Abstract
Part review, part perspective, this article examines the applications and potential of in-vivo Nuclear Magnetic Resonance (NMR) for understanding environmental toxicity. In-vivo NMR can be applied in high field NMR spectrometers using either magic angle spinning based approaches, or flow systems. Solution-state NMR in combination with a flow system provides a low stress approach to monitor dissolved metabolites, while magic angle spinning NMR allows the detection of all components (solutions, gels and solids), albeit with additional stress caused by the rapid sample spinning. With in-vivo NMR it is possible to use the same organisms for control and exposure studies (controls are the same organisms prior to exposure inside the NMR). As such individual variability can be reduced while continual data collection over time provides the temporal resolution required to discern complex interconnected response pathways. When multidimensional NMR is combined with isotopic labelling, a wide range of metabolites can be identified in-vivo providing a unique window into the living metabolome that is highly complementary to more traditional metabolomics studies employing extracts, tissues, or biofluids.
1. Introduction
Anthropogenic activity since the beginning of the industrial revolution has had increasingly negative consequences on the environment, many of which are just beginning to be discovered, with some of the largest impacts on water, soil, and air quality [1]. Environmental contamination is a source of growing concern, with wide impacts and implications for both environmental and human health [2]. This manuscript is part review and part perspective with a focus on environmental toxicity that will provide an insight into current in-vivo Nuclear Magnetic Resonance (NMR) metabolomics approaches, as well as a discussion of the future potential.
1.1. Common Sources of Contamination
1.1.1. Water Contamination
Clean water is a rapidly decreasing resource that threatens 1.1 billion people around the globe lacking sufficient access [3]. There are many sources of water contamination that are introduced through run-off from agriculture, disposal of personal care products, and industrial processes including heavy metal contamination [1]. Over the last 15 years, pharmaceuticals have been receiving increased attention as emerging contaminants in water bodies, as they have potential negative impacts on water quality and aquatic ecosystems due to the lack of regulation [4,5]. Pharmaceuticals persist in the environment in part due to their incomplete elimination through sewage treatment [5]. Current methods leave 60–90% of pharmaceuticals remaining in the water after treatment [4]. The concentrations and identity of many of these pharmaceuticals are just being discovered and even less is known about the impacts of these xenobiotics (and their transformation products) on the aquatic ecosystem, and human health [5]. Water pollution has become a major threat to ecosystem health; thus, further research into identifying which contaminants pose the largest threat, and their toxic mode of action, is needed to determine effects on individual organisms and populations [6,7].
1.1.2. Soil Contamination
In addition to the aquatic pollution, soil is also a potential sink for contaminants. Often this is related to heavy metals from industry and mining, but can also include a range of organic xenobiotics such as agrochemicals, pharmaceuticals, and surfactants. These contaminants bind to the soil and often bioaccumulate in plants [8]. Therefore, it is important to study plants to obtain information on environmental contaminant impacts, which can potentially serve as an indicator, and even predictor prior to larger scale ecosystem shifts [9,10,11,12].
1.1.3. Air and Atmospheric Pollution
Approximately 95% of the Earth’s atmosphere is in an 8–12 km range surrounding the earth, known as the troposphere [13]. The troposphere represents a delicate chemical balance that can be easily disrupted. One prominent example being the release of chlorofluorocarbons (CFCs) that can persist in the environment leading to the continual depletion of ozone [14,15]. Ozone is critically important as it represents a key source of hydroxyl radicals, which react rapidly with most air pollutants [16]. In addition, increased levels of acid rain can change the transport potential and transformation of contaminants with widespread implications for ecosystems and human respiratory health [17,18,19].
1.2. A Bottom-Up Approach
A traditional top-down approach which involves identifying all contaminants, their degradation products, and then assessing toxicities on an individual compound basis is extremely challenging given the complexity and dynamics of our environment. Conversely, a bottom-up approach focuses on the organisms themselves, and asks questions such as; is a population stressed?; what are the stressor/stressors causing the stress?; and which biochemical pathways are impacted by the stressor/stressors?
As toxic impacts manifest more rapidly in the metabolic profile, compared to the genome or proteome, the metabolome represents a key biological indictor of stress [20]. NMR spectroscopy due to its high resolution, ability to identify molecules de-novo, and non-destructive nature represents an ideal detector for metabolic profiles in-vivo.
Metabolomics
Metabolomics is the study of the biochemical changes occurring within an organism often in response to exposure to external stressors [21,22]. Although the first examples of metabolite profiling appeared in the literature in the 1950s [23], metabolomics is a relatively new field that has been increasing in popularity in recent years and has found applications across a number of disciplines including human and animal health [24], drug discovery [25], ecology [26], food chemistry [27], microbiology [23], and environmental monitoring [28,29]. Historically, metabolomics has been used as a method of examining the effects of drugs in the medical field [23]. However, since 2001 there has been an interest in environmental studies as a method to detect and explain toxicity [23,30]. Environmental metabolomics studies changes in the metabolite profile with changing environmental conditions [30], and have been used on a host of different species including: worms [31,32,33,34,35,36,37,38], Drosophila melanogaster [39], Daphnia magna [20,40,41,42,43,44,45], Hyalella azteca [46,47,48], Caenorhabditis elegans [49,50,51], rodents [52,53,54,55], and plants [56,57,58,59,60]. Species are often chosen either due to their susceptibility to external stressors or their abundance geographically. For example, earthworms are useful organisms as they will absorb chemicals both through their skin and via soil ingestion, making it possible to examine if different physical uptake routes alter the impact of contaminants [37,61,62,63,64]. Many metabolomics studies are conducted in-vitro which permits sensitive measurements but often involves sacrificing organisms or using extracted cells [65]. As such these methods provide a snapshot of the effects on the species, but lack the temporal resolution to resolve complex interconnected biochemical response pathways. Such information is critical as the biochemical pathways impacted, and their interconnectivities, describe how chemicals are toxic (toxic mode of action) and provide an insight into how the organism responds to the stressor (bioconversion, excretion, disease, adaptation, etc.) [66]. Due to the need to better understand toxic modes of action, there has been renewed interest in metabolomics studies to examine the effects of environmental contaminants on species in-vivo. For the purposes of this article in-vivo studies are defined as experiments that involve studies on the whole living organism rather than a sub component of, or extraction from, an organism. Methods have recently been developed to keep organisms alive during metabolomics experiments, allowing for longer testing, as well as long-term effects to be studied [67]. Many of these in-vivo studies examine Daphnia magna as a model organism for aquatic toxicity testing. D. magna have been used as model organisms since the 1960s due to their ability to survive in a wide range of habitats, ease of maintenance to culture, and having a relatively short life cycle (~40 days) [68]. These species are considered a keystone species in the environment as they are a primary consumer of phytoplankton and a key food source for secondary consumers thus representing a critical link in many ecosystems [69,70,71]. They cannot produce essential lipids themselves and instead assimilate them from their diet (i.e., algae), which in turn are utilized by higher order predators. In-vivo metabolomics approaches represent a powerful tool to study these transfers from algae and assimilation into D. magna and how external stressors impact their biochemical function [72,73]. The potential of in-vivo NMR to study this, and processes in other species will be discussed in more depth in the “Why in-vivo NMR?” section.
1.3. Toxicity Today
Present environmental policies are set primarily based on acute toxicity of individual species monitored mainly through death and reproduction. The Environmental Protection Agency has created standardized protocols to determine the toxicity of various contaminants [74]. These protocols are created by determining the Lethal Concentration 50% (LC50), which is the concentration of the contaminant at which 50% of the organisms die, or the Lethal Dose 50% (LD50) which is a single dose that kills 50% of all the organisms [75]. These methods are an excellent first defense to identify acutely toxic chemicals quickly, but results are often variable between species and there is no information regarding the metabolic impacts, sub-lethal impacts, or bioaccumulation [65,76].
While these methods have provided key “front-line” information on acute toxicity, the ever-increasing number of contaminants and complex mixtures at sub-lethal levels require the development of complimentary methods. A 2007 report by the National Academy of Sciences proposed numerous changes, with a large emphasis on examining the effects from a biochemical perspective. They argued this entails examining the mode of action, bioaccumulation, and biochemical impacts to the metabolome of the organisms, not only over 48 h, but also over longer periods, and after exposure to examine recovery times [65,76]. NMR spectroscopy due to its non-invasive nature is ideally suited to provide in-depth metabolic information in-vivo to better understand the sub-lethal toxicity of individual chemicals and mixtures, providing information on the mode of action, bioaccumulation, biotransformation, molecular reactivity, excretion, binding and bioavailability of chemicals [67].
1.4. NMR Spectroscopy as an Environmental Tool
Nuclear Magnetic Resonance (NMR) Spectroscopy is one of the most powerful analytical tools in modern research, providing unprecedented levels of molecular information on chemical structure and inter/intra molecular interactions [67], with minimum sample preparation. NMR targets magnetically susceptible nuclei such as 1H, 31P, 15N, and 13C, and after excitation using radio frequencies inside a magnetic field, every unique chemical environment within a molecule gives rise to a signal [77]. 1H NMR has a high natural abundance and one of the highest gyromagnetic ratios (ratio of its magnetic moment to its angular momentum) resulting in high sensitivity. As such, 1-dimensional (1D) proton NMR is the most commonly used nuclei for metabolomics studies [78]. However, 2D NMR examining the correlation between 1H-1H and 1H-13C offers additional spectral dispersion for molecular fingerprinting and connectivity information for molecular assignment [79]. NMR is a tool that can be used with varying amounts of sample (even down to sub-nL eggs) [80,81,82], is a non-destructive technique, and is highly reproducible across samples and labs [83,84,85,86]. NMR is also fully quantitative and when applied appropriately, each nuclei gives the same response in the spectrum leading to accurate quantitation of unknowns without the need for internal standards [87,88,89,90,91,92,93,94]. This makes the technique attractive for many metabolomics studies. Similarly, NMR is excellent for kinetic studies to measure chemical reaction rates and biological processes [95,96].
NMR is highly versatile and can be used to study solid, liquid, and gel samples. Recently, a new technique termed comprehensive multiphase (CMP) NMR was introduced which can examine all three states simultaneously. As such providing the ability to examine the interactions between phases, organization, layering, and transport across phases in close to real-time [46,66,97,98].
After the development of wide-bore superconducting magnets, NMR has become widely applied in the clinical field in the form of Magnetic Resonance Imaging (MRI) [99]. In addition to imaging, MRI can also provide localized spectroscopy within larger species, although magnetic susceptibility distortions limit the amount of metabolic information that can be extracted [100]. Readers interested on the applications of MRI in the medical field may consult these reviews [99,101]. This article will not cover the applications of in-vivo MRI but instead will focus on the use of high field NMR spectrometers to provide high resolution in-vivo metabolic information on small organisms in relation to environmental toxicity.
Due to its many advantages, NMR has huge potential in environmental research [102]. Unlike most analytical approaches, it can be applied in-vivo and represents an effective and reproducible method of determining which contaminants in the environment have deleterious effects on organisms. NMR can also serve as a powerful tool in providing in-depth information on the metabolic responses of plants [60,103,104,105,106] and animals [38,53,107,108], while providing information on the toxic mode of action of contaminants in the environment. Considering the broader picture, the subtle effects at sub-lethal levels are arguably more hazardous to animal, plant, and human populations, as they are often detected too late, and only after physical symptoms become widespread. As such the development of NMR approaches to detect and explain sub-lethal stress are of paramount importance to protecting both environmental and human health from a continuum of evolving environmental stressors.
2. Types of NMR
2.1. Solution-State NMR
Several types of NMR technology are available for the in-vivo study of environmental samples. Among which, solution-state provides the highest resolution and most comprehensive molecular information for soluble components [66]. Multiple nuclei can be studied using in-vivo solution-state NMR, with 1H and 31P being the two most commonly studied nuclei to date in an environmental context. Phosphorous is convenient as it is a spin half nucleus (i.e., produces sharp lines) and is relatively abundant (can be studied without enrichment) [109]. This nucleus is present in key bioenergetics molecules such as ATP/ADP and in DNA/RNA bases, both categories being important indicators of stress, such as changes in the energy cycle or DNA/RNA oxidation [110,111]. Proton NMR is also commonly studied due to its natural high abundance, its occurrence in most metabolites, and the availability of 1H NMR metabolic databases for assignments [24,78,112].
2.1.1. In-Vivo Applications
Early studies employed 31P NMR spectroscopy for pioneering in-vivo analysis for the characterization of embryogenesis in plaice and freshwater catfish [113,114]. NMR experiments looking at phosphorous in larger organisms, such as Atlantic Cod (~40 cm) provided important information on temperature-dependent changes in the metabolome [115]. In this later case, while 1D NMR spectra provided metabolic discrimination, an MRI system was used to accommodate the larger organisms. Early pioneering of in-vivo analysis, also employed 1H NMR, where valuable information on hypoxic stress in marine worms was obtained [116].
Key applications of solution-state in-vivo NMR experiments involve the integration of flow systems for aquatic organisms. The use of a flow system in NMR experiments can be seen as early as 1981 to study metabolite responses to cadmium in Chironomus tentans and D. magna, which demonstrated the potential of flow in-vivo NMR to study metabolism [110].
Later an improved flow design was developed which enabled higher sensitivity and was used to deliver stressors to molluscs [117,118]. The results of the exposure can be seen in Figure 1. Further applications of NMR flow systems supplied organisms with oxygen and/or food, allowing them to be kept alive indefinitely [69,119,120]. This provides a low-stress environment thereby owing any metabolic changes detected as a direct response to the stressors, rather than a result of anoxic stress and/or starvation. An example of this flow system is shown in Figure 2. Japanese medaka embryos were studied using the flow system to deliver oxygenated water prior to exposing them to pesticide-treated water [119,120]. By monitoring their metabolic responses, a significant relationship between dose and response was observed [119,120]. The metabolic changes observed in these conditions correlated to traditional toxic endpoints, such as reduced growth and heart rates, abnormal development, and post-exposure mortality [120]. However, these studies were performed on a 10 mm probe which are not commonly available in NMR facilities [121].
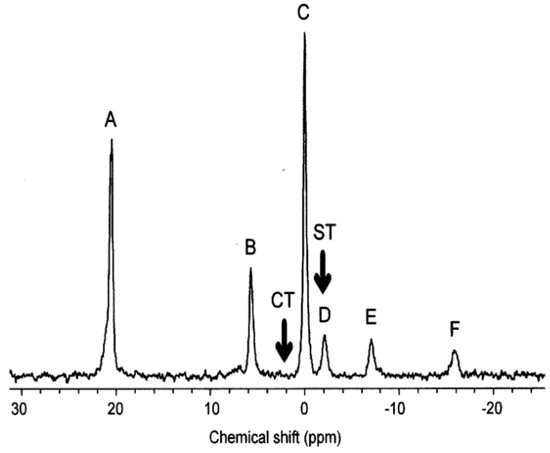
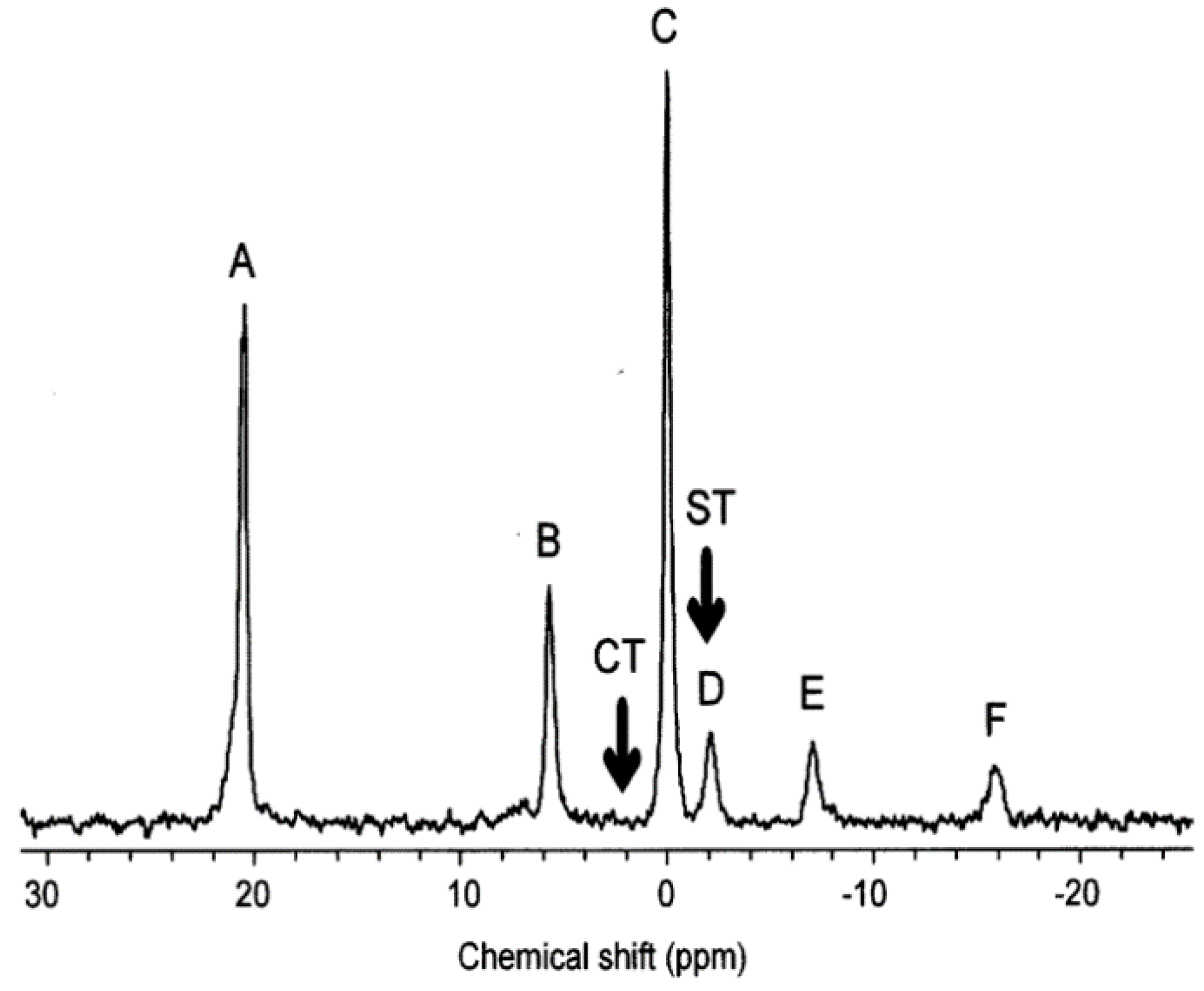
Figure 1.
An example of an early NMR exposure study. The 31P NMR spectrum of molluscs (mussels) during acute exposure to copper. The assigned peaks are: A, MDP external standard; B, inorganic phosphate; C, phosphoarginine; D, overlapping γ-ATP and β-ADP; E, overlapping α-ATP, α-ADP and NADH; and F, β-ATP [118]. Modified with permission from Elsevier.
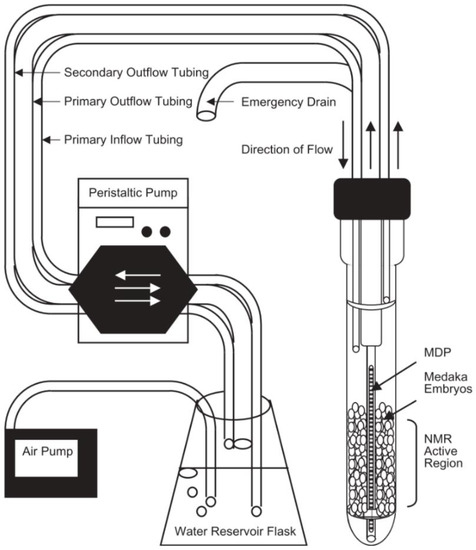
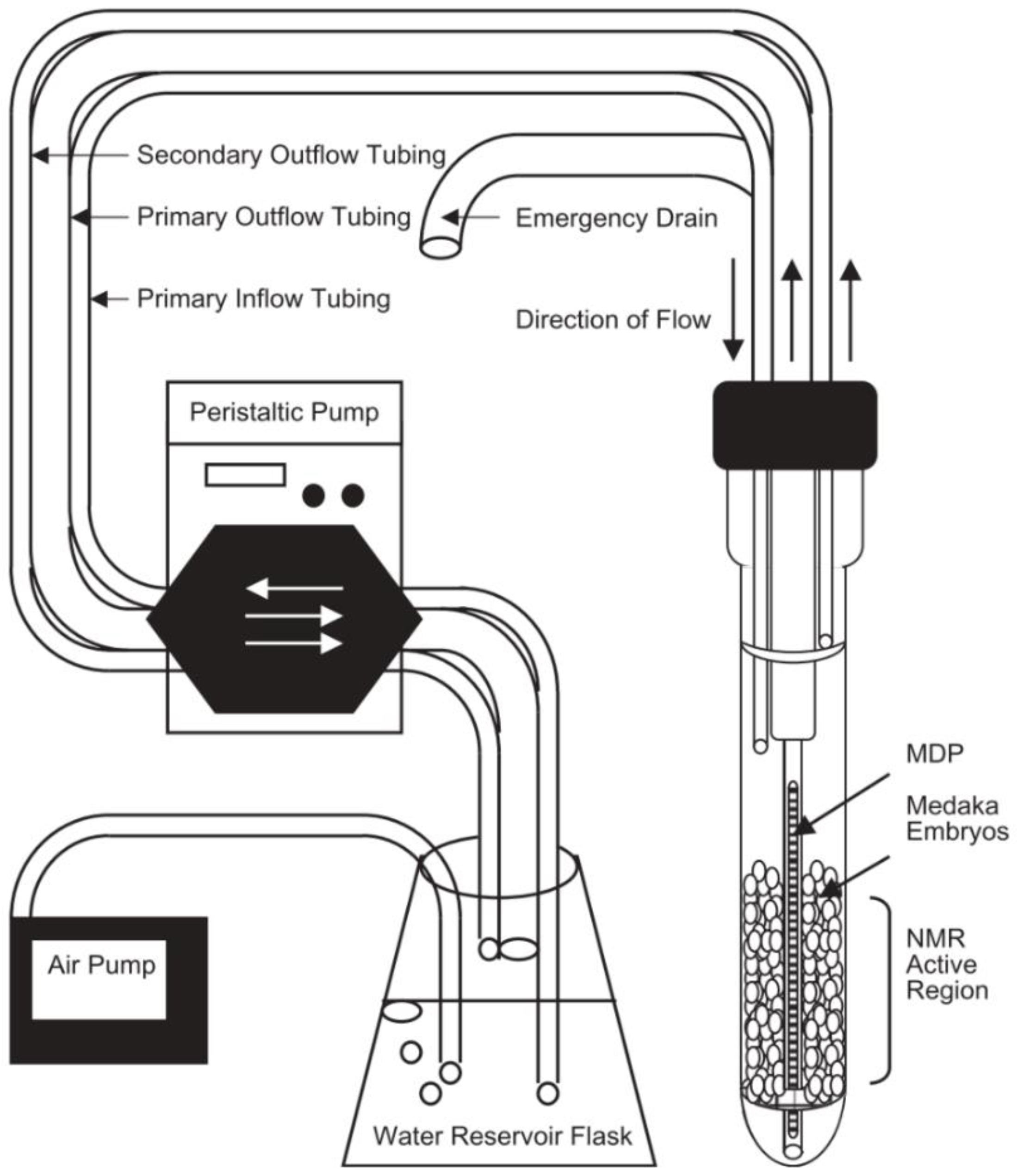
Figure 2.
Example of one of the first flow systems for solution state NMR created by Pincetich et al. to keep Medaka embryos alive during the experiment in a 10 mm probe. Reproduced with permission from Elsevier [119].
More recently Soong et al. developed a 5 mm NMR flow system that allowed for metabolic profiling of isotopically 13C labelled freely swimming small organisms through high resolution 1D 1H and 2D 1H-13C NMR [69]. In this study, 13C labelled D. magna were placed in an NMR tube where oxygenated water was continuously circulated. The results of the flow, either on or off, were compared and anoxic stress was quickly observed in the absence of water circulation. The ability to obtain 2D 1H-13C NMR correlation experiments provides considerable spectral dispersion and permits a more comprehensive assignment of the metabolome in-vivo. Due to the key role D. magna play in environmental toxicity testing, Majumdar et al. chose them as the organism of interest in a recent publication of a standardized protocol for solution-state in-vivo NMR-based metabolomics studies [121]. In addition, flow systems can be applied to plant studies explored by Roscher et al. in a detailed guide on how to perform in-vivo analysis on plant materials, including considerations for appropriate plant material, culture medium, circulation systems, and data acquisition [122].
2.1.2. Considerations
While solution-state NMR is a very useful technique and allows for NMR analysis in a low stress environment, there are a few challenges that need to be addressed. One is the low resolution obtained from 1D in-vivo NMR spectra, caused by magnetic susceptibility distortions (in simple terms the organisms body distorts the NMR magnetic field). This broadens the NMR signals causing signal overlap, and therefore, masking information from individual metabolites [94]. The simplest approach to overcome the spectral crowding is to disperse the NMR signal of interest into multiple dimensions. Novel approaches, and a more in-depth analysis will be discussed later.
Another challenge of 1H in-vivo NMR is the need to suppress the intense and broad water signal in aquatic organisms and their media. The 1H signal from water interfering with spectral resolution was once a major hurdle in solution-state in-vivo studies. Due to much of the sample being water, the signal is very broad and intense, masking important metabolite information and preventing full receiver optimization which in turn lowers sensitivity and reduces dynamics range. However, efficient water suppression methods (see later for further details) have allowed for more routine analysis using 1H in-vivo NMR.
2.2. High Resolution Magic Angle Spinning NMR
Although high resolution spectra can be acquired with solution-state NMR, information is only obtained on the truly dissolved metabolites. Species such as rigid gels and solids exhibit spectral broadening from dipolar interactions and anisotropy that make them challenging to detect without narrowing afforded by magic angle spinning. As such using solution-state NMR alone could lead to information from more rigid components such as membranes, cell walls, shells, and even bound stressors to be missed [123]. For this reason, it can be beneficial to compliment solution-state in-vivo NMR with gel-phase NMR spectroscopy, also known as high resolution-magic angle spinning (HR-MAS) NMR.
HR-MAS NMR probes allow the detection of solution and gel-like domains [124]. In HR-MAS NMR, the samples are spun at the magic angle of 54.74° to minimize the inhomogeneous broadening effects [125], while the presence of water in organisms help to reduce dipolar interactions that dominate in pure solids [126]. HR-MAS probes have a pulse field gradient, a lock channel and susceptibility matched stators. The result is that many modern solution-state NMR experiments can be implemented on HR-MAS probes and all components except true solids can be detected [127].
2.2.1. In-Vivo Applications
HR-MAS was first introduced in 1996, where researchers used a derivatized Wang resin (a linker for peptide synthesis) to demonstrate its application [124]. Since then, the approach has rapidly evolved to include in-vivo studies looking at a wide range of whole organisms, including Drosophila melanogaster, Caenorhabditis elegans, Aporrectodea caliginosa, Calanus finmarchicus, and D. magna where a range of well-resolved signals of metabolites were obtained (Figure 3) [50,128,129,130,131,132,133,134,135]. One study even applied a slow magic angle spinning approach to a whole live mouse [136]. Other studies have directed the technique towards plants leading to data on metabolic profiling during circadian cycles [137], carbon/nitrogen intracellular ratios [138], alkaloid metabolism [139], as well as assimilation of compounds and metabolite formation [140]. In-vivo samples, such as organisms and plants, can be studied with minimal to no sample preparation, since internal water acts as the solvent, allowing experiments to be done on samples in their natural state [141].
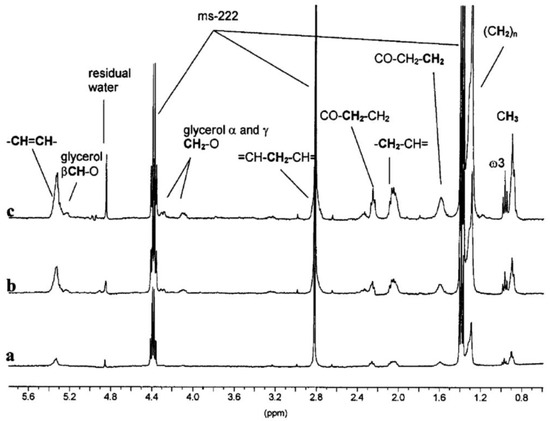
Figure 3.
Example assigned spectra obtained from in-vivo proton high resolution-magic angle spinning (HR-MAS) NMR experiments examining Daphnia magna at: (a) 24 h old, (b) 7 days old with and (c) 7 days old without eggs in the brood pouch [129]. Reproduced with permission from Royal Society of Chemistry.
2.2.2. Considerations
HR-MAS has demonstrated potential for in-vivo experiments, however, the major drawback to these studies is the unavoidable stress exerted from spinning. To determine the level of stress, Bunescu et al. tested the survival rates of D. magna following spinning under different speeds [129]. Their results showed that it was possible for the organisms to recover from certain experimental setups, especially when anesthetized. They then used these results to outline the optimal conditions for in-vivo HR-MAS NMR studies [129]. Additional improvements have been made by researchers that have focused on reducing the stress on organisms by slowing spinning through improvements of novel pulse sequences and suppression methods [94]. These results represent a promising future for in-vivo HR-MAS NMR experiments and permit studies without the use of anesthetic.
2.3. Comprehensive Multiphase NMR
In-vivo samples often encompass a range of phases, for example, in biological sections this may include liquid (blood), gel (tissue), and solid (shell/bone). It is the interactions between and the transport across these phases that ultimately give rise to the larger scale biological properties. As such it would be of great benefit to be able to study all components; liquid, gels and solids in-vivo. Although these phases can be individually studied using their respective probes (liquids, HR-MAS and solids NMR probes), very few groups have access to all three. In addition, using different probes provides data within individual phases, but lacks information on the interactions and kinetics occurring between the phases. As a solution, comprehensive multiphase (CMP) NMR spectroscopy was introduced in 2012 as a novel approach that combines a lock (for sharp lines), pulse field gradients (permits many 2D experiments and water suppression), magic angle spinning (narrows lines in gels/solids), and high power handling (required for pure solids) [127]. The technique allows for all bonds in all phases to be observed in natural, unaltered samples (see Figure 4), making it an ideal approach for materials with complex multiphase structures such as plants, air particles, sediments, and soils [56,66,97,98,142,143]. Using this approach, it has been possible to follow the penetration of contaminants into soil, as they move from solution into gel components, and finally become sequestered in the solid-phase [98]. The study was able to demonstrate the kinetic transfer between phases and identify the binding orientation and receptors in each phase, providing an in-depth insight into how and why the contaminant binds in soil. Further studies on soil have demonstrated that the approach can reveal how components organize and layer to form larger aggregate structures [97], critical information required for organisms in-vivo. Similarly, the technique applied in-vivo could be extremely important for understanding the binding and fate of contaminants and drugs [123]. In many ways it can be considered that metabolomics provides information on the rapid response of an organism to stress, whereas changes to the structural components may occur slower over time and reflect the longer-term impacts. An example, would be the altered composition in crustacean shells in polluted sites [144]. With the added ability to detect true solids, compared to HR-MAS probes, CMP-NMR probes can access all toxic impacts from the soluble metabolome through to the rigid exoskeleton, providing the potential for a complete insight into both long and short term toxic impacts.
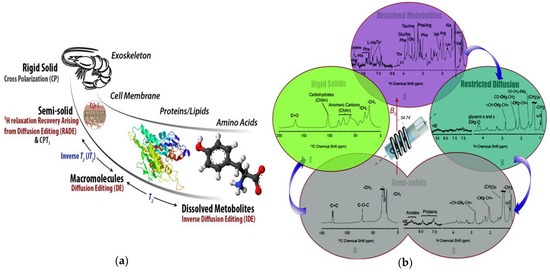
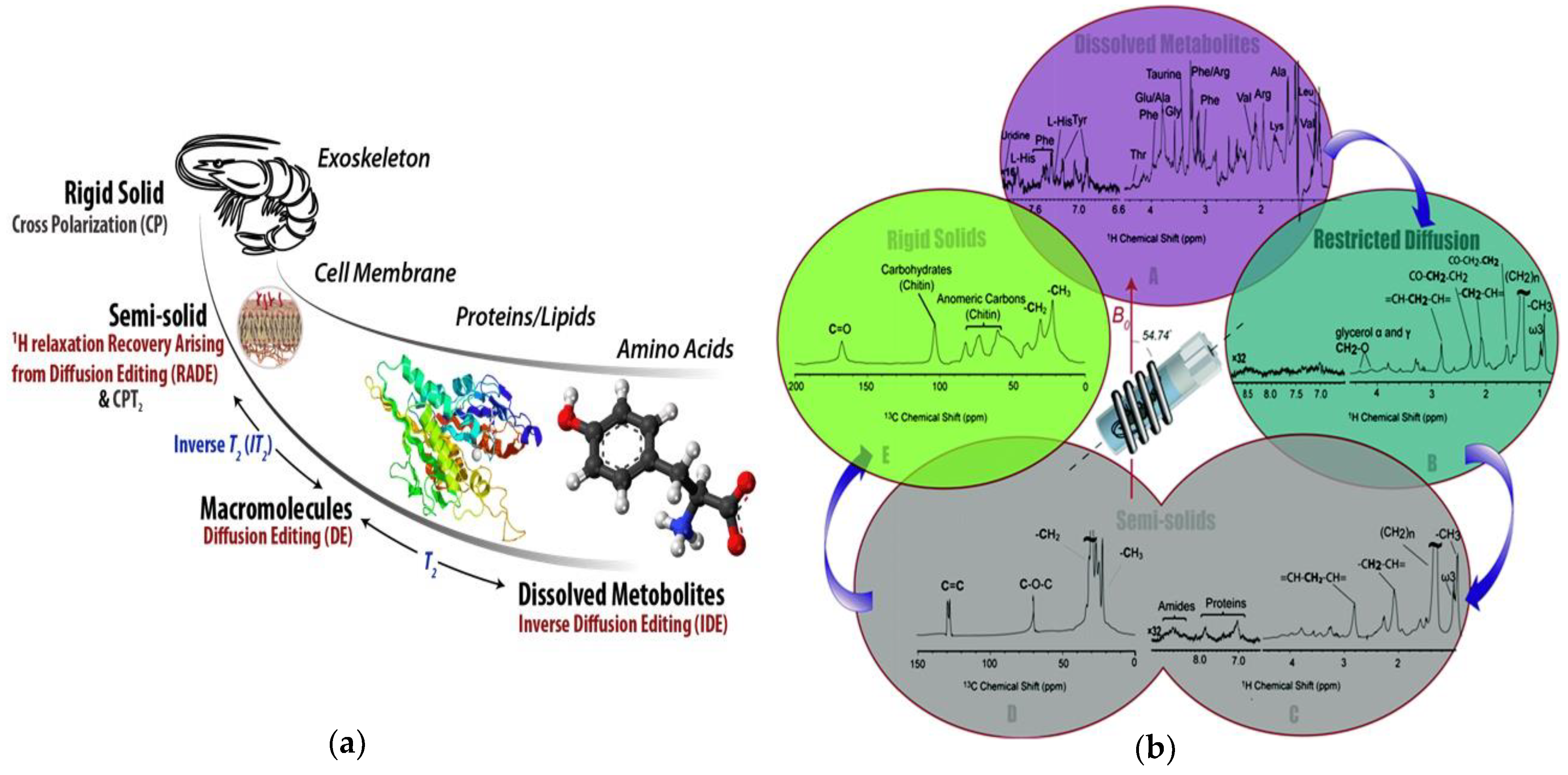
Figure 4.
(a) An example of the types of information comprehensive multiphase (CMP)-NMR can provide when applied to a living freshwater shrimp; (b) Spectra from the metabolites (most mobile) through to the shell (most rigid) could be obtained in-vivo. Reproduced from Liaghati Mobarhan et al. under the Creative Commons Attribution 3.0 Unreported License [46].
2.3.1. In-Vivo Applications
In some of its earliest applications, CMP-NMR technology has been applied to identify the complete metabolic and structural profile of intact 13C-labelled seeds [142]. This was followed by a second study which focused on the growth of the seeds and followed all the components during germination and early stages of development [145]. In 2016, CMP-NMR was applied to the living organisms H. azteca (freshwater shrimp) [46], an organism commonly used in aquatic toxicity testing, that swims and burrows into sediments, thus providing information on both water and sediment contamination [146]. CMP-NMR was able to fully differentiate the various phases, providing information on the shell and membranes, as-well as identifying a wide range of metabolites [46]. In many ways CMP-NMR can be thought of as “changing NMR technology to match the sample, rather than changing the sample to suit a specific type of NMR analysis” [66].
2.3.2. Considerations
CMP-NMR is a very powerful approach and offers a range of novel information on intact samples. The major hurdle for in-vivo analysis is the rapid spinning of the sample. In a 2016 study spinning at 2.5 KHz on H. azteca, it was noted that spinning itself causes slight changes in the metabolome, especially for the amino acid alanine [46]. More recent work has slowed the spinning to 500 Hz, which greatly reduced stress and increased survival time [48]. However, at this speed sidebands (spectral artifacts) dominate. Novel pulse sequences were presented to overcome these artifacts in 1D 1H NMR [48], but no solution has been found yet for obtaining 2D 1H-13C at lower spinning speeds (500 Hz), spectra which greatly increase spectral dispersion and aids assignment.
3. Challenges and Solutions
3.1. Water Suppression
In-vivo aquatic organisms are comprised mostly of water and are surrounded by water during experiments inside the NMR. The signal from this water needs to be suppressed for two main reasons. Firstly, the large peak can be extremely wide at the base, masking a wide range of metabolites signals [147], and secondly, and arguably more importantly, the water requires suppressing in order for the NMR receiver to be set to the maximum. The NMR receiver is somewhat analogues to a tape recorder in that if the recording sensitivity is set too high, “loud” signals, such as the intense signal from water, will lead to distortions and corrupt data. To avoid this, the receiver gain must be reduced. However, by reducing the receiver gain, the sensitivity is also reduced and low concentration signals close to the noise are lost due to the limited dynamic range of the NMR receiver. As such, in the case of in-vivo samples it is critical to reduce the water signal below that of the sample signals, permitting a maximum receiver gain and therefore allowing the in-vivo organisms to be studied without compromise.
The simplest type of water suppression is presaturation, which is easy to implement and incorporate into a range of pulse sequences [148]. Unfortunately, presaturation is ineffective in dealing will extremely large (and often) broad signals as is in the case of environmental and in-vivo samples [149]. A detailed study compared a wide range of water suppression sequences for environmental samples, concluding that a combination of shaped presaturation and W5 WATERGATE was the most effective, and in fact the only sequence that could suppress especially broad water signals [149]. The presaturation block using shaped pulses helps narrow the water signal, which is followed by two W5 blocks surrounded by gradient pairs. The W5 blocks invert all signals except the water, which is de-phased twice by the gradients. The result is excellent water suppression often below the spectrometer noise, but at the cost of loss of signal in the central window (under and adjacent to the water). The approach has been successfully applied to study a wide range of matrices including, water samples [149], Antarctic ice [150,151], soils and plant materials [147] environmental photochemistry [152], and living organisms [46,69,121,153]. In the authors’ experience SPR-W5-WATERGATE is currently the only effective solution for suppressing the extremely wide water signal encountered in living organisms and would be the recommended starting point for any group attempting in-vivo NMR.
3.2. Spectral Overlap
One of the largest challenges with in-vivo NMR is spectral broadening caused by magnetic susceptibility distortions [154,155]. Distinct parts of the organism (shell, cell walls, cell contents, membranes, etc.) all have slightly different magnetic susceptibilities’ (i.e., they all interact with the external magnetic field differently). This causes slight distortions in the magnetic field, which in turn causes spins in various parts of the sample to experience different magnetic fields and thus resonate at slightly different frequencies. The result is that the net signal from the whole sample is broadened and key splitting information is lost under a broad spectral profile. Essentially two solutions are possible to overcome the broad chemicals shift (1) spread the signals out into more than one dimension to create the dispersion required for metabolite assignments in-vivo or (2) remove the magnetic susceptibility distortions.
3.2.1. 1H-13C Multidimensional NMR
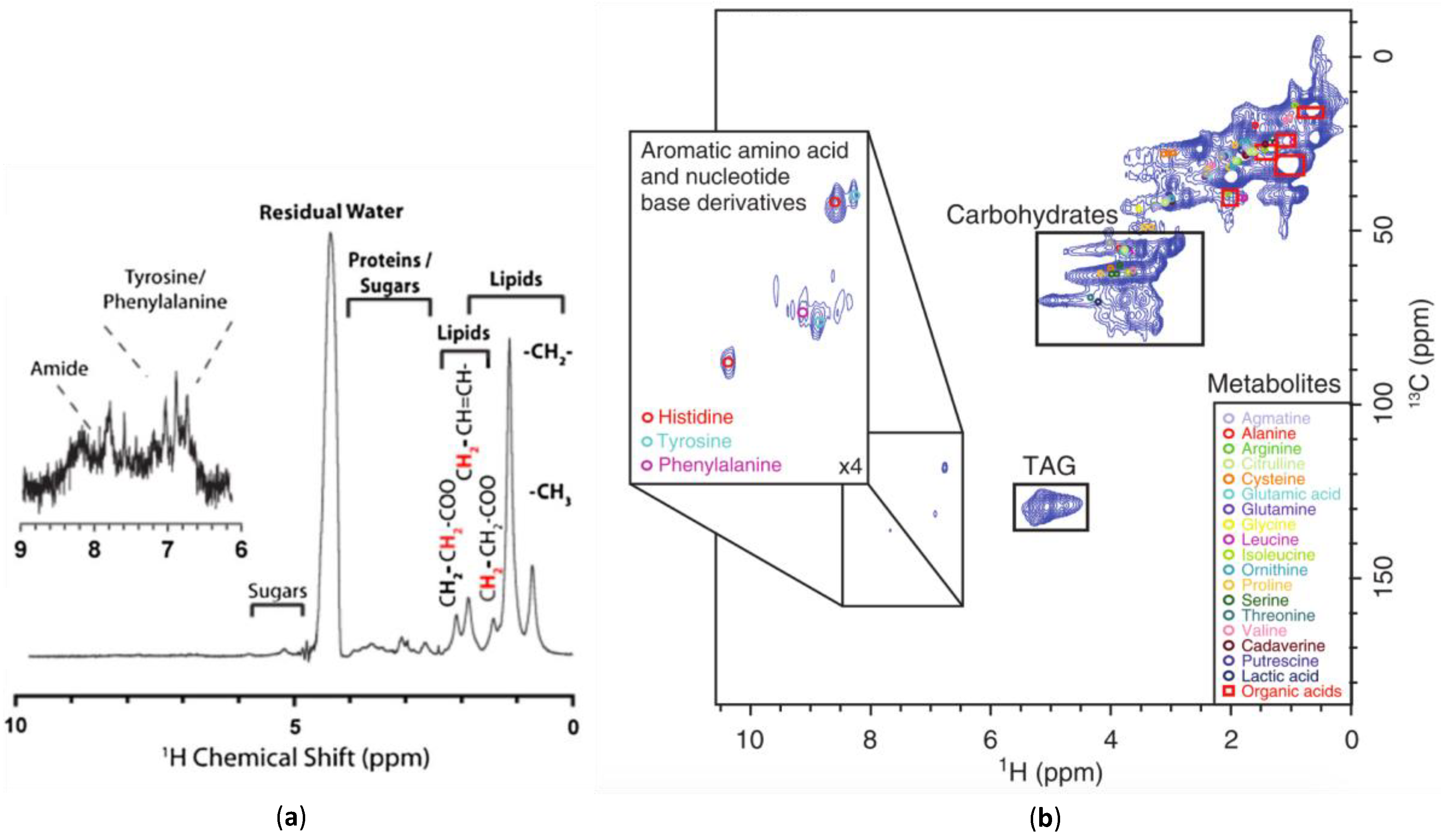
The simplest approach to increasing spectral dispersion and overcoming broad lines is to spread the signal into multiple dimensions. This can be achieved using 1H-13C HSQC NMR, which is relatively easy to obtain if the organisms are isotopically labelled with 13C to increase signal [21,156]. 1D 1H NMR has a peak capacity of 3000 peaks [157], whereas 1H-13C HSQC NMR approaches 2,000,000 providing the dispersion required for detailed metabolic assignments (see Figure 5) [80]. Modern HSQC (and HMQC) experiments use gradients for coherence selection, meaning a 1H that is not attached to a 13C is not selected. This results in the water signal being rejected by the gradient filter, allowing the experiments to have reasonable water suppression without modification. Furthermore, standard databases such as the Bruker Bioreference Databases [46,158,159] and the Human Metabolome Database [160,161,162,163] contain a wide array of assigned 1H-13C one bond correlations making assignments of HSQC relatively straight forward. Additional 1H-1H COSY data can be extremely useful to help confirm mixture assignments, which has been shown to be relatively easy to acquire under MAS conditions in-vivo. [46].
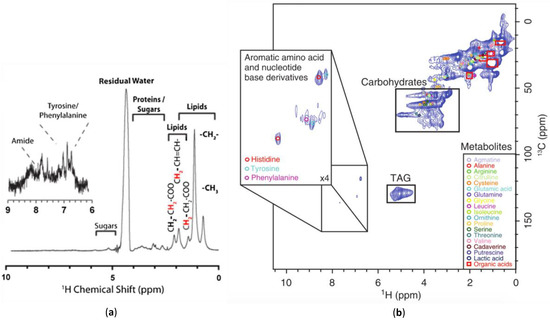
Figure 5.
A comparison of: (a) a 1D 1H NMR spectrum [69]; (b) a 2D 1H-13C HMQC NMR spectrum [121] of 13C enriched D. magna. The additional spectral dispersion afforded by 2D permits metabolite assignments not possible from the 1D data. Modified with permission from John Wiley and Sons.
3.2.2. Overcoming Magnetic Susceptibility Distortions
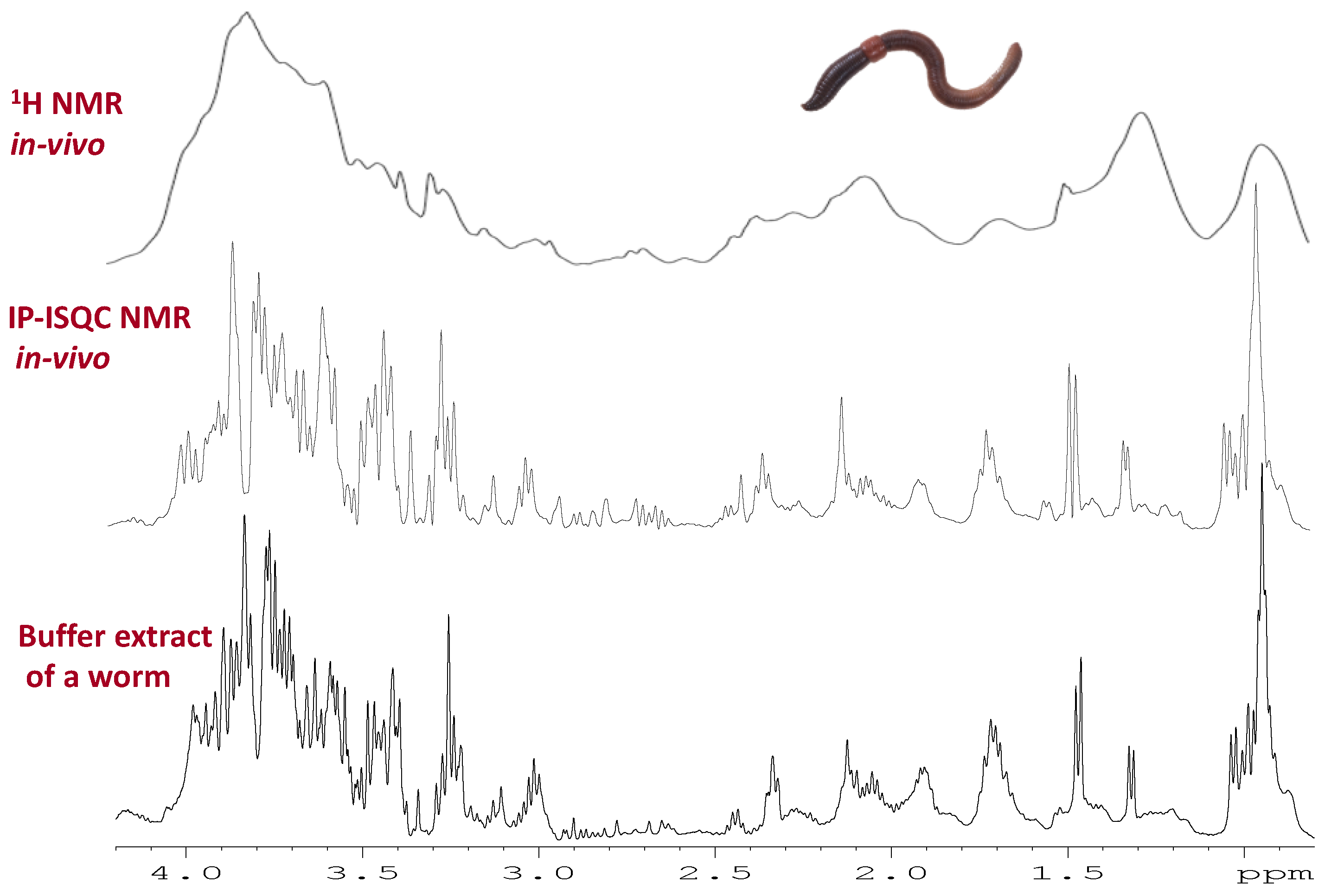
The main approach to reduce or eliminate magnetic susceptibility distortions is to take advantage of intermolecular multiple quantum coherences between water and solutes. The approach was first discovered by Warren [164] and has been further developed by various other groups [165,166]. Long range interactions between water and solutes are reintroduced in the liquid state by utilizing a pulse field gradient. The interactions build up over distances longer than the more local susceptibility distortions and the chemical shift and magnetic susceptibility distortions can be separated by means of a 2D experiment. Recently, a phase sensitive version of the experiment was introduced and optimized for in-vivo samples with fast relaxation [153]. The resulting line shape from the In Phase—Intermolecular Single Quantum Coherence (IP-ISQC) experiment applied to living organisms was near identical to that obtained from buffer extracts (see Figure 6). The primary advantage of IP-ISQC is based on 1H detection, which does not require the use of 13C enriched organisms. As such the approach opens the possibility to study organisms directly from the environment rather than being reliant on lab raised 13C enriched organisms.
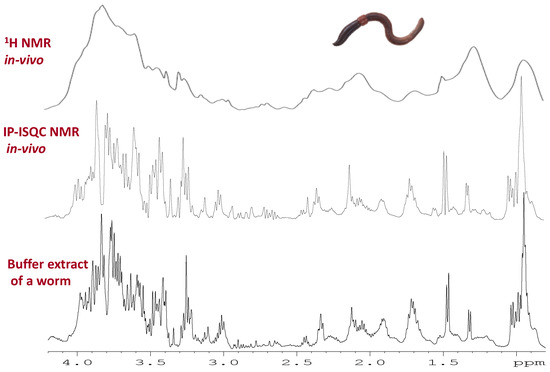
Figure 6.
Example of an IP-iSQC NMR spectrum [153] which is used to remove magnetic susceptibility distortions from in-vivo NMR. The top spectrum shows a conventional 1H in-vivo NMR spectrum of a worm. 1H IP-iSQC approach which uses a 2D sequence to remove the distortions for the same worm (in-vivo). The bottom is a buffer extract from a similar worm (ex-vivo). The results demonstrate that the in-vivo IP-iSQC experiment produced near identical line shape, integral, and spectral profile as a buffer extract. Modified with permission from John Wiley and Sons.
3.3. Sensitivity
NMR is a relatively insensitive technique. For larger organisms (>1 mm), it is not a major concern and signal is easy to detect, especially if numerous organisms are present in the sample. However, if small organisms such as eggs or individual organisms are to be studied then low amounts of biomass may make detection challenging or impossible using conventional 5 mm NMR probes. While cryoprobes can offer some improvement [167], microcoils hold greater potential for very small samples [168]. This was demonstrated in two recent studies by Fugariu et al. [81] and Grisi et al. [82]. Both studies demonstrate that small aquatic eggs could be studied. Grisi showed NMR on eggs as small as 100 pL, whereas Fugariu demonstrates analysis on the smallest coils (20 μm I.D.). These coils achieved a mass sensitivity improvement of >6000 times over a 5 mm room temperature probe, which translates into reduction in experimental time of ~36 million. These types of studies hold valuable potential for the analysis of resting eggs. Resting eggs are of great significance to environmental studies as they are produced by many planktonic organisms to ensure species survival during adverse times, and will exist in sediments up to decades until conditions improve [169]. Previous studies have indicated that eggs respond differently to toxins than young neonates [170]. Unfortunately, despite their critical role for the long-term survival of populations, very little is known about the impacts of toxic chemicals on resting eggs. Traditional toxicity tests are difficult to employ to study eggs, due to their lack of movement and reproduction, however, with the mass sensitivity improvement microcoil NMR has demonstrated, the technique is ideal for metabolic profiling in tiny samples.
4. Why In-Vivo NMR?
4.1. Metabolic Pathways and Recovery
In-vivo NMR is highly complementary to conventional metabolic approaches. Traditional sampling approaches (homogenates of biofluids) provide a snapshot in time, whereas with in-vivo studies, the organisms are kept alive in the system, and data acquisition can occur continuously, and with high temporal resolution (sensitivity permitting). This provides potential to understand complex inter-connected response pathways to better understand how organisms react to, and deal with toxic chemicals. Furthermore, in-vivo NMR provides a convenient framework to study recovery from exposure [67]. If for example, an organism can return to homeostasis after exposure, then the contaminant likely only creates a temporary “flux response” from which the organism can fully recover. However, if the metabolome never returns to homeostasis post-exposure, this suggests permanent or long-term changes in the biochemistry which could be a precursor to disease or health deterioration [67]. A permanent change in the metabolome, even if it does not lead to rapid physical symptoms, is likely an important indicator for policy makers trying to estimate the safest, but also realistic targets for anthropogenic chemicals in the environment [67]. Studies with chemical mixtures could also provide information on synergistic effects. Some chemicals may not exhibit toxic properties alone, but when combined with others, can have deleterious effects. Continuous exposure to potentially hundreds of chemicals at trace concentrations is often the norm in the environment rather than the exception and the toxic synergism as well as impacts of long term exposure are currently not well understood [65,76].
4.2. Reducing the Impacts of Natural Variation
Traditional in-vitro metabolomics studies that use separate “control” and “exposed” populations have additional variation due the natural differences between individuals in the populations. This is unavoidable if the populations are sacrificed for analysis, as is the case for small organisms such as C. elegans and D. magna [102]. It has been shown this variance can be as much as 15% of the NMR signal [61]. This makes data analysis more challenging as statistical methods must detect tiny changes caused by the contaminant in the presence of often larger random variances. In-vivo NMR offers the potential to reduce this genetic variation by using the same organisms as the control and exposed populations [94]. For example, organisms could be placed inside the NMR and studied for six hours, after which they are then exposed. Data collected during the first six hours can be used as the control, and data after this point would contain additional signatures from the exposure. Care would have to be taken to ensure that the only changes are from the contaminant, thus the organisms must be supplied oxygenated water and food to avoid complications from starvation and anoxic stress.
4.3. The Contaminant, Drug, or Nutrient
In-vivo NMR holds the potential to study not just the impacts of a contaminant, but if at high enough concentration, the chemical itself. Such studies should provide unique insights into binding mechanisms, biotransformation, bioaccumulation, and excretion. Experiments such as saturation transfer difference (STD) NMR can identify how molecules bind to receptors [171,172], whereas reverse-STD can identify which receptors do the binding [173,174]. If these approaches are combined with solid-state cross polarization then molecules that become sequestered in the most rigid components (shell, bone) can be selectively identified [98]. While studies have combined all these approaches to follow contaminant penetration into soil [98], they have yet to be applied in-vivo. The biggest hurdle at present is that many of the experiments are relatively insensitive, and contaminants are often present at low concentrations. However, the approach could have enormous impacts for understanding how nutrients (such as vitamins, phosphorous, etc.) are incorporated into living systems which are often present at much higher concentrations in-vivo.
4.4. Selective Isotopic Enrichment
Recent studies have used 13C enriched organisms to increase the signal in 1H-13C 2D NMR. This is an excellent approach as it makes metabolites in organisms easier to detect and ideal for non-targeted analysis. However, if the questions are more defined, for example “how does chemical X impact the Krebs cycle?”, it may be possible to selectively enrich precursor molecules to target pathways and extract higher resolution information on specific mechanisms [175].
In other cases, a heteronucleus specific to the contaminant, but not abundant in nature, could be highly beneficial. The most obvious nucleus being 19F as it is present in many environmental contaminants and drugs [40,176,177,178], and is a highly sensitive NMR nucleus. Perfluorinated compounds are stable in the environment and are absorbed into the body irreversibly. They have been found globally and are linked to reduced fertility, reduced birth weight, and changes in thyroid hormone levels along with many other negative consequences [67,179]. Other options for isotopic labelling may include the use of 2H in organic molecules or isotopic enrichment of heavy metals such as 113Cd, 207Pb, and 199Hg such that they are easier to detect and monitor in-vivo.
5. Conclusions
In-vivo NMR can provide a unique molecular-window into a living organism and its toxic response processes. The field of toxicity testing is changing rapidly, and with continuous chemicals entering the environment more reliable, and complementary techniques to study reaction mechanisms will be important to examine the impact on aquatic species, plants, and humans. While in-vivo NMR using high field instruments and flow systems were explored as early as the 80’s [110], it has not become routine due to complications from the intense water peak, broad signals, and low sensitivity. Using modern approaches including improved water suppression, isotopic enrichments, cryoprobes, micro-coils, and multidimensional NMR, many of these hurdles are being overcome and in the last few years in-vivo NMR is making a resurgence as a very powerful technique with immense potential. A considerable amount of work is still required, including hardware (improved coils and flow cells), experiments (targeted and higher order multidimensional experiments to extract more information), and data processing (extracting key information quickly from massive amounts of data). Both smaller detection technologies (microcoils for single cell and egg analysis) and larger bore magnets for larger organisms need to be explored. Furthermore, the potential of in-vivo low field NMR is an exciting future prospect. While low field NMRs suffer from reduced resolution and sensitivity, their low cost and portability make them ideal for field deployment with often nothing more than a power supply required for monitoring. Some examples, include a mobile NMR lab for leaf phenotyping in the field [180], a portable sensor for monitoring water and sap flow [181], a device to detect water content in trees [182,183], fast field cycling NMR in plant leaves [184], as-well as lipid and metabolites profiling in seeds [185]. Advancement in technologies such as dynamic nuclear polarization in combination with low field NMR [186], and zero/ultra-low field NMR with optical detection [187] offer potential for huge increases in sensitivity that would advance low field NMR into a powerful tool for in-vivo analysis and monitoring.
In summary, given NMR’s ability to examine inside a living organism and provide unprecedented information on the living metabolome, in-vivo NMR will undoubtedly evolve to become a central and key tool for understanding living processes and how they are impacted by toxic chemicals.
Author Contributions
M.B. and A.J. are joint first authors and drafted the manuscript; M.T.A. helped with the editing and structuring the paper; A.S. planned the article and helped with editing, formatting and reviewing.
Funding
This research was funded by Natural Sciences and Engineering Research Council (NSERC) Strategic (STPGP 494273-16) and Discovery Programs (RGPIN-2014-05423).
Acknowledgments
We would like to thank the Canada Foundation for Innovation (CFI), the Ontario Ministry of Research and Innovation (MRI), and the Krembil Foundation for providing infrastructure funding and equipment donations to the Environmental NMR Center. A.J.S. would like to thank the Government of Ontario for an Early Researcher Award.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Arias-Estévez, M.; López-Periago, E.; Martínez-Carballo, E.; Simal-Gándara, J.; Mejuto, J.-C.; García-Río, L. The mobility and degradation of pesticides in soils and the pollution of groundwater resources. Agric. Ecosyst. Environ. 2008, 123, 247–260. [Google Scholar] [CrossRef]
- Crossman, J.; Futter, M.N.; Oni, S.K.; Whitehead, P.G.; Jin, L.; Butterfield, D.; Baulch, H.M.; Dillon, P.J. Impacts of climate change on hydrology and water quality: Future proofing management strategies in the Lake Simcoe watershed, Canada. J. Great Lakes Res. 2013, 39, 19–32. [Google Scholar] [CrossRef]
- Koehler, A. Water use in LCA: Managing the planet’s freshwater resources. Int. J. Life Cycle Assess. 2008, 13, 451–455. [Google Scholar] [CrossRef]
- Sirés, I.; Brillas, E. Remediation of water pollution caused by pharmaceutical residues based on electrochemical separation and degradation technologies: A review. Environ. Int. 2012, 40, 212–229. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Utrilla, J.; Sánchez-Polo, M.; Ferro-García, M.Á.; Prados-Joya, G.; Ocampo-Pérez, R. Pharmaceuticals as emerging contaminants and their removal from water. A review. Chemosphere 2013, 93, 1268–1287. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.Q.; Loganath, A.; Chong, Y.S.; Tan, J.; Philip Obbard, J. Levels of Persistent Organic Pollutant Residues in Human Adipose and Muscle Tissues in Singapore. J. Toxicol. Environ. Heal. Part A 2006, 69, 1927–1937. [Google Scholar] [CrossRef]
- Tyagi, V.; Chopra, A.; Durgapal, N.; Kumar, A. Evaluation of Daphnia magna as an indicator of Toxicity and Treatment efficacy of Municipal Sewage Treatment Plant. J. Appl. Sci. Environ. Manag. 2007, 11, 61–67. [Google Scholar] [CrossRef]
- Shahid, M.; Xiong, T.; Masood, N.; Leveque, T.; Quenea, K.; Austruy, A.; Foucault, Y.; Dumat, C. Influence of plant species and phosphorus amendments on metal speciation and bioavailability in a smelter impacted soil: A case study of food-chain contamination. J. Soils Sediments 2013, 14, 655–665. [Google Scholar] [CrossRef]
- Nagajyoti, P.C.; Lee, K.D.; Sreekanth, T.V.M. Heavy metals, occurrence and toxicity for plants: A review. Environ. Chem. Lett. 2010, 8, 199–216. [Google Scholar] [CrossRef]
- Kumar, A.; Aery, N.C. Impact, Metabolism, and Toxicity of Heavy Metals in Plants. In Plant Responses to Xenobiotics; Springer: Singapore, 2016; pp. 141–176. [Google Scholar]
- Oukarroum, A. Alleviation of Metal-Induced Toxicity in Aquatic Plants by Exogenous Compounds: A Mini-Review. Water Air Soil Pollut. 2016, 227, 204. [Google Scholar] [CrossRef]
- Daghrir, R.; Drogui, P. Tetracycline antibiotics in the environment: A review. Environ. Chem. Lett. 2013, 11, 209–227. [Google Scholar] [CrossRef]
- Smith, W.H. Air Pollution and Forests: Interactions between Air Contaminants and Forest Ecosystems, 1st ed.; Springer Science & Business Media: Berlin, Germany, 1981; ISBN 978-1-4684-0104-2. [Google Scholar]
- Anderson, J.G.; Toohey, D.W.; Brune, W.H. Free Radicals within the Antarctic Vortex: The Role of CFCs in the Antarctic Ozone Loss. Science 1991, 251, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Haas, P.M. Banning chlorofluorocarbons: Epistemic community efforts to protect stratospheric ozone. Int. Organ. 1992, 46, 187–224. [Google Scholar] [CrossRef]
- Finlayson-Pitts, B.J.; Pitts, J.N. Tropospheric Air Pollution: Ozone, Airborne Toxics, Polycyclic Aromatic Hydrocarbons, and Particles. Science 1997, 276, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Kampa, M.; Castanas, E. Human health effects of air pollution. Environ. Pollut. 2008, 151, 362–367. [Google Scholar] [CrossRef] [PubMed]
- Simkhovich, B.Z.; Kleinman, M.T.; Kloner, R.A. Air Pollution and Cardiovascular Injury. J. Am. Coll. Cardiol. 2008, 52, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Darrall, N.M. The effect of air pollutants on physiological processes in plants. Plant Cell Environ. 1989, 12, 1–30. [Google Scholar] [CrossRef]
- Lankadurai, B.P.; Nagato, E.G.; Simpson, M.J. Environmental metabolomics: An emerging approach to study organism responses to environmental stressors. Environ. Rev. 2013, 21, 180–205. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. “Metabonomics”: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 2008, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Rochfort, S. Metabolomics Reviewed: A New “Omics” Platform Technology for Systems Biology and Implications for Natural Products Research. J. Nat. Prod. 2005, 68, 1813–1820. [Google Scholar] [CrossRef] [PubMed]
- Gowda, G.A.N.; Raftery, D. Recent Advances in NMR-Based Metabolomics. Anal. Chem. 2017, 89, 490–510. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016, 15, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Macel, M.; Van Dan, N.M.; Keurentjes, J.J.B. Metabolomics: The chemistry between ecology and genetics. Mol. Ecol. Resour. 2010, 10, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Cevallos, J.M.; Reyes-De-Corcuera, J.I.; Etxeberria, E.; Danyluk, M.D.; Rodrick, G.E. Metabolomic analysis in food science: A review. Trends Food Sci. Technol. 2009, 20, 557–566. [Google Scholar] [CrossRef]
- Miller, M.G. Environmental Metabolomics: A SWOT Analysis (Strengths, Weaknesses, Opportunities, and Threats). J. Proteome Res. 2007, 6, 540–545. [Google Scholar] [CrossRef] [PubMed]
- Bundy, J.G.; Davey, M.P.; Viant, M.R. Environmental metabolomics: A critical review and future perspectives. Metabolomics 2009, 5, 3–21. [Google Scholar] [CrossRef]
- García-Sevillano, M.Á.; García-Barrera, T.; Gómez-Ariza, J.L. Environmental metabolomics: Biological markers for metal toxicity. Electrophoresis 2015, 36, 2348–2365. [Google Scholar] [CrossRef] [PubMed]
- Bundy, J.G.; Spurgeon, D.J.; Svendsen, C.; Hankard, P.K.; Weeks, J.M.; Osborn, D.; Lindon, J.C.; Nicholson, J.K. Environmental metabonomics: Applying combination biomarker analysis in earthworms at a metal contaminated site. Ecotoxicology 2004, 13, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Gibb, J.O.T.; Svendsen, C.; Weeks, J.M.; Nicholson, J.K. 1H NMR spectroscopic investigations of tissue metabolite biomarker response to Cu II exposure in terrestrial invertebrates: Identification of free histidine as a novel biomarker of exposure to copper in earthworms. Biomarkers 1997, 2, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Bundy, J.; Osborn, D.; Weeks, J.; Lindon, J.; Nicholson, J. An NMR-based metabonomic approach to the investigation of coelomic fluid biochemistry in earthworms under toxic stress. FEBS Lett. 2001, 500, 31–35. [Google Scholar] [CrossRef]
- Lenz, E.M.; Weeks, J.M.; Lindon, J.C.; Osborn, D.; Nicholson, J.K. Qualitative high field 1H-NMR spectroscopy for the characterization of endogenous metabolites in earthworms with biochemical biomarker potential. Metabolomics 2005, 1, 123–136. [Google Scholar] [CrossRef]
- Whitfield Åslund, M.L.; Simpson, A.J.; Simpson, M.J. 1H NMR metabolomics of earthworm responses to polychlorinated biphenyl (PCB) exposure in soil. Ecotoxicology 2011, 20, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Griffith, C.M.; Williams, P.B.; Tinoco, L.W.; Dinges, M.M.; Wang, Y.; Larive, C.K. 1H NMR Metabolic Profiling of Earthworm (Eisenia fetida) Coelomic Fluid, Coelomocytes, and Tissue: Identification of a New Metabolite-Malylglutamate. J. Proteome Res. 2017, 16, 3407–3418. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A.E.; McKelvie, J.R.; Simpson, A.J.; Simpson, M.J. 1H NMR metabolomics of earthworm exposure to sub-lethal concentrations of phenanthrene in soil. Environ. Pollut. 2010, 158, 2117–2123. [Google Scholar] [CrossRef] [PubMed]
- McKelvie, J.R.; Yuk, J.; Xu, Y.; Simpson, A.J.; Simpson, M.J. 1H NMR and GC/MS metabolomics of earthworm responses to sub-lethal DDT and endosulfan exposure. Metabolomics 2009, 5, 84–94. [Google Scholar] [CrossRef]
- Schou, M.F.; Kristensen, T.N.; Pedersen, A.; Karlsson, B.G.; Loeschcke, V.; Malmendal, A. Metabolic and functional characterization of effects of developmental temperature in Drosophila melanogaster. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 312, R211–R222. [Google Scholar] [CrossRef] [PubMed]
- Kariuki, M.; Nagato, E.; Lankadurai, B.; Simpson, A.; Simpson, M. Analysis of Sub-Lethal Toxicity of Perfluorooctane Sulfonate (PFOS) to Daphnia magna Using 1H Nuclear Magnetic Resonance-Based Metabolomics. Metabolites 2017, 7, 15. [Google Scholar] [CrossRef] [PubMed]
- Nagato, E.G.; Simpson, A.J.; Simpson, M.J. Metabolomics reveals energetic impairments in Daphnia magna exposed to diazinon, malathion and bisphenol-A. Aquat. Toxicol. 2016, 170, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Wagner, N.D.; Simpson, A.J.; Simpson, M.J. Metabolomic responses to sublethal contaminant exposure in neonate and adult Daphnia magna. Environ. Toxicol. Chem. 2017, 36, 938–946. [Google Scholar] [CrossRef] [PubMed]
- Nagato, E.G.; D’eon, J.C.; Lankadurai, B.P.; Poirier, D.G.; Reiner, E.J.; Simpson, A.J.; Simpson, M.J. 1H NMR-based metabolomics investigation of Daphnia magna responses to sub-lethal exposure to arsenic, copper and lithium. Chemosphere 2013, 93, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, V.; Simpson, A.J.; Simpson, M.J. 1H NMR-based metabolomics of Daphnia magna responses after sub-lethal exposure to triclosan, carbamazepine and ibuprofen. Comp. Biochem. Physiol. Part D 2016, 19, 199–210. [Google Scholar] [CrossRef]
- Wagner, N.D.; Lankadurai, B.P.; Simpson, M.J.; Simpson, A.J.; Frost, P.C. Metabolomic differentiation of nutritional stress in an aquatic invertebrate. Physiol. Biochem. Zool. 2015, 88, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Mobarhan, Y.L.; Fortier-Mcgill, B.; Soong, R.; Maas, W.E.; Fey, M.; Monette, M.; Stronks, H.J.; Schmidt, S.; Heumann, H.; Norwood, W.; et al. Comprehensive multiphase NMR applied to a living organism. Chem. Sci. 2016, 7, 4856–4866. [Google Scholar] [CrossRef]
- Chiu, K.H.; Dong, C.D.; Chen, C.F.; Tsai, M.L.; Ju, Y.R.; Chen, T.M.; Chen, C.W. NMR-based metabolomics for the environmental assessment of Kaohsiung Harbor sediments exemplified by a marine amphipod (Hyalella azteca). Mar. Pollut. Bull. 2017, 124, 714–724. [Google Scholar] [CrossRef] [PubMed]
- Mobarhan, Y.L.; Struppe, J.; Fortier-McGill, B.; Simpson, A.J. Effective combined water and sideband suppression for low-speed tissue and in vivo MAS NMR. Anal. Bioanal. Chem. 2017, 409, 5043–5055. [Google Scholar] [CrossRef] [PubMed]
- An, Y.J.; Xu, W.J.; Jin, X.; Wen, H.; Kim, H.; Lee, J.; Park, S. Metabotyping of the C. elegans sir-2.1 Mutant Using in Vivo Labeling and 13C-Heteronuclear Multidimensional NMR Metabolomics. ACS Chem. Biol. 2012, 7, 2012–2018. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Li, X.; Molin, L.; Solari, F.; Elena-Herrmann, B.; Sakellariou, D. μHigh Resolution-Magic-Angle Spinning NMR Spectroscopy for Metabolic Phenotyping of Caenorhabditis elegans. Anal. Chem. 2014, 86, 6064–6070. [Google Scholar] [CrossRef] [PubMed]
- Korvink, J.G.; Badilita, V.; Bordonali, L.; Jouda, M.; Mager, D.; MacKinnon, N. NMR microscopy for in vivo metabolomics, digitally twinned by computational systems biology, needs a sensitivity boost. arXiv, 2017; arXiv:1707.08726. [Google Scholar]
- Xu, C.; Rezeng, C.; Li, J.; Zhang, L.; Yan, Y.; Gao, J.; Wang, Y.; Li, Z.; Chen, J. 1H NMR-Based Metabolomics Study of the Toxicological Effects in Rats Induced by “Renqing Mangjue” Pill, a Traditional Tibetan Medicine. Front. Pharmacol. 2017, 8, 602. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-A.; Choi, H.-J.; Kwon, Y.-K.; Ryu, D.H.; Kwon, T.-H.; Hwang, G.-S. 1H NMR-Based Metabolite Profiling of Plasma in a Rat Model of Chronic Kidney Disease. PLoS ONE 2014, 9, e85445. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zheng, L.; Shi, J.; Zhang, G.; Lu, L.; Zhu, L.; Zhang, J.; Liu, Z. Toxic Markers of Matrine Determined Using 1H-NMR-Based Metabolomics in Cultured Cells In Vitro and Rats In Vivo. Evid. Based Complement. Altern. Med. 2015, 2015, 598412. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.L.; Walker, L.; Shore, R.F.; Nicholson, J.K. High-resolution magic angle spinning 1H-NMR spectroscopy studies on the renal biochemistry in the bank vole Clethrionomys glareolus and the effects of arsenic (As3+) toxicity. Xenobiotica 2001, 31, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, H.L.; Soong, R.; Courtier-Murias, D.; Botana, A.; Fortier-Mcgill, B.; Maas, W.E.; Fey, M.; Hutchins, H.; Krishnamurthy, S.; Kumar, R.; et al. Comprehensive multiphase NMR: A promising technology to study plants in their native state. MRC 2015, 53, 735–744. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Huang, Y.; Hu, J.; Zhou, H.; Adeleye, A.S.; Keller, A.A. 1H NMR and GC-MS Based Metabolomics Reveal Defense and Detoxification Mechanism of Cucumber Plant under Nano-Cu Stress. Environ. Sci. Technol. 2016, 50, 2000–2010. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.-K.; Choi, Y.H.; Verberne, M.; Lefeber, A.; Erkelens, C.; Verpoote, R. Metabolic fingerprinting of wild type and transgenic tobacco plants by 1H NMR and multivariate analysis technique. Phytochemistry 2004, 65, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, P.; Kruger, N.J.; Ratcliffe, R.G. Metabolite fingerprinting and profiling in plants using NMR. J. Exp. Bot. 2004, 56, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.K.; Choi, Y.H.; Verpoorte, R. NMR-based plant metabolomics: Where do we stand, where do we go? Trends Biotechnol. 2011, 29, 267–275. [Google Scholar] [CrossRef] [PubMed]
- Whitfield Åslund, M.; Celejewski, M.; Lankadurai, B.P.; Simpson, A.J.; Simpson, M.J. Natural variability and correlations in the metabolic profile of healthy Eisenia fetida earthworms observed using 1H NMR metabolomics. Chemosphere 2011, 83, 1096–1101. [Google Scholar] [CrossRef] [PubMed]
- Yuk, J.; Simpson, M.J.; Simpson, A.J. 1-D and 2-D NMR-based metabolomics of earthworms exposed to endosulfan and endosulfan sulfate in soil. Environ. Pollut. 2013, 175, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Whitfield Åslund, M.L.; McShane, H.; Simpson, M.J.; Simpson, A.J.; Whalen, J.K.; Hendershot, W.H.; Sunahara, G.I. Earthworm Sublethal Responses to Titanium Dioxide Nanomaterial in Soil Detected by 1H NMR Metabolomics. Environ. Sci. Technol. 2012, 46, 1111–1118. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.A.E.; Simpson, A.J.; Simpson, M.J. 1H NMR metabolomics of earthworm responses to sub-lethal PAH exposure. Environ. Chem. 2009, 6, 432. [Google Scholar] [CrossRef]
- Krewski, D.; Acosta, D.; Andersen, M.; Anderson, H.; Bailar, J.C.; Boekelheide, K.; Brent, R.; Charnley, G.; Cheung, V.G.; Green, S.; et al. Toxicity Testing in the 21st Century: A Vision and a Strategy. J. Toxicol. Environ. Health Part B 2010, 13, 51–138. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.; Simpson, M.J.; Soong, R. Nuclear Magnetic Resonance Spectroscopy and Its Key Role in Environmental Research. Environ. Sci. Technol. 2012, 46, 11488–11496. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.; Liaghati, Y.; Fortier-Mcgill, B.; Soong, R.; Akhter, M. Perspective: In vivo NMR—A potentially powerful tool for environmental research. MRC 2015, 53, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Persoone, G.; Baudo, R.; Cotman, M.; Blaise, C.; Thompson, K.C.; Moreira-Santos, M.; Vollat, B.; Törökne, A.; Han, T. Review on the acute Daphnia magna toxicity test—Evaluation of the sensitivity and the precision of assays performed with organisms from laboratory cultures or hatched from dormant eggs. Knowl. Manag. Aquat. Ecosyst. 2009, 393, 1. [Google Scholar] [CrossRef]
- Soong, R.; Nagato, E.; Sutrisno, A.; Fortier-Mcgill, B.; Akhter, M.; Schmidt, S.; Heumann, H.; Simpson, A.J. In vivo NMR spectroscopy: Toward real time monitoring of environmental stress. MRC 2015, 53, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Ebert, D. Ecology, Epidemiology and Evolution of Parasitism in Daphnia; National Library of Medicine (US), National Center for Biotechnology Information: Bethesda, MD, USA, 2005; ISBN 1-932811-06-0. [Google Scholar]
- Edison, A.; Hall, R.; Junot, C.; Karp, P.; Kurland, I.; Mistrik, R.; Reed, L.; Saito, K.; Salek, R.; Steinbeck, C.; et al. The Time Is Right to Focus on Model Organism Metabolomes. Metabolites 2016, 6, 2–7. [Google Scholar] [CrossRef] [PubMed]
- Wacker, A.; Martin-Creuzburg, D. Allocation of essential lipids in Daphnia magna during exposure to poor food quality. Funct. Ecol. 2007, 21, 738–747. [Google Scholar] [CrossRef]
- Sengupta, N.; Reardon, D.C.; Gerard, P.D.; Baldwin, W.S. Exchange of polar lipids from adults to neonates in Daphnia magna: Perturbations in sphingomyelin allocation by dietary lipids and environmental toxicants. PLoS ONE 2017, 12, e0178131. [Google Scholar] [CrossRef] [PubMed]
- Weber, C.I. Methods for Measuring the Acute Toxicity of Effluents and Receiving Waters to Freshwater and Marine Organisms; US Environmental Protection Agency: Cincinnati, OH, USA, 2002.
- Guilhermino, L.; Diamantino, T.; Silva, C.; Soares, A.M.V.M. Acute Toxicity Test with Daphnia magna: An Alternative to Mammals in the Prescreening of Chemical Toxicity? Ecotoxicol. Environ. Saf. 2000, 46, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Andersen, M.E.; Krewski, D. Toxicity Testing in the 21st Century: Bringing the Vision to Life. Toxicol. Sci. 2009, 107, 324–330. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.Y.; Viant, M.R.; Tjeerdema, R.S. Metabolomics: Methodologies and applications in the environmental sciences. J. Pestic. Sci. 2006, 31, 245–251. [Google Scholar] [CrossRef]
- De Graaf, R.A. In Vivo NMR Spectroscopy: Principles and Techniques; John Wiley & Sons Ltd.: Chichester, UK, 2013; ISBN 1118681304. [Google Scholar]
- Markley, J.L.; Brüschweiler, R.; Edison, A.S.; Eghbalnia, H.R.; Powers, R.; Raftery, D.; Wishart, D.S. The future of NMR-based metabolomics. Curr. Opin. Biotechnol. 2017, 43, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Nagato, E.G.; Lankadurai, B.P.; Soong, R.; Simpson, A.J.; Simpson, M.J. Development of an NMR microprobe procedure for high-throughput environmental metabolomics of Daphnia magna. Magn. Reson. Chem. 2015, 53, 745–753. [Google Scholar] [CrossRef] [PubMed]
- Fugariu, I.; Soong, R.; Lane, D.; Fey, M.; Maas, W.; Vincent, F.; Beck, A.; Schmidig, D.; Treanor, B.; Simpson, A.J. Towards single egg toxicity screening using microcoil NMR. Analyst 2017, 142, 4812–4824. [Google Scholar] [CrossRef] [PubMed]
- Grisi, M.; Vincent, F.; Volpe, B.; Guidetti, R.; Harris, N.; Beck, A.; Boero, G. NMR spectroscopy of single sub-nL ova with inductive ultra-compact single-chip probes. Sci. Rep. 2017, 7, 44670. [Google Scholar] [CrossRef] [PubMed]
- Jaroszewski, J.W. Hyphenated NMR Methods in Natural Products Research, Part 2: HPLC-SPE-NMR and Other New Trends in NMR Hyphenation. Planta Med. 2005, 71, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.M.; Price, W.S. Common problems and artifacts encountered in solution-state NMR experiments. Concepts Magn. Reson. Part A 2017, 45A, e21387. [Google Scholar] [CrossRef]
- Bauer, M.; Bertario, A.; Boccardi, G.; Fontaine, X.; Rao, R.; Verrier, D. Reproducibility of 1H-NMR integrals: A collaborative study. J. Pharm. Biomed. Anal. 1998, 17, 419–425. [Google Scholar] [CrossRef]
- Dumas, M.-E.; Maibaum, E.C.; Teague, C.; Ueshima, H.; Zhou, B.; Lindon, J.C.; Nicholson, J.K.; Stamler, J.S.; Elliot, P.; Chan, Q.; et al. Assessment of Analytical Reproducibility of 1H NMR Spectroscopy Based Metabonomics for Large-Scale Epidemiological Research: The INTERMAP Study. Anal. Chem. 2006, 78, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Wallace, T. Quantitative analysis of a mixture by NMR spectroscopy. J. Chem. Educ. 1984, 61, 1074. [Google Scholar] [CrossRef]
- Bharti, S.K.; Roy, R. Quantitative 1H NMR spectroscopy. TrAC Trends Anal. Chem. 2012, 35, 5–26. [Google Scholar] [CrossRef]
- Fan, T.W.-M.; Lane, A.N. Applications of NMR spectroscopy to systems biochemistry. Prog. Nucl. Magn. Reson. Spectrosc. 2016, 92, 18–53. [Google Scholar] [CrossRef] [PubMed]
- Akoka, S.; Barantin, L.; Trierweiler, M. Concentration Measurement by Proton NMR Using the ERETIC Method. Anal. Chem. 1999, 71, 2554–2557. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S. Quantitative metabolomics using NMR. TrAC Trends Anal. Chem. 2008, 27, 228–237. [Google Scholar] [CrossRef]
- Barding, G.A.; Salditos, R.; Larive, C.K. Quantitative NMR for bioanalysis and metabolomics. Anal. Bioanal. Chem. 2012, 404, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Simmler, C.; Napolitano, J.G.; McAlpine, J.B.; Chen, S.-N.; Pauli, G.F. Universal quantitative NMR analysis of complex natural samples. Curr. Opin. Biotechnol. 2014, 25, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.; Simpson, M.J.; Soong, R. Environmental Nuclear Magnetic Resonance Spectroscopy: An Overview and a Primer. Anal. Chem. 2018, 90, 628–639. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, D.; Sterner, C.; Masek, B.; Svenningsen, R.; Samuelson, G. An NMR kinetics experiment. J. Chem. Educ. 1982, 59, 885. [Google Scholar] [CrossRef]
- Dobson, C.M.; Hore, P.J. Kinetic studies of protein folding using NMR spectroscopy. Nat. Struct. Biol. 1998, 5, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Masoom, H.; Courtier-Murias, D.; Farooq, H.; Soong, R.; Kelleher, B.P.; Zhang, C.; Maas, W.E.; Fey, M.; Kumar, R.; Monette, M.; et al. Soil Organic Matter in Its Native State: Unravelling the Most Complex Biomaterial on Earth. Environ. Sci. Technol. 2016, 50, 1670–1680. [Google Scholar] [CrossRef] [PubMed]
- Masoom, H.; Courtier-Murias, D.; Soong, R.; Maas, W.E.; Fey, M.; Kumar, R.; Monette, M.; Stronks, H.J.; Simpson, M.J.; Simpson, A. From Spill to Sequestration: The Molecular Journey of Contamination via Comprehensive Multiphase NMR. Environ. Sci. Technol. 2015, 49, 13983–13991. [Google Scholar] [CrossRef] [PubMed]
- Grover, V.P.B.; Tognarelli, J.M.; Crossey, M.M.E.; Cox, I.J.; Taylor-Robinson, S.D.; McPhail, M.J.W. Magnetic Resonance Imaging: Principles and Techniques: Lessons for Clinicians. J. Clin. Exp. Hepatol. 2015, 5, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Soares, D.P.; Law, M. Magnetic resonance spectroscopy of the brain: Review of metabolites and clinical applications. Clin. Radiol. 2009, 64, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Van der Graaf, M. In vivo magnetic resonance spectroscopy: Basic methodology and clinical applications. Eur. Biophys. J. 2010, 39, 527–540. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Rosenblum, E.S.; Tjeerdema, R.S. NMR-Based Metabolomics: A Powerful Approach for Characterizing the Effects of Environmental Stressors on Organism Health. Environ. Sci. Technol. 2003, 37, 4982–4989. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, R.G.; Shachar-Hill, Y. Probing Plant Metabolism with NMR. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 499–526. [Google Scholar] [CrossRef] [PubMed]
- Bligny, R.; Douce, R. NMR and plant metabolism. Curr. Opin. Plant Biol. 2001, 4, 191–196. [Google Scholar] [CrossRef]
- Fernie, A.R. Review: Metabolome characterisation in plant system analysis. Funct. Plant Biol. 2003, 30, 111–120. [Google Scholar] [CrossRef]
- Ward, J.L.; Baker, J.M.; Beale, M.H. Recent applications of NMR spectroscopy in plant metabolomics. FEBS J. 2007, 274, 1126–1131. [Google Scholar] [CrossRef] [PubMed]
- Yanshole, V.V.; Snytnikova, O.A.; Kiryutin, A.S.; Yanshole, L.V.; Sagdeev, R.Z.; Tsentalovich, Y.P. Metabolomics of the rat lens: A combined LC-MS and NMR study. Exp. Eye Res. 2014, 125, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Nagana Gowda, G.A.; Asiago, V.; Shanaiah, N.; Barbas, C.; Raftery, D. Correlative and quantitative 1H NMR-based metabolomics reveals specific metabolic pathway disturbances in diabetic rats. Anal. Biochem. 2008, 383, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Dona, A.C.; Kyriakides, M.; Scott, F.; Shephard, E.A.; Varshavi, D.; Veselkov, K.; Everett, J.R. A guide to the identification of metabolites in NMR-based metabonomics/metabolomics experiments. Comput. Struct. Biotechnol. J. 2016, 14, 135–153. [Google Scholar] [CrossRef] [PubMed]
- Waller, W.T.; Sherry, A.D. Whole Organism 31P Nuclear Magnetic Resonance Spectroscopy: A Potential Application in Aquatic Toxicity Evaluations. Bull. Environm. Contam. Toxicol. 1981, 26, 73–76. [Google Scholar] [CrossRef]
- Mckee, M.J.; Knowles, C.O. Protein, Nucleic Acid and Adenylate Levels in Daphnia magna During Chronic Exposure to Chlordecone. Environ. Pollut. Ser. A Ecol. Biol. 1986, 42, 335–351. [Google Scholar] [CrossRef]
- Shulman, R.G.; Rothman, D.L. Metabolomics by In Vivo NMR; John Wiley & Sons: Hoboken, NJ, USA, 2005; ISBN 0470011491. [Google Scholar]
- Grasdalen, H.; Jorgensen, L. 31P-NMR Studies on Developing Eggs and Larvae of Plaice. Comp. Biochem. Physiol. 1985, 81B, 291–294. [Google Scholar] [CrossRef]
- Majumdar, K.C.; Nasaruddin, K.; Ravinder, K.; Sundaram, C.S.; Manickam, P.; Shivaji, S. 31P Nuclear Magnetic Resonance Studies on the Phosphorus-Containing Metabolites of the Developing Embryos of a Freshwater Catfish, Clarias batrachus (L.). Mar. Biotechnol. 1999, 1, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Sartoris, F.J.; Bock, C.; Serendero, I.; Lannig, G.; Portner, H.O. Temperature-dependent changes in energy metabolism, intracellular pH and blood oxygen tension in the Atlantic cod. J. Fish Biol. 2003, 62, 1239–1253. [Google Scholar] [CrossRef]
- Kreutzer, U.; Jue, T. Metabolic response to oxygen limitation in Arenicola marina as determined with the 1H NMR signals of myoglobin. Comp. Biochem. Physiol. Part A 1998, 120, 127–132. [Google Scholar] [CrossRef]
- Viant, M.R.; Walton, J.H.; Tjeerdema, R.S. Comparative Sublethal Actions of 3-Trifluoromethyl-4-nitrophenol in Marine Molluscs as Measured by in Vivo31P NMR. Pestic. Biochem. Physiol. 2001, 71, 40–47. [Google Scholar] [CrossRef]
- Viant, M.R.; Walton, J.H.; TenBrook, P.L.; Tjeerdema, R.S. Sublethal actions of copper in abalone (Haliotis rufescens) as characterized by in vivo 31P NMR. Aquat. Toxicol. 2002, 57, 139–151. [Google Scholar] [CrossRef]
- Pincetich, C.A.; Viant, M.R.; Hinton, D.E.; Tjeerdema, R.S. Metabolic changes in Japanese medaka (Oryzias latipes) during embryogenesis and hypoxia as determined by in vivo 31P NMR. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2005, 140, 103–113. [Google Scholar] [CrossRef] [PubMed]
- Viant, M.R.; Pincetich, C.A.; Hinton, D.E.; Tjeerdema, R.S. Toxic actions of dinoseb in medaka (Oryzias latipes) embryos as determined by in vivo 31P NMR, HPLC-UV and 1H NMR metabolomics. Aquat. Toxicol. 2006, 76, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.D.; Akhter, M.; Fortier-McGill, B.; Soong, R.; Liaghati-Mobarhan, Y.; Simpson, A.J.; Spraul, M.; Schmidt, S.; Heumann, H. In Vivo Solution-State NMR-Based Environmental Metabolomics. eMagRes 2017, 6, 133–148. [Google Scholar] [CrossRef]
- Roscher, A.; Troufflard, S.; Taghki, A.I. In Vivo NMR for 13C metabolic Flux Analysis. In Plant Metabolic Flux Analysis; Humana Press: Totowa, NJ, USA, 2014; pp. 143–152. [Google Scholar]
- Simpson, A.J.; Courtier-Murias, D.; Longstaffe, J.G.; Masoom, H.; Soong, R.; Lam, L.; Sutrisno, A.; Farooq, H.; Simpson, M.J.; Maas, W.E.; et al. Environmental Comprehensive Multiphase NMR. eMagRes 2013, 2, 399–414. [Google Scholar] [CrossRef]
- Maas, W.E.; Laukien, F.H.; Cory Gradient, D.G. High Resolution, Magic Angle Sample Spinning NMR. J. Am. Chem. Soc. 1996, 118, 13085–13086. [Google Scholar] [CrossRef]
- Farooq, H.; Courtier-Murias, D.; Soong, R.; Bermel, W.; Kingery, W.M.; Simpson, A.J. HR-MAS NMR Spectroscopy: A Practical Guide for Natural Samples. Curr. Org. Chem. 2013, 17, 3013–3031. [Google Scholar] [CrossRef]
- Stark, R.E.; Yu, B.; Zhong, J.; Yan, B.; Wu, G.; Tian, S. Environmental NMR: High-resolution Magic-angle Spinning. eMagRes 2013, 2, 377–388. [Google Scholar] [CrossRef]
- Courtier-Murias, D.; Farooq, H.; Masoom, H.; Botana, A.; Soong, R.; Longstaffe, J.G.; Simpson, M.J.; Maas, W.E.; Fey, M.; Andrew, B.; et al. Comprehensive multiphase NMR spectroscopy: Basic experimental approaches to differentiate phases in heterogeneous samples. J. Magn. Reson. 2012, 217, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Bon, D.; Gilard, V.; Massou, S.; Pérès, G.; Malet-Martino, M.; Martino, R.; Desmoulin, F. In vivo 31P and 1H HR-MAS NMR spectroscopy analysis of the unstarved Aporrectodea caliginosa (Lumbricidae). Biol. Fertil. Soils 2006, 43, 191–198. [Google Scholar] [CrossRef]
- Bunescu, A.; Garric, J.; Vollat, B.; Canet-Soulas, E.; Graveron-Demilly, D.; Fauvelle, F. In vivo proton HR-MAS NMR metabolic profile of the freshwater cladoceran Daphnia magna. Mol. Biosyst. 2009, 6, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Constantinou, C.; Apidianakis, Y.; Psychogios, N.; Righi, V.; Mindrinos, M.N.; Khan, N.; Swartz, H.M.; Szeto, H.H.; Tompkins, R.G.; Rahme, L.G.; et al. In vivo high-resolution magic angle spinning magnetic and electron paramagnetic resonance spectroscopic analysis of mitochondria-targeted peptide in Drosophila melanogaster with trauma-induced thoracic injury. Int. J. Mol. Med. 2016, 37, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Righi, V.; Apidianakis, Y.; Mintzopoulos, D.; Astrakas, L.; Rahme, L.G.; Tzika, A.A. In vivo high-resolution magic angle spinning magnetic resonance spectroscopy of Drosophila melanogaster at 14.1 T shows trauma in aging and in innate immune-deficiency is linked to reduced insulin signaling. Int. J. Mol. Med. 2010, 26, 175–184. [Google Scholar] [PubMed]
- Righi, V.; Apidianakis, Y.; Psychogios, N.; Rahme, L.G.; Tompkins, R.G.; Tzika, A.A. In vivo high-resolution magic angle spinning proton NMR spectroscopy of Drosophila melanogaster flies as a model system to investigate mitochondrial dysfunction in Drosophila GST2 mutants. Int. J. Mol. Med. 2014, 34, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Sarou-Kanian, V.; Joudiou, N.; Louat, F.; Yon, M.; Szeremeta, F.; Même, S.; Massiot, D.; Decoville, M.; Fayon, F.; Beloeil, J.-C. Metabolite localization in living drosophila using High Resolution Magic Angle Spinning NMR. Sci. Rep. 2015, 5, 9872. [Google Scholar] [CrossRef] [PubMed]
- Blaise, B.J.; Giacomotto, J.; Bénédicte, E.; Dumas, M.-E.; Toulhoat, P.; Ségalat, L.; Emsley, L. Metabotyping of Caenorhabditis elegans reveals latent phenotypes. Proc. Natl. Acad. Sci. USA 2007, 104, 19808–19812. [Google Scholar] [CrossRef] [PubMed]
- Hansen, B.H.; Altin, D.; Booth, A.; Vang, S.-H.; Frenzel, M.; Sørheim, K.R.; Brakstad, O.G.; Størseth, T.R. Molecular effects of diethanolamine exposure on Calanus finmarchicus (Crustacea: Copepoda). Aquat. Toxicol. 2010, 99, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Wind, R.A.; Hu, J.Z.; Rommereim, D.N. High-resolution 1H NMR spectroscopy in a live mouse subjected to 1.5 Hz magic angle spinning. Magn. Reson. Med. 2003, 50, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Augustijn, D.; Roy, U.; van Schadewijk, R.; De Groot, H.J.M.; Alia, A. Metabolic Profiling of Intact Arabidopsis thaliana Leaves during Circadian Cycle Using 1H High Resolution Magic Angle Spinning NMR. PLoS ONE 2016, 11, e0163258. [Google Scholar] [CrossRef] [PubMed]
- Bondu, S.; Kervarec, N.; Deslandes, E.; Pichon, R. The use of HRMAS NMR spectroscopy to study the in vivo intra–cellular carbon/nitrogen ratio of Solieria chordalis (Rhodophyta). J. Appl. Phycol. 2008, 20, 673–679. [Google Scholar] [CrossRef]
- Hinse, C.; Richter, C.; Provenzani, A.; Stöckigt, J. In vivo monitoring of alkaloid metabolism in hybrid plant cell cultures by 2D cryo-NMR without labelling. Bioorg. Med. Chem. 2003, 11, 3913–3919. [Google Scholar] [CrossRef]
- Singh, H.; Shukla, M.R.; Chary, K.V.R.; Rao, B.J. Acetate and bicarbonate assimilation and metabolite formation in Chlamydomonas reinhardtii: A 13C-NMR study. PLoS ONE 2014, 9, e106457. [Google Scholar] [CrossRef] [PubMed]
- Simpson, A.J.; Mcnally, D.J.; Simpson, M.J. NMR spectroscopy in environmental research: From molecular interactions to global processes. Prog. Nucl. Magn. Reson. Spectrosc. 2011, 58, 97–175. [Google Scholar] [CrossRef] [PubMed]
- Lam, L.; Soong, R.; Sutrisno, A.; De Visser, R.; Simpson, M.J.; Wheeler, H.L.; Campbell, M.; Maas, W.E.; Fey, M.; Gorissen, A.; et al. Comprehensive Multiphase NMR Spectroscopy of Intact 13C-Labeled Seeds. J. Agric. Food Chem. 2014, 62, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Farooq, H.; Courtier-Murias, D.; Simspon, M.J.; Maas, W.E.; Fey, M.; Andrew, B.; Struppe, J.; Hutchins, H.; Krishnamurthy, S.; Kumar, R.; et al. Characterisation of oil contaminated soils by comprehensive multiphase NMR spectroscopy. Environ. Chem. 2015, 12, 227–235. [Google Scholar] [CrossRef]
- Gagné, F.; Blaise, C.; Pellerin, J. Altered exoskeleton composition and vitellogenesis in the crustacean Gammarus sp. collected at polluted sites in the Saguenay Fjord, Quebec, Canada. Environ. Res. 2005, 98, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Fortier-McGill, B.E.; Dutta Majumdar, R.; Lam, L.; Soong, R.; Liaghati-Mobarhan, Y.; Sutrisno, A.; de Visser, R.; Simpson, M.J.; Wheeler, H.L.; Campbell, M.; et al. Comprehensive Multiphase (CMP) NMR Monitoring of the Structural Changes and Molecular Flux Within a Growing Seed. J. Agric. Food Chem. 2017, 65, 6779–6788. [Google Scholar] [CrossRef] [PubMed]
- Science and Technology Branch. Biological Test Method: Test for Survival and Growth in Sediment and Water Using the Freshwater Amphipod Hyalella Azteca, 2nd ed.; Environment Canada: Gatineau, QC, Canada, 2013; ISBN 978-1-10-021674-4. [Google Scholar]
- Masoom, H.; Adamo, A.A.; Andr, A.; Simpson, J. From the environment to NMR: Water suppression for whole samples in their native state. Environ. Chem. 2016, 13, 767–775. [Google Scholar] [CrossRef]
- Giraudeau, P.; Silvestre, V.; Akoka, S. Optimizing water suppression for quantitative NMR-based metabolomics: A tutorial review. Metabolomics 2015, 11, 1041–1055. [Google Scholar] [CrossRef]
- Lam, B.; Simpson, A.J. Direct 1H NMR spectroscopy of dissolved organic matter in natural waters. Analyst 2008, 133, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Pautler, B.G.; Simpson, A.J.; Simpson, M.J.; Tseng, L.-H.; Spraul, M.; Dubnick, A.; Sharp, M.J.; Fitzsimons, S.J. Detection and Structural Identification of Dissolved Organic Matter in Antarctic Glacial Ice at Natural Abundance by SPR-W5-WATERGATE 1H NMR Spectroscopy. Environ. Sci. Technol. 2011, 45, 4710–4717. [Google Scholar] [CrossRef] [PubMed]
- Pautler, B.G.; Woods, G.C.; Dubnick, A.; Simpson, A.J.; Sharp, M.J.; Fitzsimons, S.J.; Simpson, M.J. Molecular Characterization of Dissolved Organic Matter in Glacial Ice: Coupling Natural Abundance 1H NMR and Fluorescence Spectroscopy. Environ. Sci. Technol. 2012, 46, 3753–3761. [Google Scholar] [CrossRef] [PubMed]
- Dutta Majumdar, R.; Bliumkin, L.; Lane, D.; Soong, R.; Simpson, M.; Simpson, A.J. Analysis of DOM phototransformation using a looped NMR system integrated with a sunlight simulator. Water Res. 2017, 120, 64–76. [Google Scholar] [CrossRef] [PubMed]
- Fugariu, I.; Bermel, W.; Lane, D.; Soong, R.; Simpson, A.J. In-Phase Ultra High-Resolution In Vivo NMR. Angew. Chem. Int. Ed. 2017, 56, 6324–6328. [Google Scholar] [CrossRef] [PubMed]
- Vathyam, S.; Lee, S.; Warren, W.S. Homogeneous NMR Spectra in Inhomogeneous Fields. Science 1996, 272, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Richter, W.; Warren, W.S. Intermolecular multiple quantum coherences in liquids. Concepts Magn. Reson. 2000, 12, 396–409. [Google Scholar] [CrossRef]
- Sekiyama, Y.; Chikayama, E.; Kikuchi, J. Evaluation of a Semipolar Solvent System as a Step toward Heteronuclear Multidimensional NMR-Based Metabolomics for 13C-Labeled Bacteria, Plants, and Animals. Anal. Chem. 2011, 83, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Hertkorn, N.; Ruecker, C.; Meringer, M.; Gugisch, R.; Frommberger, M.; Perdue, E.M.; Witt, M.; Schmitt-Kopplin, P. High-precision frequency measurements: Indispensable tools at the core of the molecular-level analysis of complex systems. Anal. Bioanal. Chem. 2007, 389, 1311–1327. [Google Scholar] [CrossRef] [PubMed]
- Woods, G.C.; Simpson, M.J.; Koerner, P.J.; Napoli, A.; Simpson, A.J. HILIC-NMR: Toward the Identification of Individual Molecular Components in Dissolved Organic Matter. Environ. Sci. Technol. 2011, 45, 3880–3886. [Google Scholar] [CrossRef] [PubMed]
- Ellinger, J.J.; Chylla, R.A.; Ulrich, E.L.; Markley, J.L. Databases and Software for NMR-Based Metabolomics. Curr. Metabol. 2013, 1. [Google Scholar] [CrossRef]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Eisner, R.; Young, N.; Gautam, B.; Hau, D.D.; Psychogios, N.; Dong, E.; Bouatra, S.; et al. HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 2009, 37, D603–D610. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2012, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Feunang, Y.D.; Marcu, A.; Guo, A.C.; Liang, K.; Vázquez-Fresno, R.; Sajed, T.; Johnson, D.; Li, C.; Karu, N.; et al. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 2018, 46, D608–D617. [Google Scholar] [CrossRef] [PubMed]
- Warren, W.S.; Richter, W.; Andreotti, A.H.; Farmer, B.T. Generation of impossible cross-peaks between bulk water and biomolecules in solution NMR. Science 1993, 262, 2005–2009. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, Z.; Zhong, J. High-Resolution NMR Spectra in Inhomogeneous Fields via IDEAL (Intermolecular Dipolar-Interaction Enhanced All Lines) Method. J. Am. Chem. Soc. 2004, 126, 446–447. [Google Scholar] [CrossRef] [PubMed]
- Branca, R.T. MRI using intermolecular multiple-quantum coherences. Methods Mol. Biol. 2011, 771, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.-M.; Lane, A.N. Structure-based profiling of metabolites and isotopomers by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2008, 52, 69–117. [Google Scholar] [CrossRef]
- Li, Y.; Wolters, A.M.; Malawey, P.V.; Sweedler, J.V.; Webb, A.G. Multiple Solenoidal Microcoil Probes for High-Sensitivity, High-Throughput Nuclear Magnetic Resonance Spectroscopy. Anal. Chem. 1999, 71, 4815–4820. [Google Scholar] [CrossRef] [PubMed]
- Alekseev, V.; Lampert, W. Maternal control of resting-egg production in Daphnia. Nature 2001, 414, 899–901. [Google Scholar] [CrossRef] [PubMed]
- Navis, S.; Waterkeyn, A.; Voet, T.; De, L.; bullet, M.; Brendonck, L. Pesticide exposure impacts not only hatching of dormant eggs, but also hatchling survival and performance in the water flea Daphnia magna. Ecotoxicology 2013, 22, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Shirzadi, A.; Simpson, M.J.; Xu, Y.; Simpson, A.J. Application of Saturation Transfer Double Difference NMR to Elucidate the Mechanistic Interactions of Pesticides with Humic Acid. Environ. Sci. Technol. 2008, 42, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Meyer, B. Characterization of Ligand Binding by Saturation Transfer Difference NMR Spectroscopy. Angew. Chem. Int. Ed. 1999, 38, 1784–1788. [Google Scholar] [CrossRef]
- D’eon, J.C.; Simpson, A.J.; Kumar, R.; Baer, A.J.; Mabury, S.A. Determining the molecular interactions of perfluorinated carboxylic acids with human sera and isolated human serum albumin using nuclear magnetic resonance spectroscopy. Environ. Toxicol. Chem. 2010, 29, 1678–1688. [Google Scholar] [CrossRef] [PubMed]
- Longstaffe, J.G.; Simpson, M.J.; Maas, W.; Simpson, A.J. Identifying Components in Dissolved Humic Acid That Bind Organofluorine Contaminants using 1H{19F} Reverse Heteronuclear Saturation Transfer Difference NMR Spectroscopy. Environ. Sci. Technol. 2010, 44, 5476–5482. [Google Scholar] [CrossRef] [PubMed]
- Krüger, M.; Moser, M.; Ussar, S.; Thievessen, I.; Luber, C.A.; Forner, F.; Schmidt, S.; Zanivan, S.; Fässler, R.; Mann, M. SILAC Mouse for Quantitative Proteomics Uncovers Kindlin-3 as an Essential Factor for Red Blood Cell Function. Cell 2008, 134, 353–364. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Lee, J.-W.; Kim, K.; Shin, Y.-J.; Kim, J.; Kim, S.; Kim, H.; Kim, P.; Park, K. PFOA-induced metabolism disturbance and multi-generational reproductive toxicity in Oryzias latipes. J. Hazard. Mater. 2017, 340, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Niu, J.; Li, Y.; Wang, Y.; Sun, D. Evaluating the sub-lethal toxicity of PFOS and PFOA using rotifer Brachionus calyciflorus. Environ. Pollut. 2013, 180, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Li, M.H. Toxicity of perfluorooctane sulfonate and perfluorooctanoic acid to plants and aquatic invertebrates. Environ. Toxicol. 2009, 24, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Webster, G. Potential Human Health Effects of Perfluorinated Chemicals (PFCs); National Collaborating Centre for Environmental Health: Winnipeg, MB, Canada, 2010. [Google Scholar]
- Musse, M.; Leport, L.; Cambert, M.; Debrandt, W.; Sorin, C.; Bouchereau, A.; Mariette, F. A mobile NMR lab for leaf phenotyping in the field. Plant Methods 2017, 13, 53. [Google Scholar] [CrossRef] [PubMed]
- Windt, C.W.; Blümler, P. A portable NMR sensor to measure dynamic changes in the amount of water in living stems or fruit and its potential to measure sap flow. Tree Physiol. 2015, 35, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Yoder, J.; Malone, M.W.; Espy, M.A.; Sevanto, S. Low-field nuclear magnetic resonance for the in vivo study of water content in trees. Rev. Sci. Instrum. 2014, 85, 95110. [Google Scholar] [CrossRef] [PubMed]
- Malone, M.W.; Yoder, J.; Hunter, J.F.; Espy, M.A.; Dickman, L.T.; Nelson, R.O.; Vogel, S.C.; Sandin, H.J.; Sevanto, S. In vivo Observation of Tree Drought Response with Low-Field NMR and Neutron Imaging. Front. Plant Sci. 2016, 7, 564. [Google Scholar] [CrossRef] [PubMed]
- Berns, A.E.; Bubici, S.; De Pasquale, C.; Alonzo, G.; Conte, P. Applicability of solid state fast field cycling NMR relaxometry in understanding relaxation properties of leaves and leaf-litters. Org. Geochem. 2011, 42, 978–984. [Google Scholar] [CrossRef]
- Borisjuk, L.; Rolletschek, H.; Fuchs, J.; Melkus, G.; Neuberger, T. Low and High Field Magnetic Resonance for in Vivo Analysis of Seeds. Materials 2011, 4, 1426–1439. [Google Scholar] [CrossRef] [PubMed]
- Tonthat, D.M.; Augustine, M.P.; Pines, A.; Clarke, J. Low magnetic field dynamic nuclear polarization using a single-coil two-channel probe. Rev. Sci. Instrum. 1997, 68, 1527–1531. [Google Scholar] [CrossRef]
- Blanchard, J.W.; Ledbetter, M.P.; Theis, T.; Butler, M.C.; Budker, D.; Pines, A. High-Resolution Zero-Field NMR J-Spectroscopy of Aromatic Compounds. J. Am. Chem. Soc. 2013, 135, 3607–3612. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).