An Overview of the Bacterial Carbonic Anhydrases
Abstract
1. Introduction
2. Classification and Structure
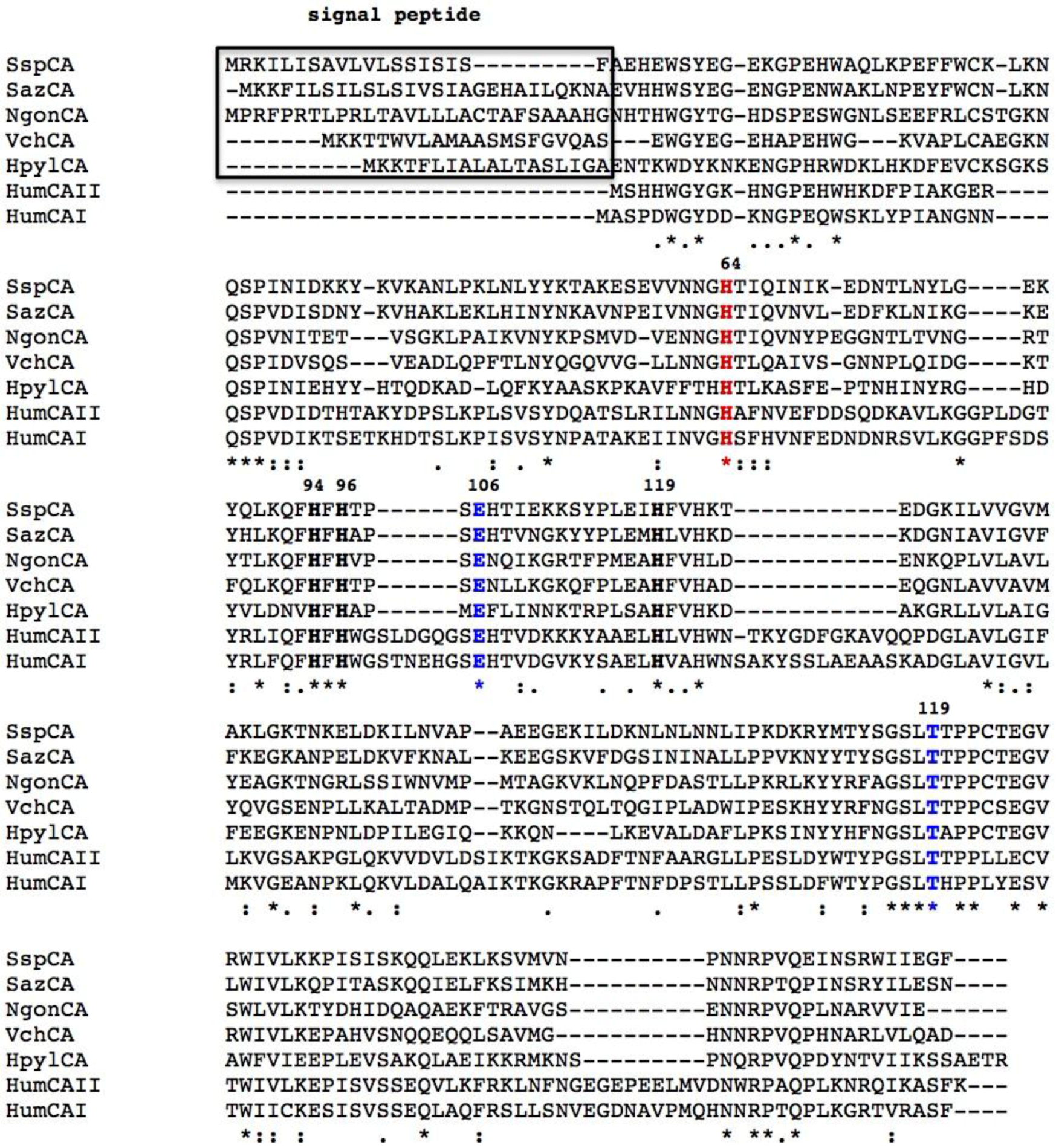
2.1. CA-Classes
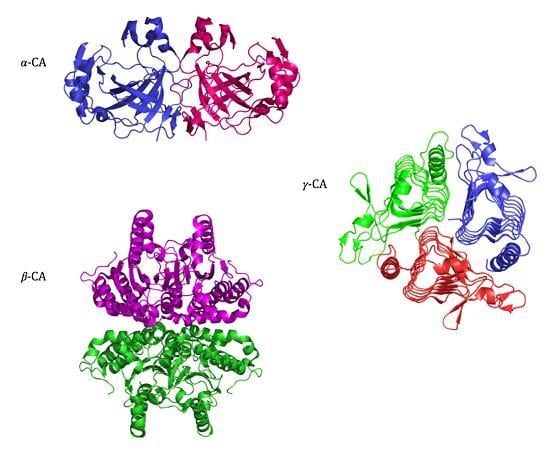
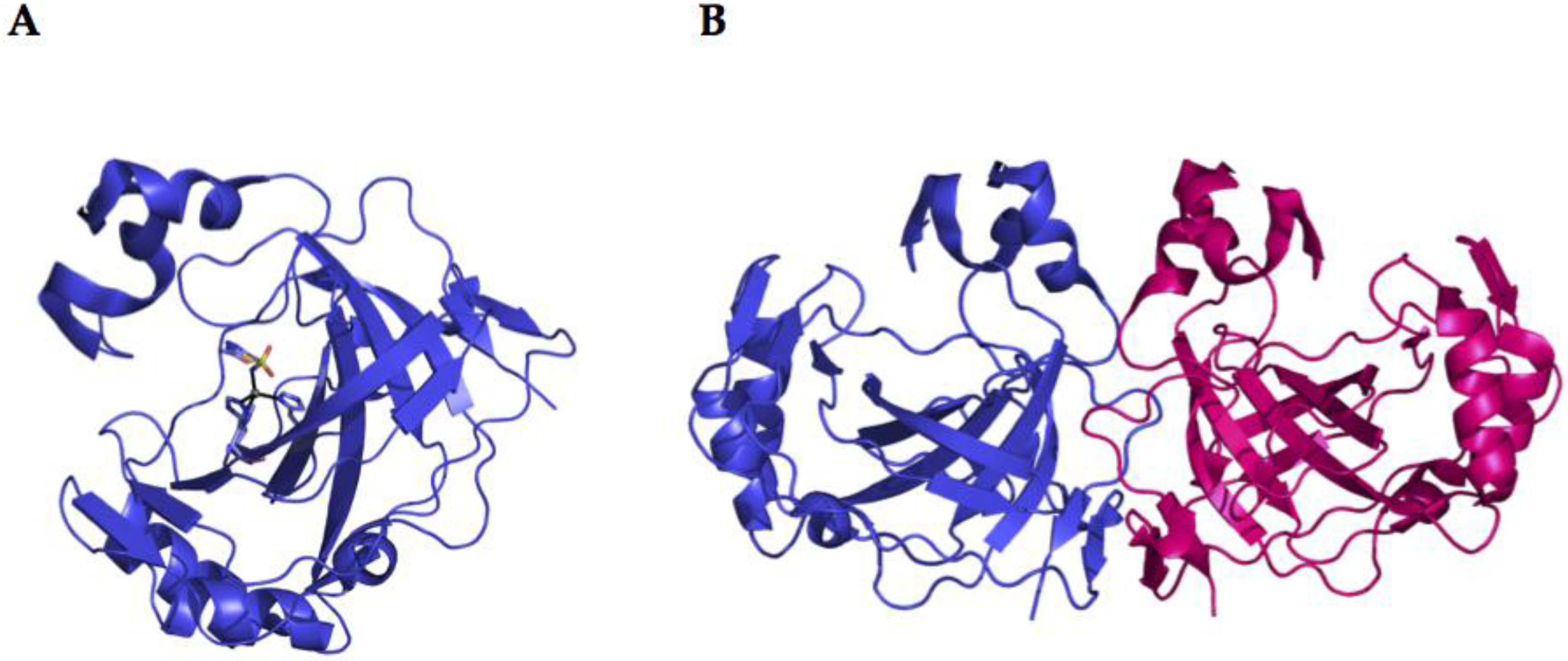
2.2. α-CA Structure
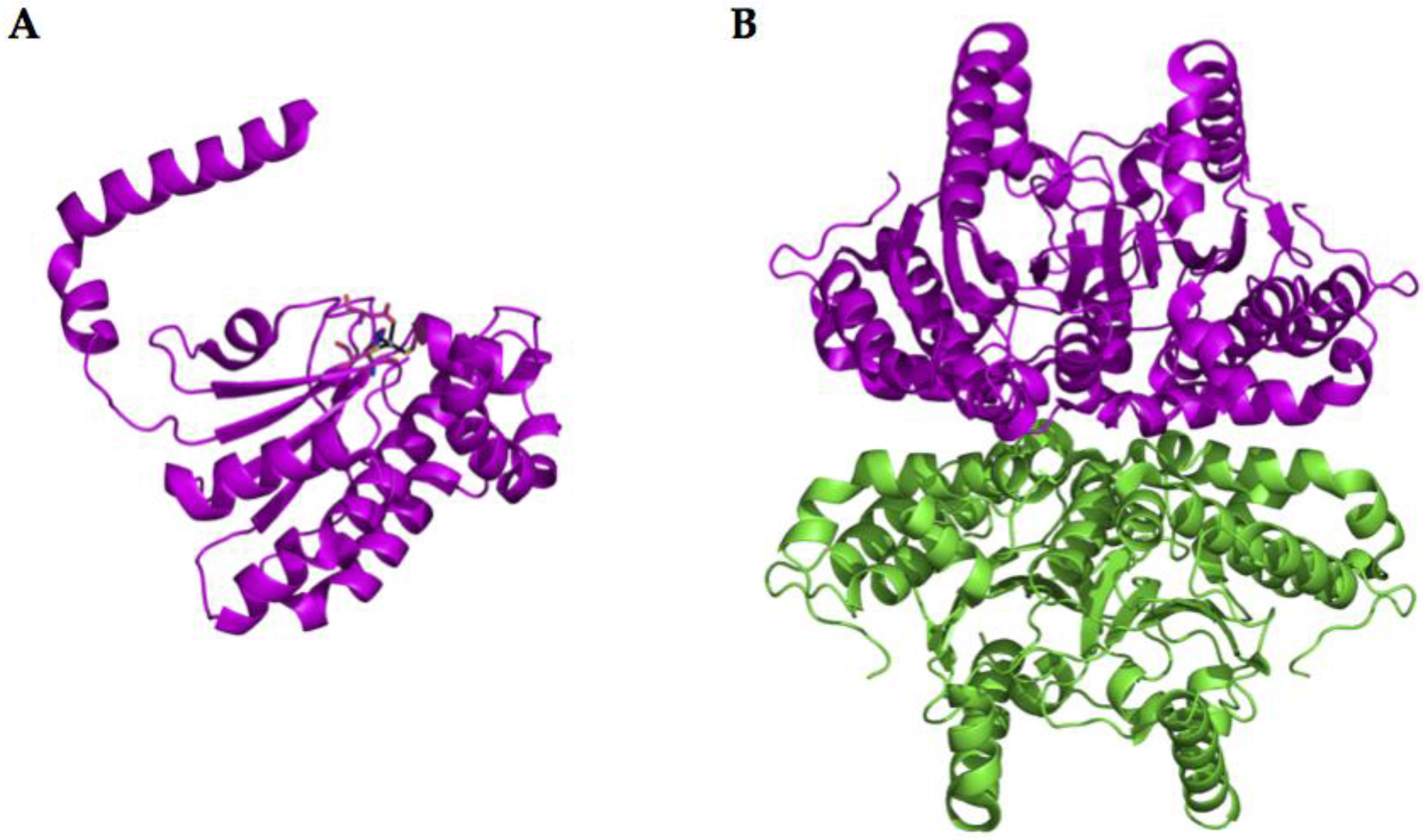
2.3. β-CA Structure
2.4. γ-CA Structure
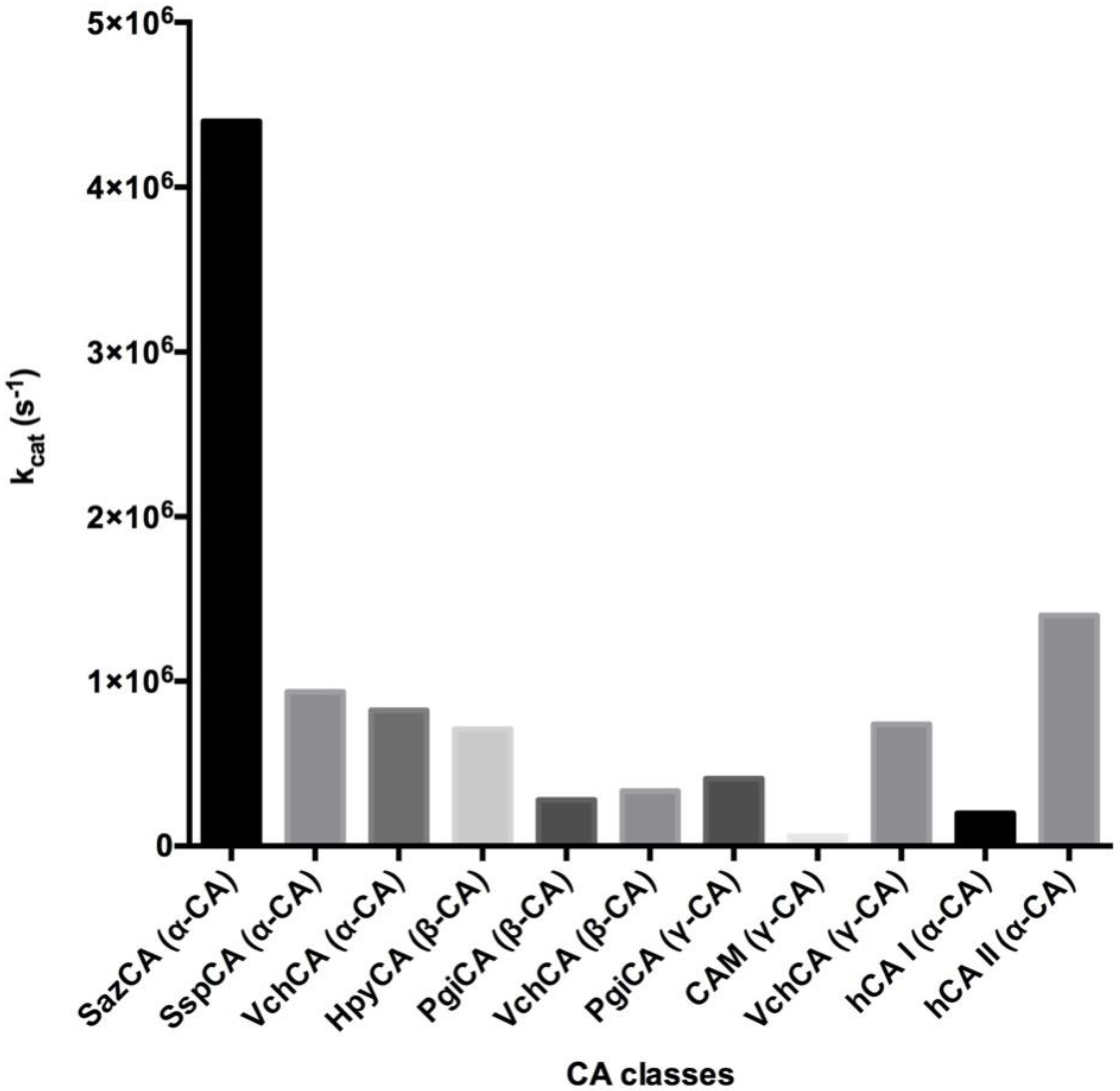
3. Catalytic Activity
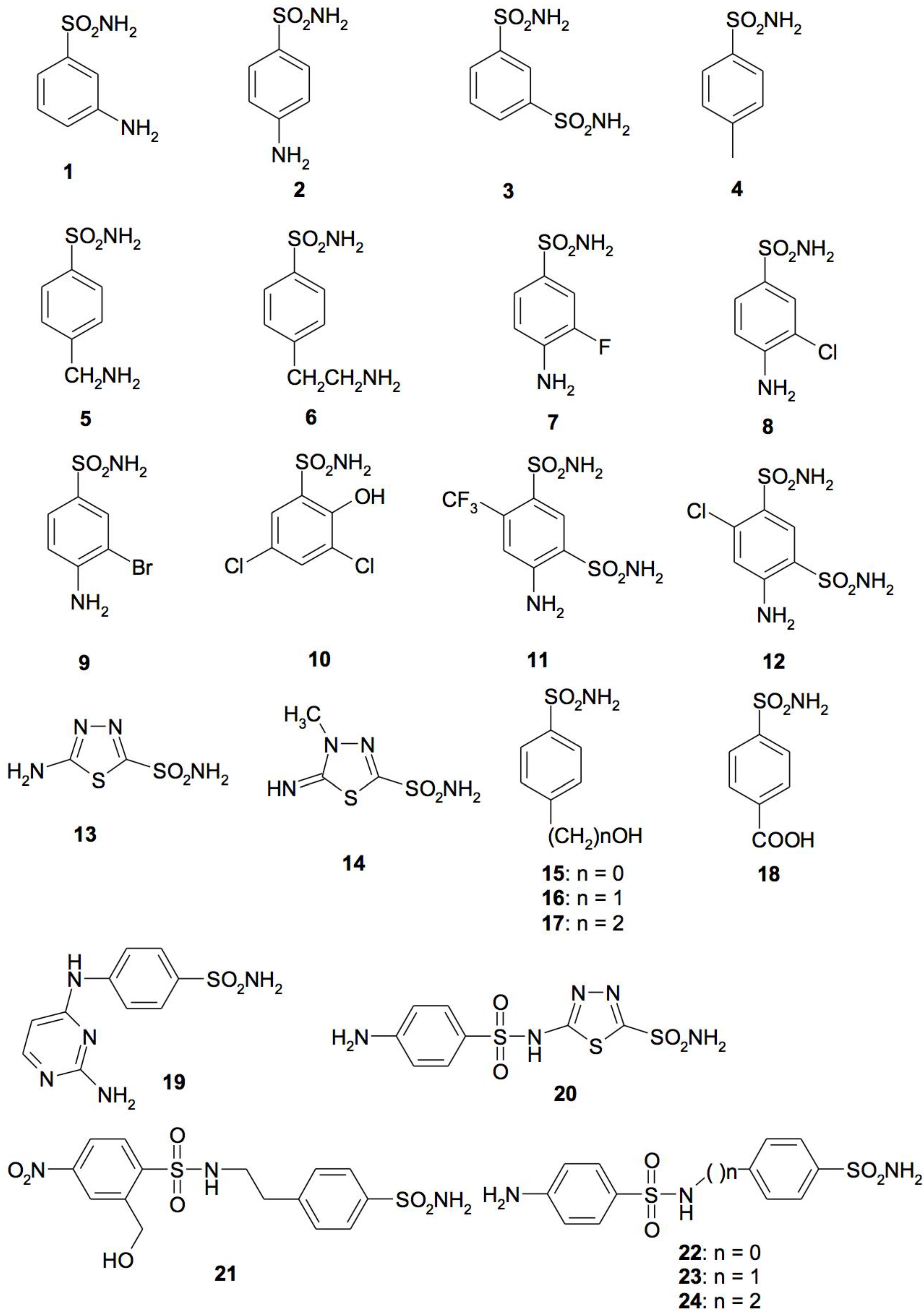
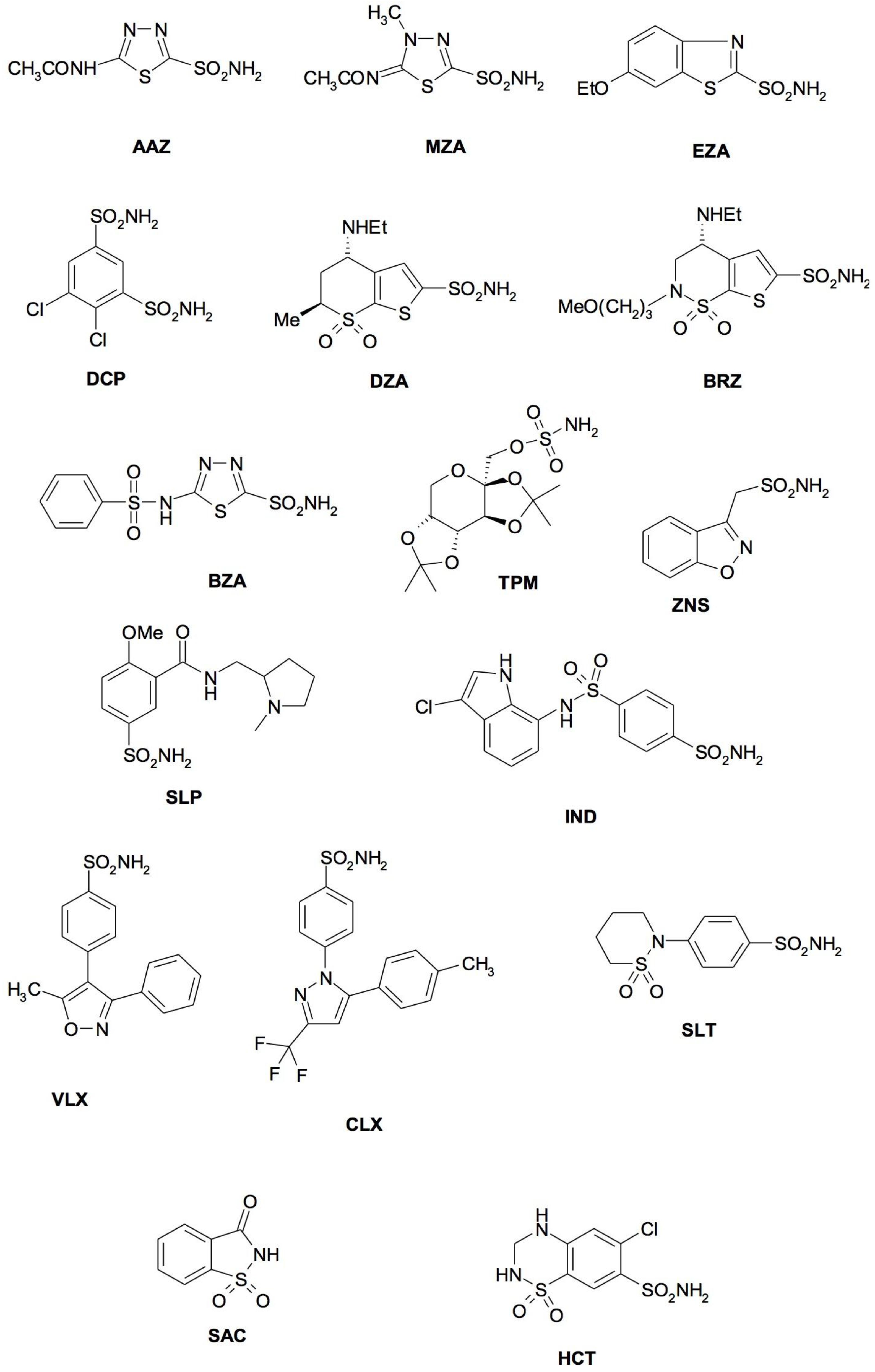
4. CA Inhibitors
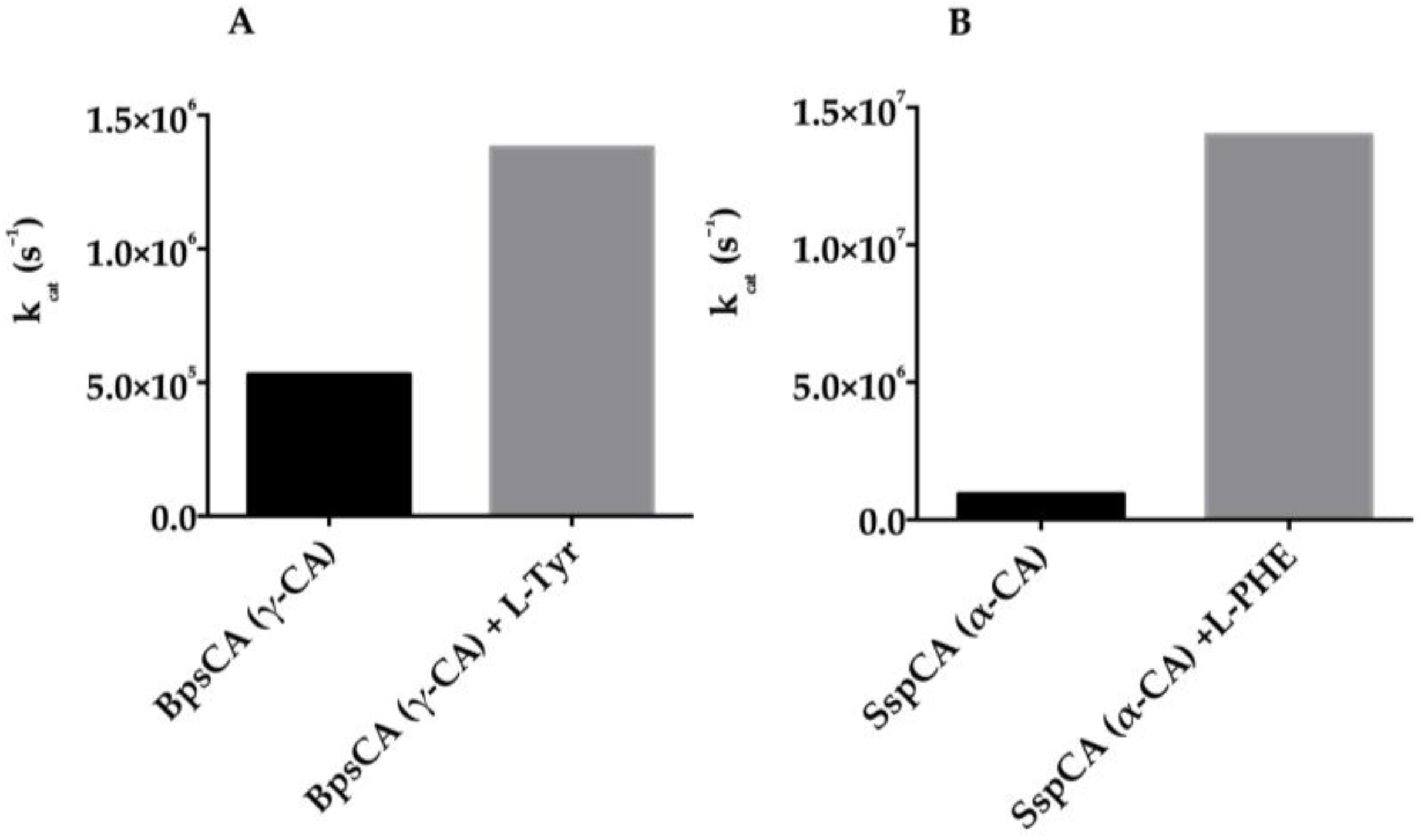
5. Activators
6. Phylogenetic Analysis
7. Localization and Physiological Role
8. Engineered Bacteria with a Thermostable CA for CO2 Capture
9. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Turk, V.E.; Simic, I.; Likic, R.; Makar-Ausperger, K.; Radacic-Aumiler, M.; Cegec, I.; Juricic Nahal, D.; Kraljickovic, I. New drugs for bad bugs: What’s new and what’s in the pipeline? Clin. Ther. 2016, 38, e9. [Google Scholar] [CrossRef] [PubMed]
- Decker, B.; Masur, H. Bad bugs, no drugs: Are we part of the problem, or leaders in developing solutions? Crit. Care Med. 2015, 43, 1153–1155. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug. Dis. 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Annunziato, G.; Angeli, A.; D’Alba, F.; Bruno, A.; Pieroni, M.; Vullo, D.; De Luca, V.; Capasso, C.; Supuran, C.T.; Costantino, G. Discovery of new potential anti-infective compounds based on carbonic anhydrase inhibitors by rational target-focused repurposing approaches. Chem. Med. Chem. 2016, 11, 1904–1914. [Google Scholar] [CrossRef] [PubMed]
- Ozensoy Guler, O.; Capasso, C.; Supuran, C.T. A magnificent enzyme superfamily: Carbonic anhydrases, their purification and characterization. J. Enzyme Inhibi. Med. Chem. 2016, 31, 689–694. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; Ferraroni, M.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Sulfonamide inhibition studies of the beta-carbonic anhydrase from the Pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. 2016, 24, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; De Luca, V.; De Simone, G.; Supuran, C.T.; Capasso, C. Cloning, expression and purification of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. J. Enzyme Inhib. Med. Chem. 2016, 31, 54–59. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr. Top. Med. Chem. 2016, 16, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibitors. Bioorg. Med. Chem. Lett. 2010, 20, 3467–3474. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Cloning, expression, purification and sulfonamide inhibition profile of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg. Med. Chem. Lett. 2016, 26, 4184–4190. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; Di Fonzo, P.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Anion inhibition profiles of the complete domain of the η-carbonic anhydrase from Plasmodium falciparum. Bioorg. Med. Chem. 2016, 24, 4410–4414. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; di Fonzo, P.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Anion inhibition profiles of α-, β- and γ-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. 2016, 24, 3413–3417. [Google Scholar] [CrossRef] [PubMed]
- Abdel Gawad, N.M.; Amin, N.H.; Elsaadi, M.T.; Mohamed, F.M.; Angeli, A.; De Luca, V.; Capasso, C.; Supuran, C.T. Synthesis of 4-(thiazol-2-ylamino)-benzenesulfonamides with carbonic anhydrase I, II and IX inhibitory activity and cytotoxic effects against breast cancer cell lines. Bioorg. Med. Chem. 2016, 24, 3043–3051. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; De Luca, V.; Carginale, V.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Comparison of the sulfonamide inhibition profiles of the α-, β- and γ -carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg. Med Chem. Lett. 2016, 26, 1941–1946. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, I.; Onishi, S.; Takeuchi, H.; Supuran, C.T. The α and β classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr. Pharm. Des. 2008, 14, 622–630. [Google Scholar] [PubMed]
- Morishita, S.; Nishimori, I.; Minakuchi, T.; Onishi, S.; Takeuchi, H.; Sugiura, T.; Vullo, D.; Scozzafava, A.; Supuran, C.T. Cloning, polymorphism, and inhibition of β-carbonic anhydrase of Helicobacter pylori. J. Gastroenterol. 2008, 43, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Cheah, W.Y.; Ling, T.C.; Juan, J.C.; Lee, D.J.; Chang, J.S.; Show, P.L. Biorefineries of carbon dioxide: From carbon capture and storage (CCS) to bioenergies production. Bioresour. Technol. 2016, 215, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Migliardini, F.; De Luca, V.; Carginale, V.; Rossi, M.; Corbo, P.; Supuran, C.T.; Capasso, C. Biomimetic CO2 capture using a highly thermostable bacterial α-carbonic anhydrase immobilized on a polyurethane foam. J. Enzyme Inhib. Med. Chem. 2014, 29, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Perfetto, R.; Del Prete, S.; Vullo, D.; Sansone, G.; Barone, C.; Rossi, M.; Supuran, C.T.; Capasso, C. Biochemical characterization of the native α-carbonic anhydrase purified from the mantle of the Mediterranean mussel, Mytilus galloprovincialis. J. Enzyme Inhib. Med. Chem. 2017, 32, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Perfetto, R.; Del Prete, S.; Vullo, D.; Sansone, G.; Barone, C.M.A.; Rossi, M.; Supuran, C.T.; Capasso, C. Production and covalent immobilisation of the recombinant bacterial carbonic anhydrase (SspCA) onto magnetic nanoparticles. J. Enzyme Inhib. Med. Chem. 2017, 32, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; De Luca, V.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. The first activation study of a bacterial carbonic anhydrase (CA). The thermostable α-CA from Sulfurihydrogenibium yellowstonense YO3AOP1 is highly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2012, 22, 6324–6327. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin. Ther. Targets 2015, 19, 1689–1704. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; Capasso, C. The η-class carbonic anhydrases as drug targets for antimalarial agents. Exp. Opin. Ther. Targets 2015, 19, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. An Overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr. Med. Chem. 2015, 22, 2130–2139. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. An overview of the α-, β- and γ-carbonic anhydrases from Bacteria: Can bacterial carbonic anhydrases shed new light on evolution of bacteria? J. Enzyme Inhib. Med. Chem. 2015, 30, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Sulfa and trimethoprim-like drugs-antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J. Enzyme Inhib. Med. Chem. 2014, 29, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Anti-infective carbonic anhydrase inhibitors: A patent and literature review. Exp. Opin. Ther. Pat. 2013, 23, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Pinard, M.A.; Lotlikar, S.R.; Boone, C.D.; Vullo, D.; Supuran, C.T.; Patrauchan, M.A.; McKenna, R. Structure and inhibition studies of a type II beta-carbonic anhydrase psCA3 from Pseudomonas aeruginosa. Bioorg. Med. Chem. 2015, 23, 4831–4838. [Google Scholar] [CrossRef] [PubMed]
- Ferraroni, M.; Del Prete, S.; Vullo, D.; Capasso, C.; Supuran, C.T. Crystal structure and kinetic studies of a tetrameric type II beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr. Sect. D Biolog. Crystallogr. 2015, 71, 2449–2456. [Google Scholar] [CrossRef] [PubMed]
- De Simone, G.; Monti, S.M.; Alterio, V.; Buonanno, M.; De Luca, V.; Rossi, M.; Carginale, V.; Supuran, C.T.; Capasso, C.; Di Fiore, A. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg. Med. Chem. Lett. 2015, 25, 2002–2006. [Google Scholar] [CrossRef] [PubMed]
- Zolnowska, B.; Slawinski, J.; Pogorzelska, A.; Chojnacki, J.; Vullo, D.; Supuran, C.T. Carbonic anhydrase inhibitors. synthesis, and molecular structure of novel series N-substituted N’-(2-arylmethylthio-4-chloro-5-methylbenzenesulfonyl)guanidines and their inhibition of human cytosolic isozymes I and II and the transmembrane tumor-associated isozymes IX and XII. Euro. J. Med. Chem. 2014, 71, 135–147. [Google Scholar]
- De Luca, L.; Ferro, S.; Damiano, F.M.; Supuran, C.T.; Vullo, D.; Chimirri, A.; Gitto, R. Structure-based screening for the discovery of new carbonic anhydrase VII inhibitors. Euro. J. Med. Chem. 2014, 71, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; Capasso, C.; De Luca, V.; Monti, S.M.; Carginale, V.; Supuran, C.T.; Scozzafava, A.; Pedone, C.; Rossi, M.; De Simone, G. X-ray structure of the first ‘extremo-alpha-carbonic anhydrase’, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Structure-based drug discovery of carbonic anhydrase inhibitors. J. Enzyme Inhib. Med. Chem. 2012, 27, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases—An overview. Curr. Pharm. Des. 2008, 14, 603–614. [Google Scholar] [CrossRef]
- Supuran, C.T. Structure and function of carbonic anhydrases. Biochem. J. 2016, 473, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Buzas, G.M.; Supuran, C.T. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puscas (1932–2015). J. Enzyme Inhib. Med. Chem. 2016, 31, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Carta, F.; Supuran, C.T.; Scozzafava, A. Sulfonamides and their isosters as carbonic anhydrase inhibitors. Future Med. Chem. 2014, 6, 1149–1165. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Dis. 2008, 7, 168–181. [Google Scholar] [CrossRef] [PubMed]
- James, P.; Isupov, M.N.; Sayer, C.; Saneei, V.; Berg, S.; Lioliou, M.; Kotlar, H.K.; Littlechild, J.A. The structure of a tetrameric alpha-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides that contribute to its thermostability. Acta Crystallogr. Sect. D Biol. Crystallogr. 2014, 70, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Xue, Y.; Sauer-Eriksson, E.; Chirica, L.; Lindskog, S.; Jonsson, B.H. Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide. J. Mol. Biol. 1998, 283, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Cronk, J.D.; O’Neill, J.W.; Cronk, M.R.; Endrizzi, J.A.; Zhang, K.Y. Cloning, crystallization and preliminary characterization of a β-carbonic anhydrase from Escherichia coli. Acta Crystallogr. Sec. D Biol. Crystallogr. 2000, 56, 1176–1179. [Google Scholar] [CrossRef]
- Rowlett, R.S.; Tu, C.; Lee, J.; Herman, A.G.; Chapnick, D.A.; Shah, S.H.; Gareiss, P.C. Allosteric site variants of Haemophilus influenzae β-carbonic anhydrase. Biochemistry 2009, 48, 6146–6156. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias, A.S.; Bergfors, T.; Jones, T.A.; Hogbom, M. Structural mechanics of the pH-dependent activity of β-carbonic anhydrase from Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 4993–4999. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, I.; Minakuchi, T.; Vullo, D.; Scozzafava, A.; Supuran, C.T. Inhibition studies of the β-carbonic anhydrases from the bacterial pathogen Salmonella enterica serovar Typhimurium with sulfonamides and sulfamates. Bioorg. Med. Chem. 2011, 19, 5023–5030. [Google Scholar] [CrossRef] [PubMed]
- Kisker, C.; Schindelin, H.; Alber, B.E.; Ferry, J.G.; Rees, D.C. A left-hand β-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 1996, 15, 2323–2330. [Google Scholar] [PubMed]
- Supuran, C.T. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Exp. Opin. Drug Dis. 2017, 12, 61–88. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. How many carbonic anhydrase inhibition mechanisms exist? J. Enzyme Inhib. Med. Chem. 2016, 31, 345–360. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; De Luca, V.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. Anion inhibition studies of the fastest carbonic anhydrase (CA) known, the extremo-CA from the bacterium Sulfurihydrogenibium azorense. Bioorg. Med. Chem. Lett. 2012, 22, 7142–7145. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Inhibition of bacterial carbonic anhydrases as a novel approach to escape drug resistance. Curr. Top. Med. Chem. 2017, 17, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibition and the management of neuropathic pain. Exp. Rev. Neurother 2016, 16, 961–968. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Drug interaction considerations in the therapeutic use of carbonic anhydrase inhibitors. Exp. Opin. Drug Metab. Toxicol. 2016, 12, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Otten, H. Domagk and the development of the sulphonamides. J. Antimicrob. Chem. 1986, 17, 689–696. [Google Scholar] [CrossRef]
- Achari, A.; Somers, D.O.; Champness, J.N.; Bryant, P.K.; Rosemond, J.; Stammers, D.K. Crystal structure of the anti-bacterial sulfonamide drug target dihydropteroate synthase. Nat. Struct. Biol. 1997, 4, 490–497. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Del Prete, S.; Fisher, G.M.; Andrews, K.T.; Poulsen, S.A.; Capasso, C.; Supuran, C.T. Sulfonamide inhibition studies of the η-class carbonic anhydrase from the malaria pathogen Plasmodium falciparum. Bioorg. Med. Chem. 2015, 23, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; De Luca, V.; Del Prete, S.; Carginale, V.; Scozzafava, A.; Capasso, C.; Supuran, C.T. Sulfonamide inhibition studies of the γ-carbonic anhydrase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg. Med. Chem. Lett. 2015, 25, 3550–3555. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; De Luca, V.; Del Prete, S.; Carginale, V.; Scozzafava, A.; Capasso, C.; Supuran, C.T. Sulfonamide inhibition studies of the γ-carbonic anhydrase from the Antarctic cyanobacterium Nostoc commune. Bioorg. Med. Chem. 2015, 23, 1728–1734. [Google Scholar] [CrossRef] [PubMed]
- Dedeoglu, N.; DeLuca, V.; Isik, S.; Yildirim, H.; Kockar, F.; Capasso, C.; Supuran, C.T. Sulfonamide inhibition study of the β-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorg. Med. Chem. Lett. 2015, 25, 2291–2297. [Google Scholar] [CrossRef] [PubMed]
- Alafeefy, A.M.; Ceruso, M.; Al-Tamimi, A.M.; Del Prete, S.; Supuran, C.T.; Capasso, C. Inhibition studies of quinazoline-sulfonamide derivatives against the γ-CA (PgiCA) from the pathogenic bacterium, Porphyromonas gingivalis. J. Enzyme Inhib. Med. Chem. 2015, 30, 592–596. [Google Scholar] [CrossRef] [PubMed]
- Alafeefy, A.M.; Abdel-Aziz, H.A.; Vullo, D.; Al-Tamimi, A.M.; Awaad, A.S.; Mohamed, M.A.; Capasso, C.; Supuran, C.T. Inhibition of human carbonic anhydrase isozymes I, II, IX and XII with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. J. Enzyme Inhib. Med. Chem. 2015, 30, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Diaz, J.R.; Fernandez Baldo, M.; Echeverria, G.; Baldoni, H.; Vullo, D.; Soria, D.B.; Supuran, C.T.; Cami, G.E. A substituted sulfonamide and its Co (II), Cu (II), and Zn (II) complexes as potential antifungal agents. J. Enzyme Inhib. Med. Chem. 2016, 31, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Legionella pneumophila carbonic anhydrases: Underexplored antibacterial drug targets. Pathogens 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, I.; Vullo, D.; Minakuchi, T.; Scozzafava, A.; Capasso, C.; Supuran, C.T. Sulfonamide inhibition studies of two β-carbonic anhydrases from the bacterial pathogen Legionella pneumophila. Bioorg. Med. Chem. 2014, 22, 2939–2946. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Sai Kumar, R.S.; Scozzafava, A.; Capasso, C.; Ferry, J.G.; Supuran, C.T. Anion inhibition studies of a β-carbonic anhydrase from Clostridium perfringens. Bioorg. Med. Chem. Lett. 2013, 23, 6706–6710. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, I.; Minakuchi, T.; Maresca, A.; Carta, F.; Scozzafava, A.; Supuran, C.T. The β-carbonic anhydrases from Mycobacterium tuberculosis as drug targets. Curr. Pharm. Des. 2010, 16, 3300–3309. [Google Scholar] [CrossRef]
- Carta, F.; Maresca, A.; Covarrubias, A.S.; Mowbray, S.L.; Jones, T.A.; Supuran, C.T. Carbonic anhydrase inhibitors. Characterization and inhibition studies of the most active β-carbonic anhydrase from Mycobacterium tuberculosis, Rv3588c. Bioorg. Med. Chem. Lett. 2009, 19, 6649–6654. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Acetazolamide for the treatment of idiopathic intracranial hypertension. Exp. Rev. Neurother 2015, 15, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Vullo, D.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. Sulfonamide inhibition profiles of the β-carbonic anhydrase from the pathogenic bacterium Francisella tularensis responsible of the febrile illness tularemia. Bioorg. Med. Chem. 2017, 25, 3555–3561. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Del Prete, S.; Di Fonzo, P.; Carginale, V.; Donald, W.A.; Supuran, C.T.; Capasso, C. Comparison of the sulfonamide inhibition profiles of the β- and γ-carbonic anhydrases from the pathogenic bacterium Burkholderia pseudomallei. Molecules 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Dedeoglu, N.; DeLuca, V.; Isik, S.; Yildirim, H.; Kockar, F.; Capasso, C.; Supuran, C.T. Cloning, characterization and anion inhibition study of a β-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorg. Med. Chem. 2015, 23, 2995–3001. [Google Scholar] [CrossRef] [PubMed]
- Cau, Y.; Mori, M.; Supuran, C.T.; Botta, M. Mycobacterial carbonic anhydrase inhibition with phenolic acids and esters: Kinetic and computational investigations. Org. Biomol. Chem. 2016, 14, 8322–8330. [Google Scholar] [CrossRef] [PubMed]
- Modak, J.K.; Liu, Y.C.; Supuran, C.T.; Roujeinikova, A. Structure-activity relationship for sulfonamide inhibition of Helicobacter pylori α-carbonic anhydrase. J. Med. Chem. 2016, 59, 11098–11109. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Bortezomib inhibits bacterial and fungal β-carbonic anhydrases. Bioorg. Med. Chem. 2016, 24, 4406–4409. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Kumar, R.S.S.; Scozzafava, A.; Ferry, J.G.; Supuran, C.T. Sulphonamide inhibition studies of the β-carbonic anhydrase from the bacterial pathogen Clostridium perfringens. J. Enzyme Inhib. Med. Chem. 2018, 33, 31–36. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Shahidzadeh, R.; Opekun, A.; Shiotani, A.; Graham, D.Y. Effect of the carbonic anhydrase inhibitor, acetazolamide, on Helicobacter pylori infection in vivo: A pilot study. Helicobacter 2005, 10, 136–138. [Google Scholar] [CrossRef] [PubMed]
- Capasso, C.; Supuran, C.T. Carbonic anhydrase from Porphyromonas Gingivalis as a drug target. Pathogens 2017, 6, 30–42. [Google Scholar]
- Supuran, C.T.; Capasso, C. New light on bacterial carbonic anhydrases phylogeny based on the analysis of signal peptide sequences. J. Enzyme Inhib. Med. Chem. 2016, 31, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Licsandru, E.; Tanc, M.; Kocsis, I.; Barboiu, M.; Supuran, C.T. A class of carbonic anhydrase I—Selective activators. J. Enzyme Inhib. Med. Chem. 2017, 32, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibitors and activators for novel therapeutic applications. Future Med. Chem. 2011, 3, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases: From biomedical applications of the inhibitors and activators to biotechnological use for CO(2) capture. J. Enzyme Inhib. Med. Chem. 2013, 28, 229–230. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Del Prete, S.; Capasso, C.; Supuran, C.T. Carbonic anhydrase activators: Activation of the β-carbonic anhydrase from Malassezia globosa with amines and amino acids. Bioorg. Med. Chem. Lett. 2016, 26, 1381–1385. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Del Prete, S.; Osman, S.M.; AlOthman, Z.; Capasso, C.; Donald, W.A.; Supuran, C.T. Burkholderia pseudomallei γ-carbonic anhydrase is strongly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2017, 27, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Akdemir, A.; Vullo, D.; De Luca, V.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. The extremo-alpha-carbonic anhydrase (CA) from Sulfurihydrogenibium azorense, the fastest CA known, is highly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2013, 23, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- Forterre, P. Looking for the most “primitive” organism(s) on Earth today: The state of the art. Planet Space Sci. 1995, 43, 167–177. [Google Scholar] [CrossRef]
- Gupta, R.S. Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. MMBR 1998, 62, 1435–1491. [Google Scholar] [PubMed]
- Zimmerman, S.; Innocenti, A.; Casini, A.; Ferry, J.G.; Scozzafava, A.; Supuran, C.T. Carbonic anhydrase inhibitors. Inhibition of the prokariotic β and γ-class enzymes from Archaea with sulfonamides. Bioorg. Med. Chem. Lett. 2004, 14, 6001–6006. [Google Scholar] [CrossRef] [PubMed]
- Tripp, B.C.; Bell, C.B., 3rd; Cruz, F.; Krebs, C.; Ferry, J.G. A role for iron in an ancient carbonic anhydrase. J. Biol. Chem. 2004, 279, 6683–6687. [Google Scholar] [CrossRef] [PubMed]
- Tripp, B.C.; Smith, K.; Ferry, J.G. Carbonic anhydrase: New insights for an ancient enzyme. J. Biol. Chem. 2001, 276, 48615–48618. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.S.; Jakubzick, C.; Whittam, T.S.; Ferry, J.G. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc. Natl. Acad. Sci. USA 1999, 96, 15184–15189. [Google Scholar] [CrossRef]
- Torrance, J.W.; Bartlett, G.J.; Porter, C.T.; Thornton, J.M. Using a library of structural templates to recognise catalytic sites and explore their evolution in homologous families. J. Mol. Biol. 2005, 347, 565–581. [Google Scholar] [CrossRef] [PubMed]
- Torrance, J.W.; Holliday, G.L.; Mitchell, J.B.; Thornton, J.M. The geometry of interactions between catalytic residues and their substrates. J. Mol. Biol. 2007, 369, 1140–1152. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Kusian, B.; Sultemeyer, D.; Bowien, B. Carbonic anhydrase is essential for growth of Ralstonia eutropha at ambient CO(2) concentrations. J. Bact. 2002, 184, 5018–5026. [Google Scholar] [CrossRef] [PubMed]
- Merlin, C.; Masters, M.; McAteer, S.; Coulson, A. Why is carbonic anhydrase essential to Escherichia coli? J. Bact. 2003, 185, 6415–6424. [Google Scholar] [CrossRef] [PubMed]
- Cobaxin, M.; Martinez, H.; Ayala, G.; Holmgren, J.; Sjoling, A.; Sanchez, J. Cholera toxin expression by El Tor Vibrio cholerae in shallow culture growth conditions. Microb. Pathog. 2014, 66, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Abuaita, B.H.; Withey, J.H. Bicarbonate induces Vibrio cholerae virulence gene expression by enhancing ToxT activity. Infect. Immun. 2009, 77, 4111–4120. [Google Scholar] [CrossRef] [PubMed]
- Joseph, P.; Ouahrani-Bettache, S.; Montero, J.L.; Nishimori, I.; Minakuchi, T.; Vullo, D.; Scozzafava, A.; Winum, J.Y.; Kohler, S.; Supuran, C.T. A new β-carbonic anhydrase from Brucella suis, its cloning, characterization, and inhibition with sulfonamides and sulfamates, leading to impaired pathogen growth. Bioorg. Med. Chem. 2011, 19, 1172–1178. [Google Scholar] [CrossRef] [PubMed]
- Modak, J.K.; Liu, Y.C.; Machuca, M.A.; Supuran, C.T.; Roujeinikova, A. Structural basis for the inhibition of Helicobacter pylori α-carbonic anhydrase by sulfonamides. PLoS ONE 2015, 10, e0127149. [Google Scholar] [CrossRef] [PubMed]
- Nishimori, I.; Vullo, D.; Minakuchi, T.; Morimoto, K.; Onishi, S.; Scozzafava, A.; Supuran, C.T. Carbonic anhydrase inhibitors: Cloning and sulfonamide inhibition studies of a carboxyterminal truncated α-carbonic anhydrase from Helicobacter pylori. Bioorg. Med. Chem. Lett. 2006, 16, 2182–2188. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Isik, S.; Vullo, D.; De Luca, V.; Carginale, V.; Scozzafava, A.; Supuran, C.T.; Capasso, C. DNA cloning, characterization, and inhibition studies of an α-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. J. Med. Chem. 2012, 55, 10742–10748. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; Del Prete, S.; De Luca, V.; Carginale, V.; Ferraroni, M.; Dedeoglu, N.; Osman, S.M.; AlOthman, Z.; Capasso, C.; Supuran, C.T. Anion inhibition studies of the β-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg. Med. Chem. Lett. 2016, 26, 1406–1410. [Google Scholar] [CrossRef] [PubMed]
- Kohler, S.; Ouahrani-Bettache, S.; Winum, J.Y. Brucella suis carbonic anhydrases and their inhibitors: Towards alternative antibiotics? J. Enzyme Inhibit Med. Chem. 2017, 32, 683–687. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Supuran, C.T. 3D-QSAR CoMFA studies on sulfonamide inhibitors of the Rv3588c β-carbonic anhydrase from Mycobacterium tuberculosis and design of not yet synthesized new molecules. J. Enzyme Inhib. Med. Chem. 2014, 29, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Ceruso, M.; Vullo, D.; Scozzafava, A.; Supuran, C.T. Sulfonamides incorporating fluorine and 1,3,5-triazine moieties are effective inhibitors of three beta-class carbonic anhydrases from Mycobacterium tuberculosis. J. Enzyme Inhib. Med. Chem. 2014, 29, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Del Prete, S.; Perfetto, R.; Rossi, M.; Alasmary, F.A.S.; Osman, S.M.; AlOthman, Z.; Supuran, C.T.; Capasso, C. A one-step procedure for immobilising the thermostable carbonic anhydrase (SspCA) on the surface membrane of Escherichia coli. J. Enzyme Inhib. Med. Chem. 2017, 32, 1120–1128. [Google Scholar] [CrossRef] [PubMed]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Supuran, C.T.; Capasso, C. An Overview of the Bacterial Carbonic Anhydrases. Metabolites 2017, 7, 56. https://doi.org/10.3390/metabo7040056
Supuran CT, Capasso C. An Overview of the Bacterial Carbonic Anhydrases. Metabolites. 2017; 7(4):56. https://doi.org/10.3390/metabo7040056
Chicago/Turabian StyleSupuran, Claudiu T., and Clemente Capasso. 2017. "An Overview of the Bacterial Carbonic Anhydrases" Metabolites 7, no. 4: 56. https://doi.org/10.3390/metabo7040056
APA StyleSupuran, C. T., & Capasso, C. (2017). An Overview of the Bacterial Carbonic Anhydrases. Metabolites, 7(4), 56. https://doi.org/10.3390/metabo7040056