0.15) | 0.76 | 0.85 | −1.04 | 0.002 | 0.01 |
| HMDB0000506 | Alpha-Muricholic acid | 0.83 (1.42) | 0.65 (1.23) | 0.06 | 0.19 | 1.28 | 0.007 | 0.01 |
| HMDB0000415 | Beta-Muricholic acid | 7.49 (10.54) | 5.72 (8.760) | 0.09 | 0.23 | 1.31 | 0.008 | 0.02 |
| HMDB0000364 | Omega-Murichoclic acid | 4.58 (2.04) | 2.00 (2.03) | <0.0001 | 0.01 | 2.28 | 0.007 | 0.01 |
| HMDB0000036 | Taurocholic acid | 11.02 (17.81) | 9.20 (20.59) | 0.93 | 0.98 | 1.20 | 0.008 | 0.02 |
| HMDB0000951 | Taurochenodeoxycholic acid | 0.75 (1.22) | 0.79 (1.56) | 0.99 | 0.99 | −1.05 | 0.005 | 0.01 |
| HMDB0000896 | Taurodeoxycholic acid | 0.29 (0.23) | 0.35 (0.42) | 0.74 | 0.85 | −1.22 | 0.001 | 0.01 |
| HMDB0000722 | Taurolithocholic acid | 0.01 (0.02) | 0.02 (0.03) | 0.40 | 0.55 | −1.41 | 0.001 | 0.01 |
| HMDB0000932 | Tauromuricholic acid (sum of α and β) | 1.07 (1.85) | 0.42 (0.96) | 0.22 | 0.39 | 2.52 | 0.001 | 0.01 |
| HMDB0000874 | Tauroursodeoxycholic acid | 1.67 (2.71) | 0.12 (0.12) | <0.0001 | <0.001 | 14.14 | 0.001 | 0.01 |
| HMDB0000946 | Ursodeoxycholic acid | 0.55 (0.58) | 0.03 (0.05) | <0.0001 | <0.0001 | 17.55 | 0.001 | 0.02 |
| Estimate | Std. Error | z Value | Pr (>|z|) | Odds | |
|---|---|---|---|---|---|
| (Intercept) | −0.893 | 2.857 | −0.313 | 0.755 | - |
| TLCA | 11.152 | 7.264 | 1.535 | 0.125 | 69,675.46 |
| GCDCA | 8.917 | 9.571 | 0.932 | 0.352 | 7455.77 |
| TUDCA | −18.221 | 7.762 | −2.347 | 0.019 | 0 |
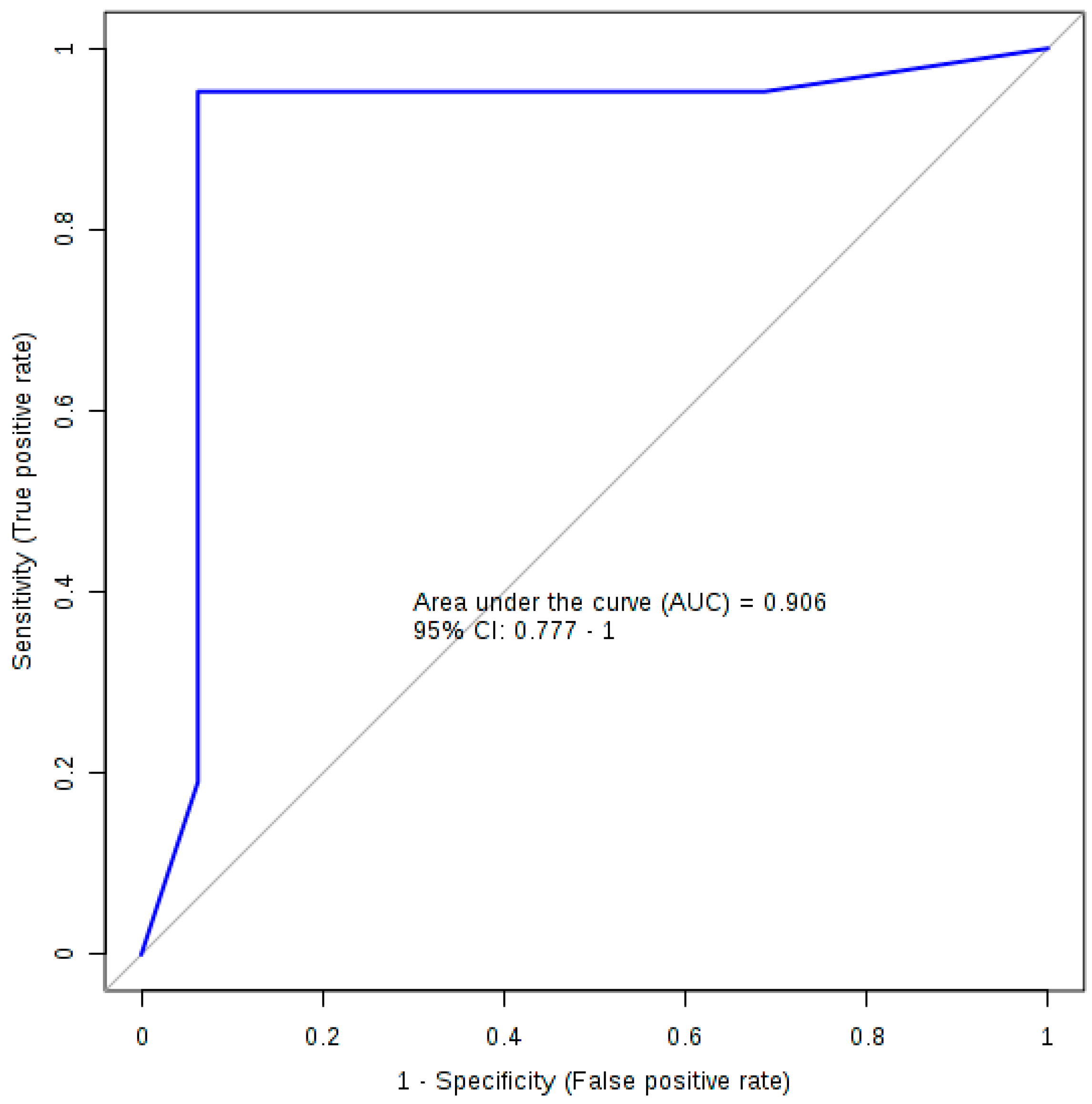
| AUC | Sensitivity | Specificity | |
|---|---|---|---|
| Training/Discovery | 0.992 (0.985~0.998) | 0.958 (0.929~0.986) | 0.944 (0.907~0.982) |
| 10-fold Cross-Validation | 0.906 (0.777~1.000) | 0.952 (0.952~1.000) | 0.938 (0.819~1.000) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Graham, S.F.; Rey, N.L.; Ugur, Z.; Yilmaz, A.; Sherman, E.; Maddens, M.; Bahado-Singh, R.O.; Becker, K.; Schulz, E.; Meyerdirk, L.K.; et al. Metabolomic Profiling of Bile Acids in an Experimental Model of Prodromal Parkinson’s Disease. Metabolites 2018, 8, 71. https://doi.org/10.3390/metabo8040071
Graham SF, Rey NL, Ugur Z, Yilmaz A, Sherman E, Maddens M, Bahado-Singh RO, Becker K, Schulz E, Meyerdirk LK, et al. Metabolomic Profiling of Bile Acids in an Experimental Model of Prodromal Parkinson’s Disease. Metabolites. 2018; 8(4):71. https://doi.org/10.3390/metabo8040071
Chicago/Turabian StyleGraham, Stewart F., Nolwen L. Rey, Zafer Ugur, Ali Yilmaz, Eric Sherman, Michael Maddens, Ray O. Bahado-Singh, Katelyn Becker, Emily Schulz, Lindsay K. Meyerdirk, and et al. 2018. "Metabolomic Profiling of Bile Acids in an Experimental Model of Prodromal Parkinson’s Disease" Metabolites 8, no. 4: 71. https://doi.org/10.3390/metabo8040071
APA StyleGraham, S. F., Rey, N. L., Ugur, Z., Yilmaz, A., Sherman, E., Maddens, M., Bahado-Singh, R. O., Becker, K., Schulz, E., Meyerdirk, L. K., Steiner, J. A., Ma, J., & Brundin, P. (2018). Metabolomic Profiling of Bile Acids in an Experimental Model of Prodromal Parkinson’s Disease. Metabolites, 8(4), 71. https://doi.org/10.3390/metabo8040071








