Effects of Early Intervention with Maternal Fecal Microbiota and Antibiotics on the Gut Microbiota and Metabolite Profiles of Piglets
Abstract
1. Introduction
2. Results
2.1. Gastrointestinal Microbial Community
2.2. Metabolite Profiles
3. Discussion
3.1. Effects of Early Maternal Fecal Microbiota and Antibiotics Intervention on the Microbiota of the Gastrointestinal Tract
3.2. Effects of Early Maternal Fecal Microbiota and Antibiotics Intervention on the Metabolite Profiles
4. Materials and Methods
4.1. Ethics Statement
4.2. Preparation of Maternal Fecal Microbiota Suspension
4.3. Experimental Design
4.4. Sampling
4.5. Illumina MiSeq Sequencing and Bioinformatics Analysis
4.6. Real-Time PCR Quantification of Total Bacteria
4.7. Sample Preparation and GC-MS Analysis
4.8. GC-MS Data Acquisition and Processing
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflict of Interest
References
- Tanaka, M.; Nakayama, J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol. Int. 2017, 66, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Penders, J.; Gerhold, K.; Stobberingh, E.E.; Thijs, C.; Zimmermann, K.; Lau, S.; Hamelmann, E. Establishment of the intestinal microbiota and its role for atopic dermatitis in early childhood. J. Allergy Clin. Immunol. 2013, 132. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef]
- Kostic, A.D.; Howitt, M.R.; Garrett, W.S. Exploring host-microbiota interactions in animal models and humans. Genes Dev. 2013, 27, 701–718. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.-C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7. [Google Scholar] [CrossRef] [PubMed]
- Arnal, M.E.; Zhang, J.; Messori, S.; Bosi, P.; Smidt, H.; Lallès, J.P. Early changes in microbial colonization selectively modulate intestinal enzymes, but not inducible heat shock proteins in young adult swine. PLoS ONE 2014, 9, e87967. [Google Scholar] [CrossRef]
- Gronlund, M.M.; Lehtonen, O.P.; Eerola, E.; Kero, P. Fecal microflora in healthy infants born by different methods of delivery: Permanent changes in intestinal flora after cesarean delivery. J. Pediatr. Gastroenterol. Nutr. 1999, 28, 19–25. [Google Scholar] [CrossRef]
- Chen, X.; Xu, J.M.; Ren, E.D.; Su, Y.; Zhu, W.Y. Co-occurrence of early gut colonization in neonatal piglets with microbiota in the maternal and surrounding delivery environments. Anaerobe 2018, 49, 30–40. [Google Scholar] [CrossRef]
- Youngster, I.; Russell, G.H.; Pindar, C.; Ziv-Baran, T.; Sauk, J.; Hohmann, E.L. Oral, Capsulized, Frozen Fecal Microbiota Transplantation for Relapsing Clostridium difficile Infection. JAMA-J. Am. Med. Assoc. 2014, 312, 1772–1778. [Google Scholar] [CrossRef]
- Pigneur, B.; Sokol, H. Fecal microbiota transplantation in inflammatory bowel disease: The quest for the holy grail. Mucosal Immunol. 2016, 9, 1360–1365. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.W.; Adams, J.B.; Gregory, A.C.; Borody, T.; Chittick, L.; Fasano, A.; Khoruts, A.; Geis, E.; Maldonado, J.; McDonough-Means, S.; et al. Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: An open-label study. Microbiome 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Vrieze, A.; Van Nood, E.; Holleman, F.; Salojarvi, J.; Kootte, R.S.; Bartelsman, J.; Dallinga-Thie, G.M.; Ackermans, M.T.; Serlie, M.J.; Oozeer, R.; et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012, 143, 913. [Google Scholar] [CrossRef] [PubMed]
- Stripling, J.; Kumar, R.; Baddley, J.W.; Nellore, A.; Dixon, P.; Howard, D.; Ptacek, T.; Lefkowitz, E.J.; Tallaj, J.A.; Benjamin, W.H.; et al. Loss of vancomycin-resistant enterococcus fecal dominance in an organ transplant patient with Clostridium difficile colitis after fecal microbiota transplant. Open Forum Infect. Dis. 2015, 2, 4. [Google Scholar] [CrossRef] [PubMed]
- Manichanh, C.; Reeder, J.; Gibert, P.; Varela, E.; Llopis, M.; Antolin, M.; Guigo, R.; Knight, R.; Guarner, F. Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res. 2010, 20, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Lawley, T.D.; Clare, S.; Walker, A.W.; Stares, M.D.; Connor, T.R.; Raisen, C.; Goulding, D.; Rad, R.; Schreiber, F.; Brandt, C.; et al. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, S.K.; Xu, B.; Nyachoti, C.M.; Giesting, D.W.; Krause, D.O. Evaluation of alternatives to antibiotics using an Escherichia coli K88(+) model of piglet diarrhea: Effects on gut microbial ecology. J. Anim. Sci. 2008, 86, 836–847. [Google Scholar] [CrossRef] [PubMed]
- Nathan, C. Antibiotics at the crossroads. Nature 2004, 431, 899–902. [Google Scholar] [CrossRef] [PubMed]
- Antonopoulos, D.A.; Huse, S.M.; Morrison, H.G.; Schmidt, T.M.; Sogin, M.L.; Young, V.B. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009, 77, 2367–2375. [Google Scholar] [CrossRef]
- Morel, F.B.; Oozeer, R.; Piloquet, H.; Moyon, T.; Pagniez, A.; Knol, J.; Darmaun, D.; Michel, C. Preweaning modulation of intestinal microbiota by oligosaccharides or amoxicillin can contribute to programming of adult microbiota in rats. Nutrition 2015, 31, 515–522. [Google Scholar] [CrossRef]
- Rauch, M.; Lynch, S.V. Probiotic manipulation of the gastrointestinal microbiota. Gut Microbes 2010, 1, 335–338. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Nie, Y.F.; Chen, J.W.; Zhang, Y.; Wang, Z.C.; Fan, Q.W.; Yan, X.H. Gradual changes of gut microbiota in weaned miniature piglets. Front. Microbiol. 2016, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Gophna, U.; Konikoff, T.; Nielsen, H.B. Oscillospira and related bacteria-from metagenomic species to metabolic features. Environ. Microbiol. 2017, 19, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, K.E. The occurrence, virulence, and antimicrobial resistance of anaerobes in polymicrobial infections. Am. J. Surg. 1995, 169, S2–S7. [Google Scholar]
- Shahi, S.K.; Freedman, S.N.; Mangalam, A.K. Gut microbiome in multiple sclerosis: The players involved and the roles they play (volume 8, p. 607, 2017). Gut Microbes 2017, 8, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Kverka, M.; Zakostelska, Z.; Klimesova, K.; Sokol, D.; Hudcovic, T.; Hrncir, T.; Rossmann, P.; Mrazek, J.; Kopecny, J.; Verdu, E.F.; et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin. Exp. Immunol. 2011, 163, 250–259. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Finegold, S.M.; Song, Y.J.; Lawson, P.A. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov and description of Blautia wexlerae sp nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58, 1896–1902. [Google Scholar] [CrossRef]
- Scanlan, P.D.; Shanahan, F.; Clune, Y.; Collins, J.K.; O’Sullivan, G.C.; O’Riordan, M.; Holmes, E.; Wang, Y.L.; Marchesi, J.R. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ. Microbiol. 2008, 10, 789–798. [Google Scholar] [CrossRef]
- Janczyk, P.; Pieper, R.; Souffrant, W.B.; Bimczok, D.; Rothkotter, H.J.; Smidt, H. Parenteral long-acting amoxicillin reduces intestinal bacterial community diversity in piglets even 5 weeks after the administration. ISME J. 2007, 1, 180–183. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Huse, S.; Sogin, M.L.; Relman, D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008, 6, 2383–2400. [Google Scholar] [CrossRef]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5. [Google Scholar] [CrossRef]
- Sullivan, A.; Edlund, C.; Nord, C.E. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 2001, 1, 101–114. [Google Scholar] [CrossRef]
- Geng, S.; Cheng, S.; Li, Y.; Wen, Z.; Ma, X.; Jiang, X.; Wang, Y.; Han, X. Fecal microbiota transplantation reduces susceptibility to epithelial injury and modulates tryptophan metabolism of microbial community in a piglet model. J. Crohns Colitis 2018, 1, 16. [Google Scholar] [CrossRef]
- Zhang, Q.; Widmer, G.; Tzipori, S. A pig model of the human gastrointestinal tract. Gut Microbes 2013, 4, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, H.; Zhao, X.Q.; Knudsen, K.E.B.; Eggum, B.O. The influence of dietary fibre source and level on the development of the gastrointestinal tract, digestibility and energy metabolism in broiler chickens. Br. J. Nutr. 1996, 75, 379–395. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.Y. Functional amino acids in frowth, reproduction, and health. Adv. Nutr. 2010, 1, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Magistretti, P.J.; Pellerin, L.; Rothman, D.L.; Shulman, R.G. Neuroscience—Energy on demand. Science 1999, 283, 496–497. [Google Scholar] [CrossRef]
- Davila, A.M.; Blachier, F.; Gotteland, M.; Andriamihaja, M.; Benetti, P.H.; Sanz, Y.; Tome, D. Intestinal luminal nitrogen metabolism: Role of the gut microbiota and consequences for the host. Pharmacol. Res. 2013, 68, 95–107. [Google Scholar] [CrossRef]
- Holmes, E.; Li, J.V.; Athanasiou, T.; Ashrafian, H.; Nicholson, J.K. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol. 2011, 19, 349–359. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Yu, K.; Yu, M.; Zhang, C.; Su, Y.; Zhu, W. Alteration of metabolomic markers of amino-acid metabolism in piglets with in-feed antibiotics. Amino Acids 2017, 49, 771–781. [Google Scholar] [CrossRef]
- Mu, C.L.; Yang, Y.X.; Su, Y.; Zoetendal, E.G.; Zhu, W.Y. Differences in microbiota membership along the gastrointestinal tract of piglets and their differential alterations following an early-life antibiotic intervention. Front. Microbiol. 2017, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Lackey, D.E.; Lynch, C.J.; Olson, K.C.; Mostaedi, R.; Ali, M.; Smith, W.H.; Karpe, F.; Humphreys, S.; Bedinger, D.H.; Dunn, T.N.; et al. Regulation of adipose branched-chain amino acid catabolism enzyme expression and cross-adipose amino acid flux in human obesity. Am. J. Physiol.-Endocrinol. Metab. 2013, 304, E1175–E1187. [Google Scholar] [CrossRef] [PubMed]
- Ormerod, K.L.; Wood, D.L.A.; Lachner, N.; Gellatly, S.L.; Daly, J.N.; Parsons, J.D.; Dal’Molin, C.G.O.; Palfreyman, R.W.; Nielsen, L.K.; Cooper, M.A.; et al. Genomic characterization of the uncultured Bacteroidales family247 S–S inhabiting the guts of homeothermic animals. Microbiome 2016, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Nobel, Y.R.; Cox, L.M.; Kirigin, F.F.; Bokulich, N.A.; Yamanishi, S.; Teitler, I.; Chung, J.; Sohn, J.; Barber, C.M.; Goldfarb, D.S.; et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.J.; Yu, M.; Yang, Y.X.; Mu, C.L.; Su, Y.; Zhu, W.Y. Effect of early antibiotic administration on cecal bacterial communities and their metabolic profiles in pigs fed diets with different protein levels. Anaerobe 2016, 42, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, C.; Yang, Y.; Mu, C.; Su, Y.; Yu, K.; Zhu, W. Long-term effects of early antibiotic intervention on blood parameters, apparent nutrient digestibility, and fecal microbial fermentation profile in pigs with different dietary protein levels. J. Anim. Sci. Biotechnol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Bian, G.; Ma, S.; Zhu, Z.; Su, Y.; Zoetendal, E.G.; Mackie, R.; Liu, J.; Mu, C.; Huang, R.; Smidt, H.; et al. Age, introduction of solid feed and weaning are more important determinants of gut bacterial succession in piglets than breed and nursing mother as revealed by a reciprocal cross-fostering model. Environ. Microbiol. 2016, 18, 1566–1577. [Google Scholar] [CrossRef]
- Hamilton, M.J.; Weingarden, A.R.; Sadowsky, M.J.; Khoruts, A. Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am. J. Gastroenterol. 2012, 107, 761–767. [Google Scholar] [CrossRef]
- Sun, Y.; Zhou, L.P.; Fang, L.D.; Su, Y.; Zhu, W.Y. Responses in colonic microbial community and gene expression of pigs to a long-term high resistant starch diet. Front. Microbiol. 2015, 6, 10. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5 ‘-nuclease assays. Appl. Environ. Microbiol. 2000, 66, 4605–4614. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yu, K.F.; Mu, C.L.; Hang, S.Q.; Che, L.Q.; Zhu, W.Y. Progressive response of large intestinal bacterial community and fermentation to the stepwise decrease of dietary crude protein level in growing pigs. Appl. Microbiol. Biotechnol. 2017, 101, 5415–5426. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.Y.; Huo, W.J.; Zhu, W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Jiye, A.; Wang, G.; Hao, H.; Huang, Q.; Yan, B.; Zha, W.; Gu, S.; Ren, H.; Zhang, Y.; et al. Gas chromatography/time-of-flight mass spectrometry based metabonomic approach to differentiating hypertension- and age-related metabolic variation in spontaneously hypertensive rats. Rapid Commun. Mass Spectrom. 2008, 22, 2882–2888. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.G.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
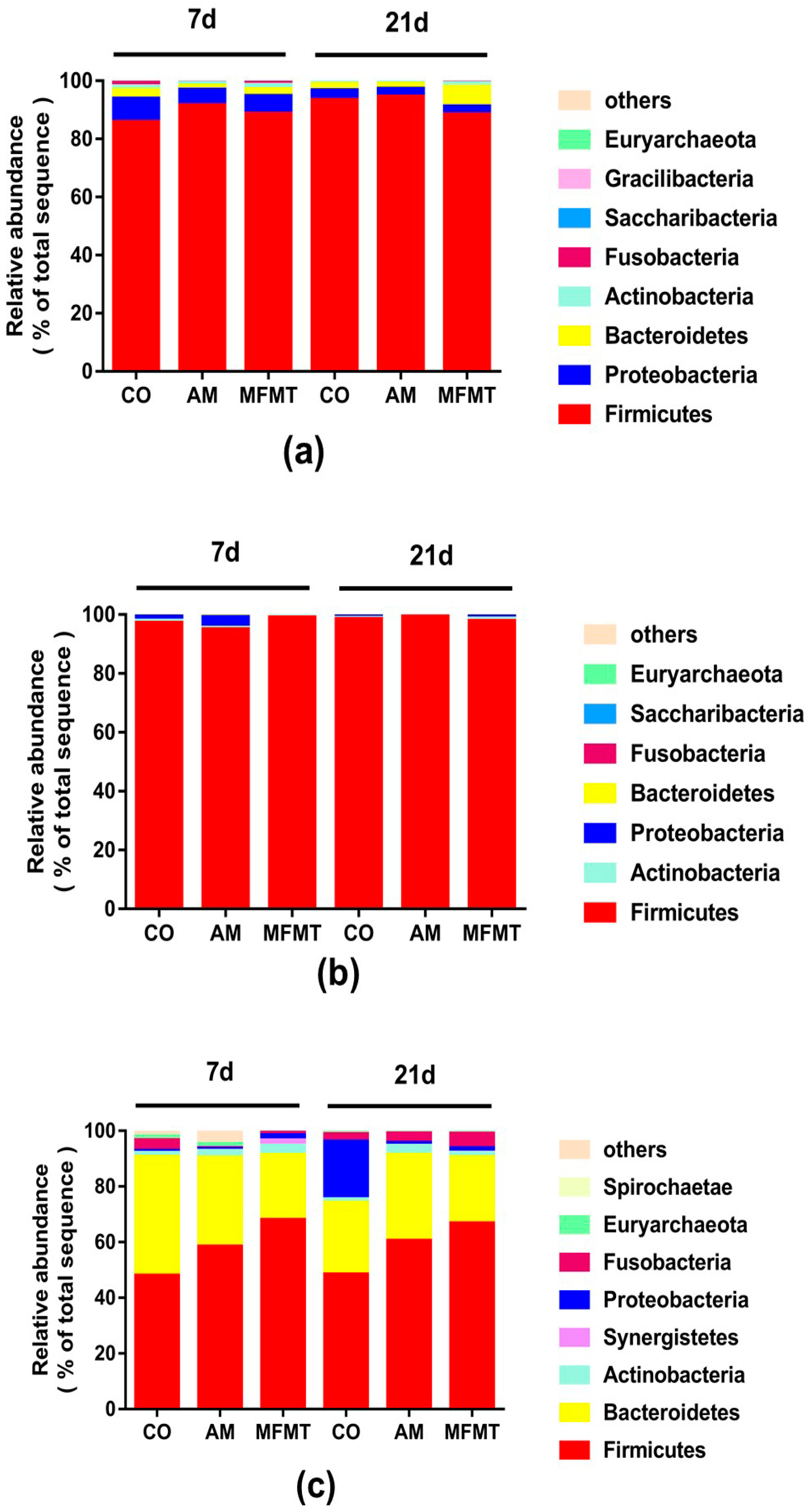
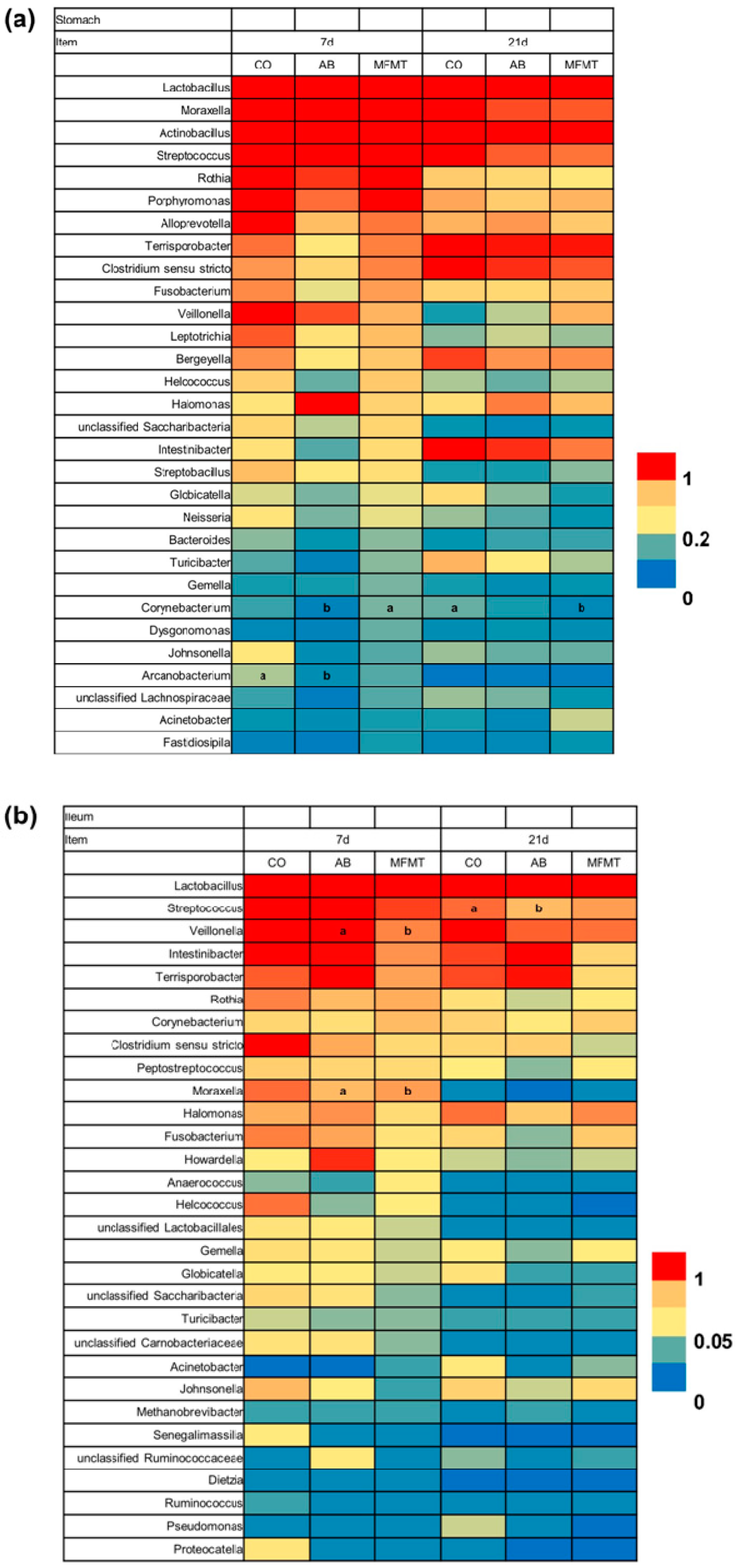
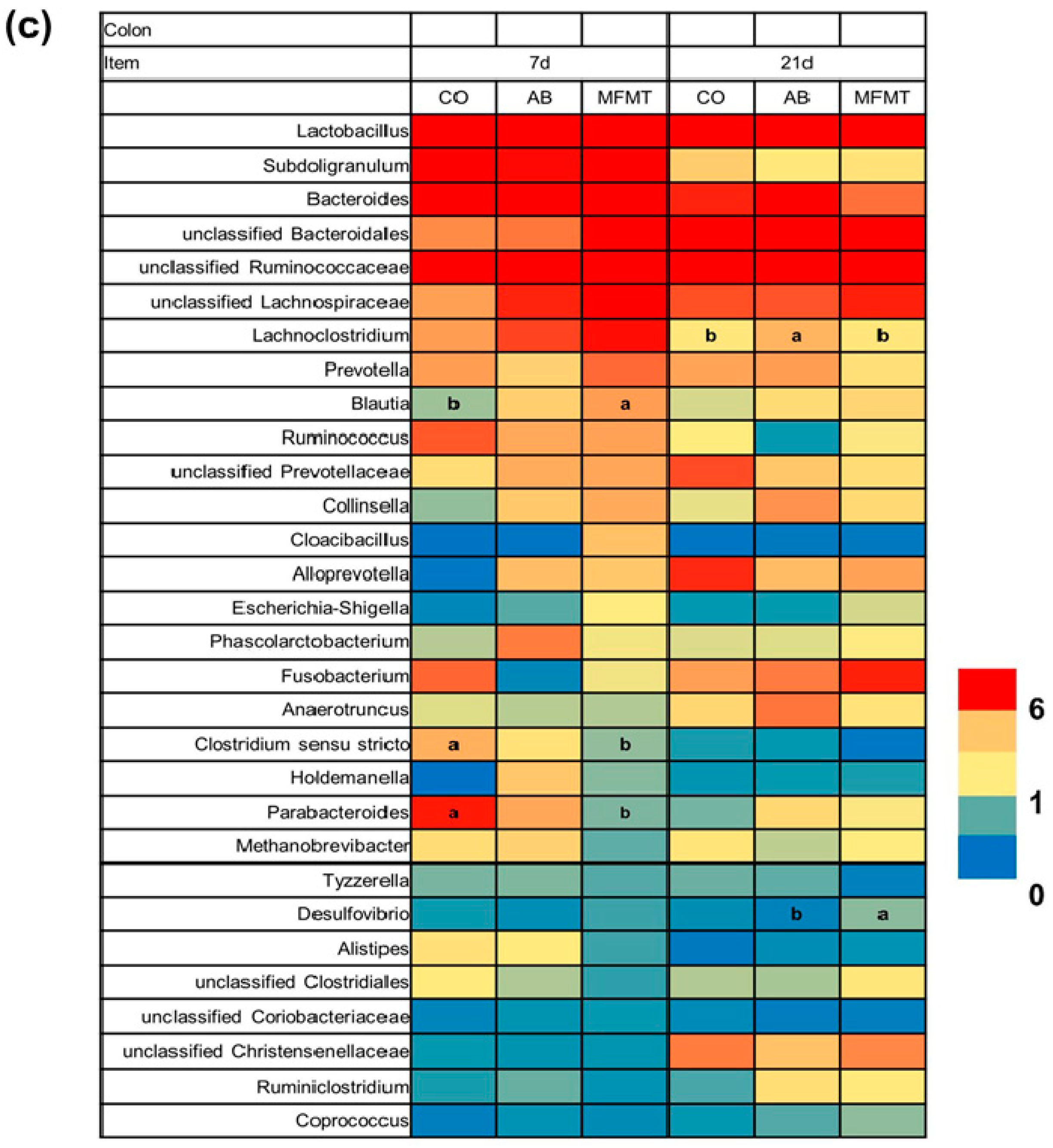
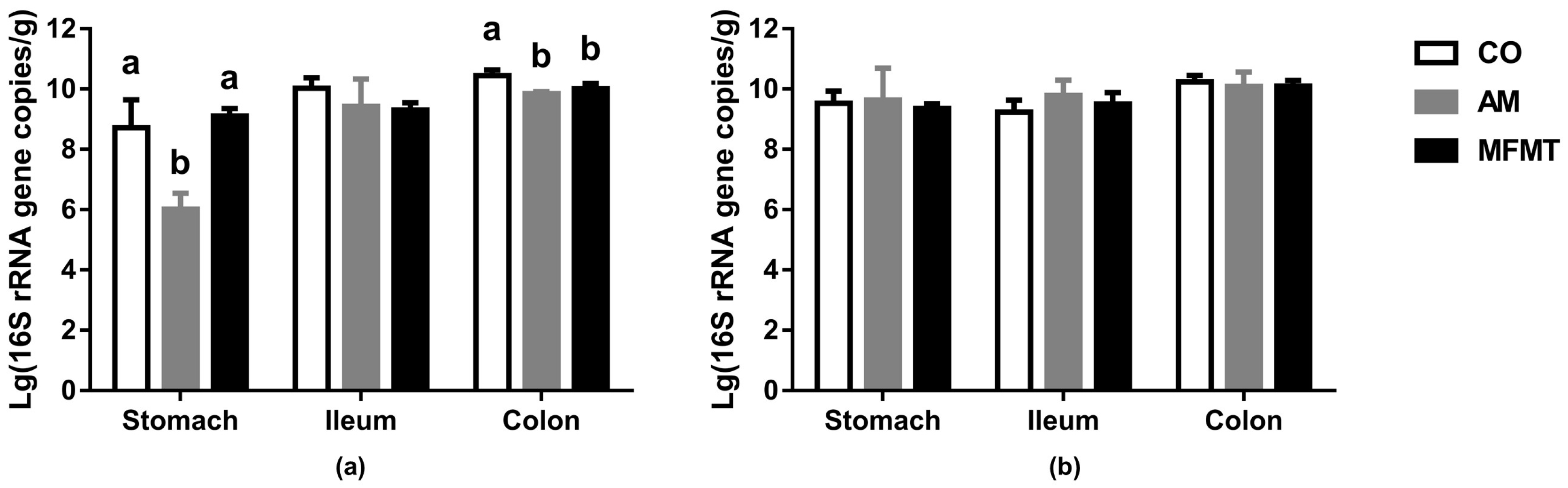
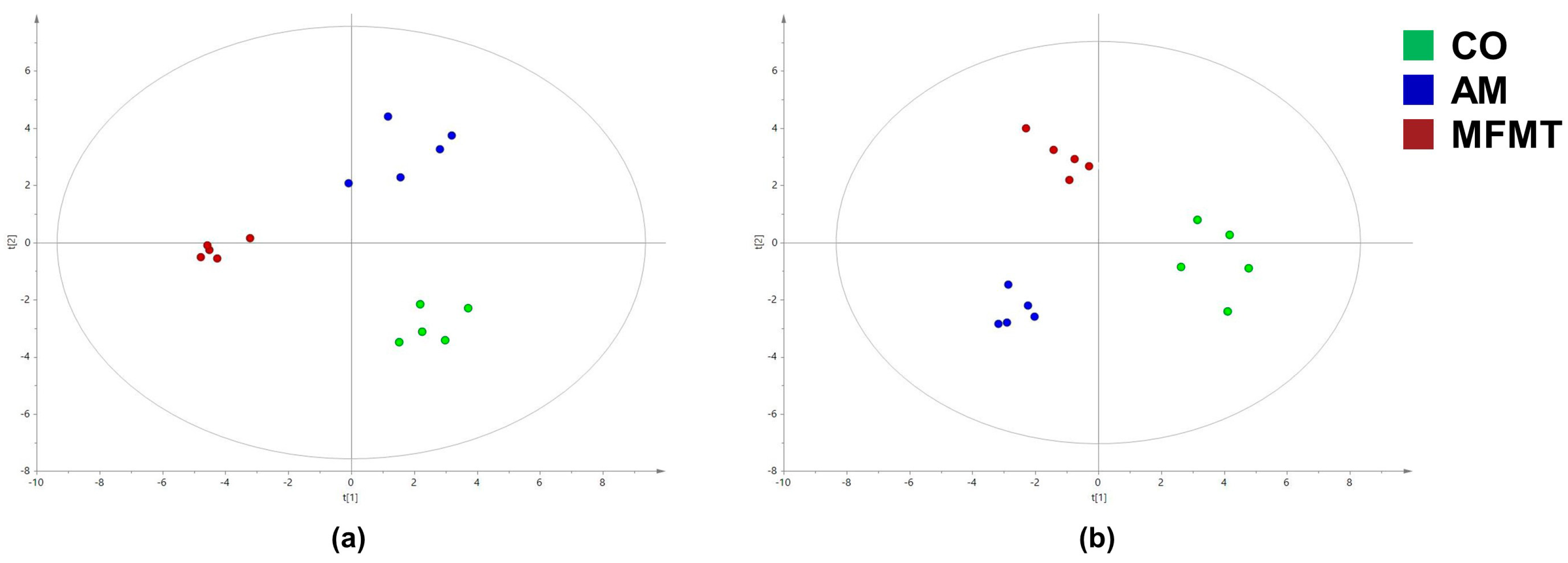
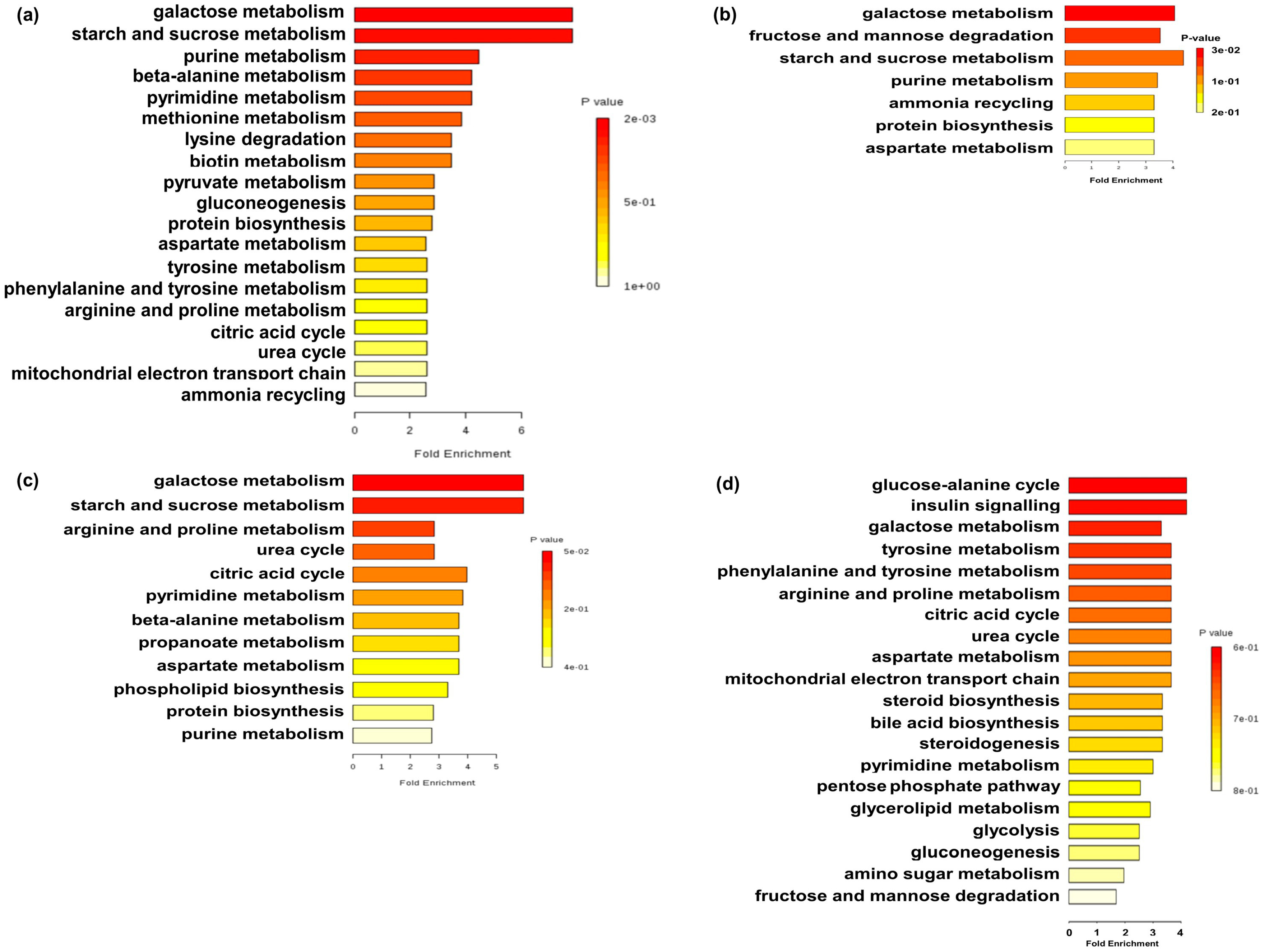
| Item | 7 Days | 21 Days | ||||
|---|---|---|---|---|---|---|
| CO | AM | MFMT | CO | AM | MFMT | |
| Stomach | ||||||
| ACE | 312.12 ± 28.82 b | 259.54 ± 16.80 b | 412.82 ± 30.39 a | 428.91 ± 26.10 | 413.41 ± 47.68 | 344.95 ± 34.48 |
| Chao | 287.15 ± 24.33 | 241.77 ± 17.79 b | 352.51 ± 30.25 a | 410.98 ± 23.96 | 390.91 ± 28.42 | 339.19 ± 42.13 |
| Shannon | 2.38 ± 0.35 | 1.93 ± 0.18 | 2.36 ± 0.06 | 2.21 ± 0.22 | 1.92 ± 0.04 | 2.33 ± 0.10 |
| Simpson | 0.21 ± 0.05 | 0.28 ± 0.04 | 0.22 ± 0.02 | 0.23 ± 0.03 | 0.31 ± 0.03 a | 0.21 ± 0.02 b |
| Ileum | ||||||
| ACE | 226.27 ± 39.13 | 240.86 ± 24.35 | 247.66 ± 30.30 | 255.18 ± 10.87 | 222.05 ± 26.10 | 234.52 ± 29.94 |
| Chao | 194.19 ± 28.53 | 218.32 ± 27.09 | 201.50 ± 14.65 | 241.54 ± 11.05 | 200.68 ± 25.40 | 214.92 ± 30.62 |
| Shannon | 2.19 ± 0.25 | 2.28 ± 0.26 | 2.10 ± 0.07 | 1.92 ± 0.05 | 1.80 ± 0.07 | 2.06 ± 0.18 |
| Simpson | 0.20 ± 0.04 | 0.20 ± 0.05 | 0.21 ± 0.01 | 0.28 ± 0.02 | 0.28 ± 0.03 | 0.25 ± 0.06 |
| Colon | ||||||
| ACE | 363.86 ± 27.30 | 349.31 ± 32.34 | 372.66 ± 27.10 | 385.5 ± 88.51 | 545.61 ± 28.07 | 466.36 ± 22.04 |
| Chao | 348.04 ± 18.91 | 341.52 ± 29.45 | 379.41 ± 32.03 | 385.23 ± 88.94 b | 563.07 ± 33.34 a | 472.59 ± 20.42 |
| Shannon | 3.58 ± 0.07 | 3.63 ± 0.13 | 3.38 ± 0.18 | 3.20 ± 0.72 | 4.03 ± 0.13 | 3.88 ± 0.19 |
| Simpson | 0.06 ± 0.01 | 0.05 ± 0.01 | 0.09 ± 0.02 | 0.21 ± 0.16 | 0.04 ± 0.01 | 0.06 ± 0.02 |
| Item | Metabolite | Biological Roles | Metabolic Subpathway | FC 1 | p2 | VIP 3 | FDR 4 |
|---|---|---|---|---|---|---|---|
| Day 7 | |||||||
| MFMT vs. CO | |||||||
| Carbohydrates | Sucrose | Disaccharides | Galactose metabolism | 0.38 | 0.008 | 2.44 | 0.001 |
| Others | 1-Monohexadecanoylglycerol | Others | Others | 0.72 | 0.032 | 1.84 | 0.005 |
| Amino acids | Asparagine | Amino acids | Alanine, aspartate, and glutamate metabolism | 4.15 | 0.032 | 1.05 | 0.007 |
| Alkaloids | Hypoxanthine | Purine alkaloids | Purine metabolism | 2.36 | 0.032 | 1.64 | 0.009 |
| Amino acids | Lysine | Amino acids | Lysine metabolism | 0.58 | 0.032 | 1.84 | 0.011 |
| Others | 1,3-Di-tert-butylbenzene | Others | Others | 0.67 | 0.056 | 1.81 | 0.024 |
| Lipids | Eicosanoic acid | Saturated fatty acids | Biosynthesis of unsaturated fatty acids | 0.50 | 0.056 | 1.77 | 0.028 |
| Lipids | Heptanoic acid | Straight chain fatty acids | Others | 0.66 | 0.056 | 2.06 | 0.032 |
| Nucleic acids | Uracil | Pyrimidines | Pantothenate and CoA biosynthesis | 1.77 | 0.056 | 1.94 | 0.036 |
| Others | 2,4,6-Tri-tert-butylbenzenethiol | Others | Others | 0.74 | 0.095 | 1.66 | 0.068 |
| Carboxylic acid | Fumaric acid | Others | TCA cycle | 1.44 | 0.095 | 1.42 | 0.075 |
| Organic acids | Lactic acid | Hydroxycarboxylic acids | Glycolysis/gluconeogenesis | 4.92 | 0.095 | 1.04 | 0.081 |
| Alkaloids | Pipecolic acid | Piperidine alkaloids | Lysine degradation | 0.31 | 0.095 | 1.45 | 0.088 |
| Peptides | Putrescine | Amines | Arginine and proline metabolism | 0.64 | 0.095 | 1.80 | 0.095 |
| AM vs. CO | |||||||
| Carbohydrates | Sucrose | Disaccharides | Galactose metabolism | 0.57 | 0.008 | 2.49 | 0.001 |
| Carbohydrates | Glucaric acid | Carbohydrates | Ascorbate and aldarate metabolism | 2.35 | 0.056 | 1.79 | 0.016 |
| Organic acids | Oxalic acid | Dicarboxylic acids | Glyoxylate and dicarboxylate metabolism | 0.72 | 0.056 | 1.99 | 0.024 |
| Carbohydrates | Sorbitol | Sugar alcohols | Polyol metabolism | 1.90 | 0.056 | 2.11 | 0.032 |
| Amino acids | Asparagine | Amino acids | Alanine, aspartate, and glutamate metabolism | 1.87 | 0.095 | 2.01 | 0.068 |
| Carbohydrate | Fructose | Ketoses | Fructose and mannose degradation | 1.78 | 0.095 | 1.73 | 0.081 |
| Alkaloids | Hypoxanthine | Purine alkaloids | Purine metabolism | 1.71 | 0.095 | 1.72 | 0.095 |
| Day 21 | |||||||
| MFMT vs. CO | |||||||
| Carbohydrates | Sucrose | Disaccharides | Galactose metabolism | 0.56 | 0.008 | 2.33 | 0.001 |
| Nucleic acids | Uridine | Nucleosides | Pyrimidine metabolism | 1.38 | 0.008 | 2.38 | 0.002 |
| Others | 2-Hydroxyglutaric acid | Others | Others | 2.77 | 0.016 | 1.80 | 0.005 |
| Organic acids | Citric acid | Tricarboxylic acid | TCA cycle | 0.34 | 0.032 | 1.72 | 0.013 |
| Peptides | Ethanolamine | Biogenic amines | Glycerophospholipid metabolism | 2.28 | 0.056 | 1.55 | 0.028 |
| Amino acids | Arginine | Amino acids | Arginine biosynthesis | 1.54 | 0.095 | 1.68 | 0.057 |
| Amino acids | Beta-alanine | Amino acids | Beta-alanine metabolism | 2.48 | 0.095 | 1.86 | 0.067 |
| Nucleic acids | Guanine | Purines | Purine metabolism | 1.54 | 0.095 | 1.77 | 0.076 |
| Amino acids | Ornithine | Other amino acids | Arginine biosynthesis | 0.53 | 0.095 | 1.73 | 0.086 |
| Amino acids | Urea | Amino acids metabolism relatives | Ornithine cycle | 1.29 | 0.095 | 1.74 | 0.095 |
| AM vs. CO | |||||||
| Others | Sorbitol-6-phosphate | Others | Fructose and mannose metabolism | 1.83 | 0.008 | 2.29 | 0.001 |
| Nucleic acids | Uridine | Nucleosides | Pyrimidine metabolism | 1.71 | 0.032 | 1.38 | 0.006 |
| Steroids | Cholesterol | Cholestane derivatives | Steroid biosynthesis | 0.47 | 0.056 | 1.53 | 0.015 |
| Carboxylic acid | Fumaric acid | Others | TCA cycle | 1.65 | 0.056 | 1.87 | 0.020 |
| Carbohydrates | Glucose | Aldoses | Glycolysis/gluconeogenesis | 1.78 | 0.056 | 1.94 | 0.025 |
| Carbohydrates | Glycerol | Sugar alcohols | Glycerolipid metabolism | 0.39 | 0.056 | 1.40 | 0.031 |
| Carbohydrate | Ribose | Aldoses | Pentose phosphate pathway | 2.08 | 0.056 | 1.91 | 0.036 |
| Carbohydrates | Xylose | Aldoses | Pentose and glucuronate interconversions | 1.57 | 0.056 | 1.58 | 0.041 |
| Carbohydrates | Fructose-6-phosphate | Ketose | Glycolysis/gluconeogenesis | 2.01 | 0.095 | 1.78 | 0.078 |
| Others | Galactose-6-phosphate | Others | Others | 1.23 | 0.095 | 1.01 | 0.086 |
| Others | Mannose-6-phosphate | Others | Fructose and mannose metabolism | 1.51 | 0.095 | 1.36 | 0.095 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, C.; Wan, J.; Su, Y.; Zhu, W. Effects of Early Intervention with Maternal Fecal Microbiota and Antibiotics on the Gut Microbiota and Metabolite Profiles of Piglets. Metabolites 2018, 8, 89. https://doi.org/10.3390/metabo8040089
Lin C, Wan J, Su Y, Zhu W. Effects of Early Intervention with Maternal Fecal Microbiota and Antibiotics on the Gut Microbiota and Metabolite Profiles of Piglets. Metabolites. 2018; 8(4):89. https://doi.org/10.3390/metabo8040089
Chicago/Turabian StyleLin, Chunhui, Jiajia Wan, Yong Su, and Weiyun Zhu. 2018. "Effects of Early Intervention with Maternal Fecal Microbiota and Antibiotics on the Gut Microbiota and Metabolite Profiles of Piglets" Metabolites 8, no. 4: 89. https://doi.org/10.3390/metabo8040089
APA StyleLin, C., Wan, J., Su, Y., & Zhu, W. (2018). Effects of Early Intervention with Maternal Fecal Microbiota and Antibiotics on the Gut Microbiota and Metabolite Profiles of Piglets. Metabolites, 8(4), 89. https://doi.org/10.3390/metabo8040089





