Metabolic Fingerprinting of Pseudomonas putida DOT-T1E Strains: Understanding the Influence of Divalent Cations in Adaptation Mechanisms Following Exposure to Toluene
Abstract
:1. Introduction
2. Material and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Growth in Response to Toluene in the Absence and Presence of Divalent Cations, Sample Collection and Analysis
2.2.1. Growth Curve Monitoring
2.2.2. Analysis of Bacterial Cells by Fourier Transform Infrared (FT-IR) Spectroscopy
Sample Preparation
Instrument Setup
Data Analysis
3. Results and Discussion
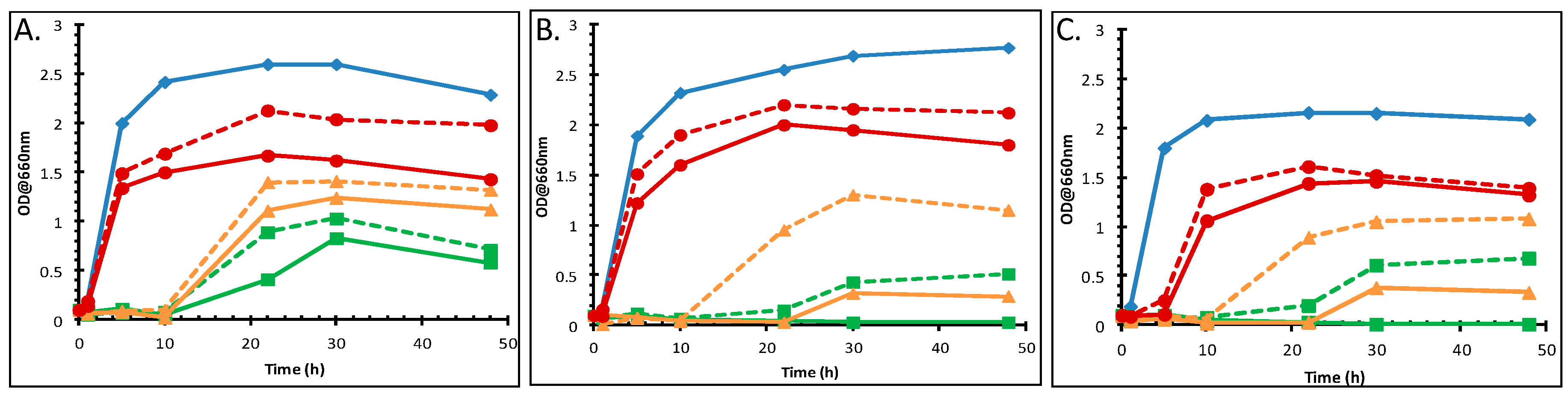
3.1. Effect of Toluene on the Growth of P. putida DOT-T1E Cells
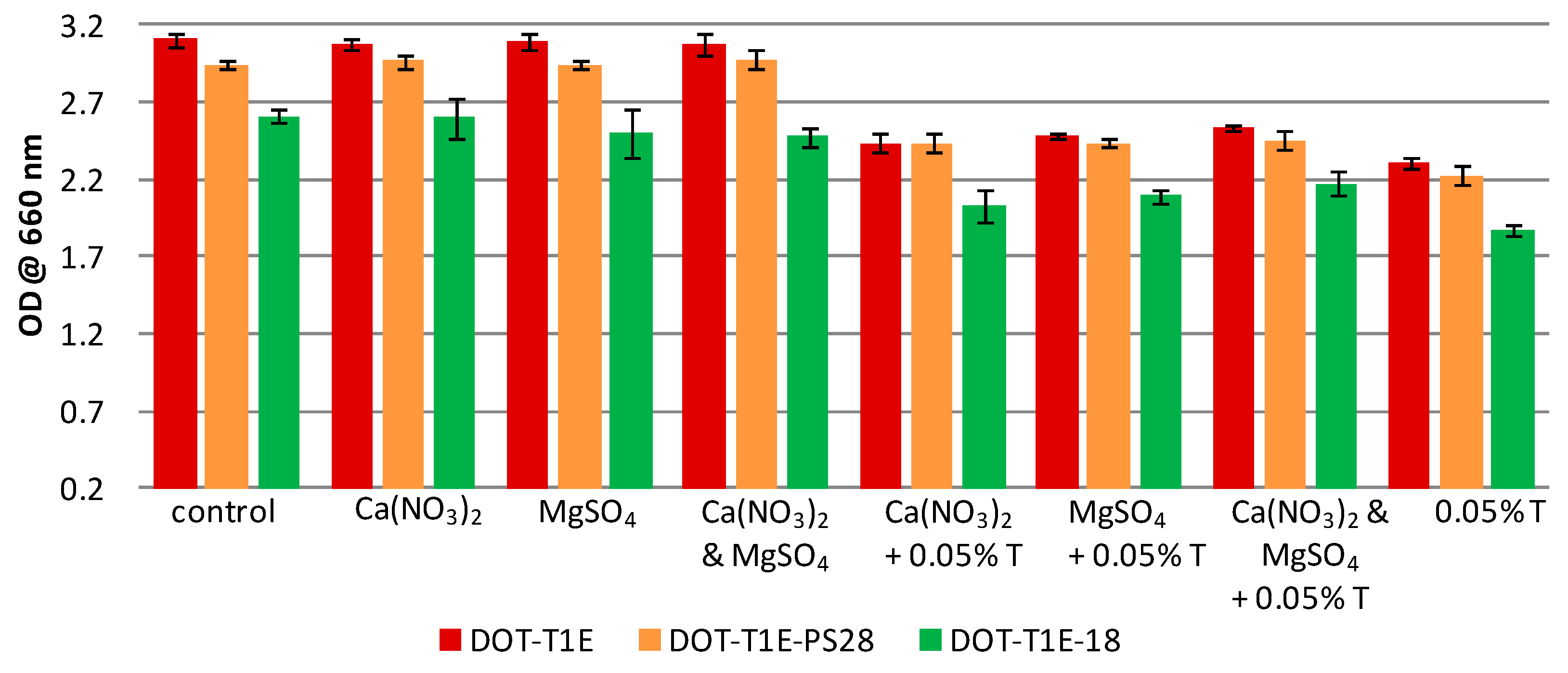
3.2. The Effect of Divalent Cations on the Growth of P. putida DOT-T1E Cultures in the Absence and Presence of Toluene
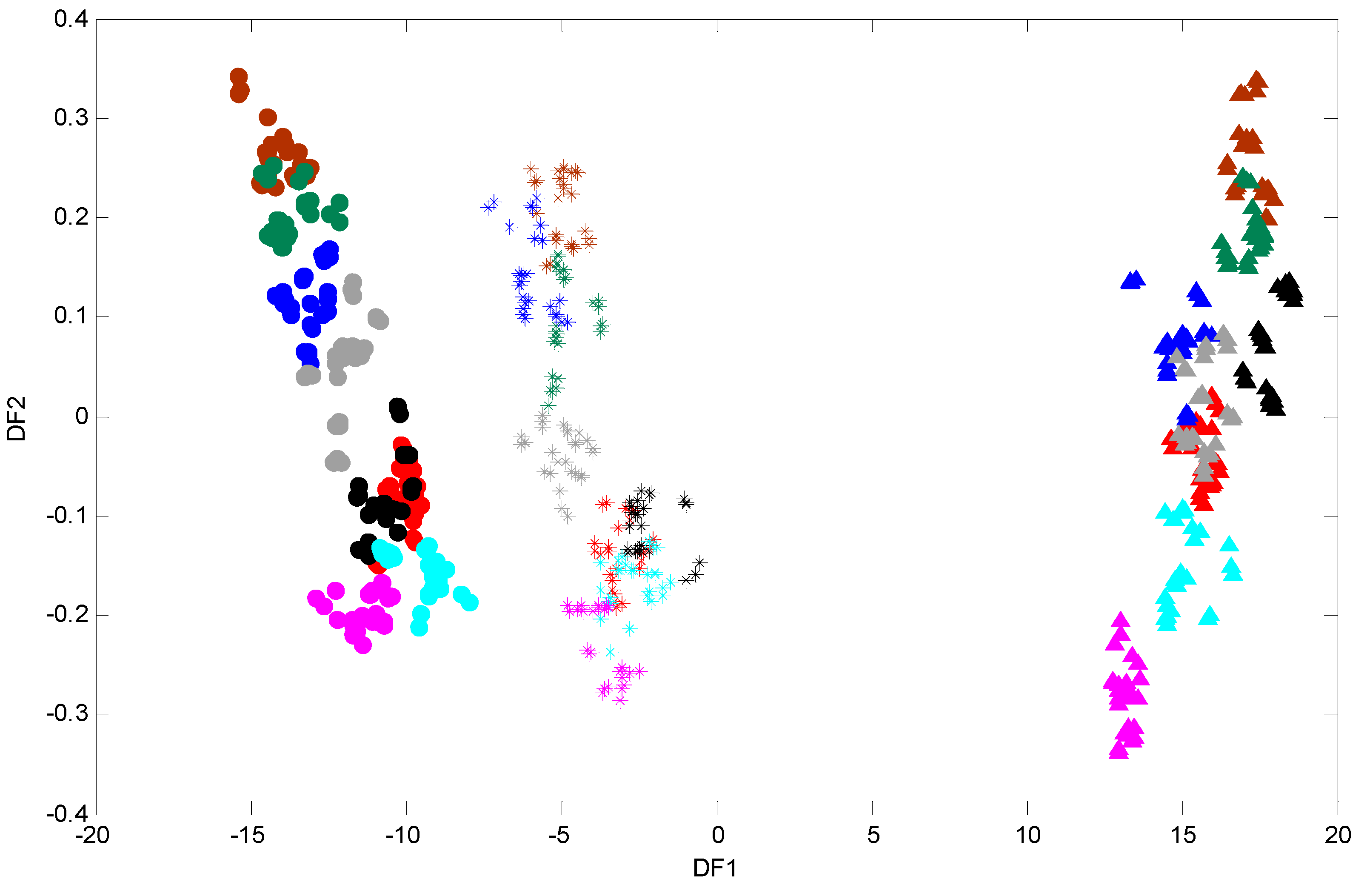
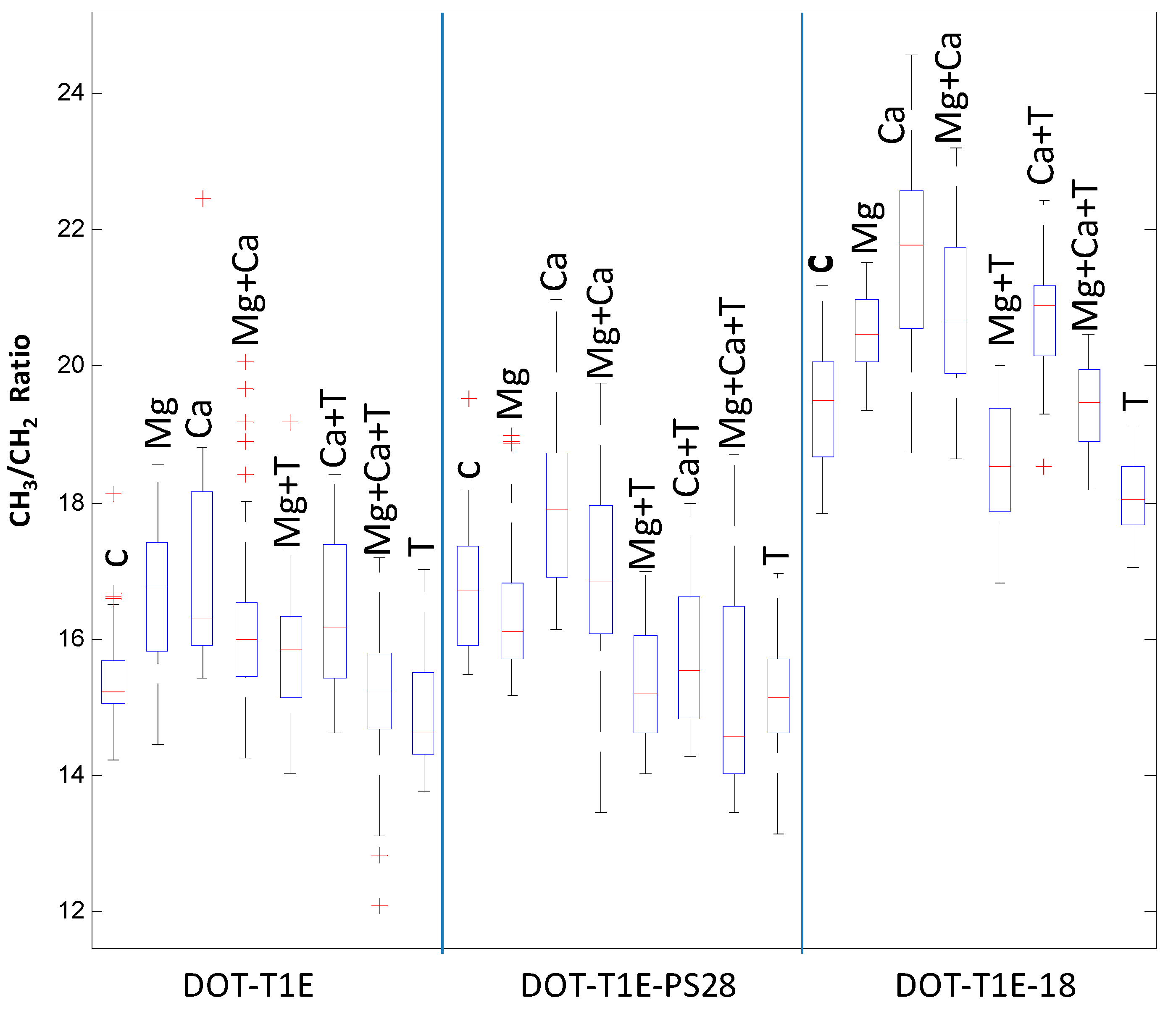
3.3. FT-IR Fingerprinting of P. putida DOT-T1E Cultures
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Inoue, A.; Horikoshi, K. A Pseudomonas thrives in high-concentrations of toluene. Nature 1989, 338, 264–266. [Google Scholar] [CrossRef]
- Sikkema, J.; de Bont, J.A.M.; Poolman, B. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 1995, 59, 201–222. [Google Scholar] [PubMed]
- Isken, S.; de Bont, J.A.M. Bacteria tolerant to organic solvents. Extremophiles 1998, 2, 229–238. [Google Scholar] [CrossRef] [PubMed]
- De Smet, M.J.; Kingma, J.; Witholt, B. The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia-coli. Biochim. Biophys. Acta 1978, 506, 64–80. [Google Scholar] [CrossRef]
- Cruden, D.L.; Wolfram, J.H.; Rogers, R.D.; Gibson, D.T. Physiological-properties of a Pseudomonas strain which grows with paraxylene in a 2-phase (organic-aqueous) medium. Appl. Environ. Microbiol. 1992, 58, 2723–2729. [Google Scholar] [PubMed]
- Kim, K.; Lee, S.J.; Lee, K.H.; Lim, D.B. Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J. Bacteriol. 1998, 180, 3692–3696. [Google Scholar] [PubMed]
- Li, X.Z.; Zhang, L.; Poole, K. Role of the multidrug efflux systems of Pseudomonas aeruginosa in organic solvent tolerance. J. Bacteriol. 1998, 180, 2987–2991. [Google Scholar] [PubMed]
- Ramos, J.L.; Duque, E.; Huertas, M.J.; Haidour, A. Isolation and expansion of the catabolic potential of a Pseudomonas-putida strain able to grow in the presence of high-concentrations of aromatic-hydrocarbons. J. Bacteriol. 1995, 177, 3911–3916. [Google Scholar] [PubMed]
- Weber, F.J.; Ooijkaas, L.P.; Schemen, R.M.W.; Hartmans, S.; de Bont, J.A.M. Adaptation of Pseudomonas putida S12 to high-concentrations of styrene and other organic-solvents. Appl. Environ. Microbiol. 1993, 59, 3502–3504. [Google Scholar] [PubMed]
- Garikipati, S.V.B.J.; Peeples, T.L. Solvent resistance pumps of Pseudomonas putida S12: Applications in 1-naphthol production and biocatalyst engineering. J. Biotechnol. 2015, 210, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Duque, E.; Mosqueda, G.; Golden, G.; Hurtado, A.; Ramos, J.L.; Segura, A. Three efflux pumps are required to provide efficient tolerance to toluene in Pseudomonas putida DOT-T1E. J. Bacteriol. 2001, 183, 3967–3973. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Yamamoto, M.; Horikoshi, K. Pseudomonas-putida which can grow in the presence of toluene. Appl. Environ. Microbiol. 1991, 57, 1560–1562. [Google Scholar] [PubMed]
- Pinkart, H.C.; Wolfram, J.W.; Rogers, R.; White, D.C. Cell envelope changes in solvent-tolerant and solvent-sensitive Pseudomonas putida strains following exposure to o-xylene. Appl. Environ. Microbiol. 1996, 62, 1129–1132. [Google Scholar] [PubMed]
- Rodriguez-Herva, J.J.; Garcia, V.; Hurtado, A.; Segura, A.; Ramos, J.L. The ttgGHI solvent efflux pump operon of Pseudomonas putida DOT-T1E is located on a large self-transmissible plasmid. Environ. Microbiol. 2007, 9, 1550–1561. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.L.; Duque, E.; Gallegos, M.T.; Godoy, P.; Ramos-Gonzalez, M.I.; Rojas, A.; Teran, W.; Segura, A. Mechanisms of solvent tolerance in Gram-negative bacteria. Annu. Rev. Microbiol. 2002, 56, 743–768. [Google Scholar] [CrossRef] [PubMed]
- Rojas, A.; Duque, E.; Schmid, A.; Hurtado, A.; Ramos, J.L.; Segura, A. Biotransformation in double-phase systems: Physiological responses of Pseudomonas putida DOT-T1E to a double phase made of aliphatic alcohols and biosynthesis of substituted catechols. Appl. Environ. Microbiol. 2004, 70, 3637–3643. [Google Scholar] [CrossRef] [PubMed]
- Nicolaou, S.A.; Gaida, S.M.; Papoutsakis, E.T. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: From biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 2010, 12, 307–331. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Godoy, P.; Daniels, C.; Hurtado, A.; Ramos, J.-L.; Segura, A. Functional analysis of new transporters involved in stress tolerance in Pseudomonas putida DOT-T1E. Environ. Microbiol. Rep. 2010, 2, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Pandey, J.; Chauhan, A.; Jain, R.K. Integrative approaches for assessing the ecological sustainability of in situ bioremediation. FEMS Microbiol. Rev. 2009, 33, 324–375. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.-C.; Wu, X.-Y.; Sun, J.-Z.; Cao, Y.-X.; Song, H. Engineering quorum sensing signaling of Pseudomonas for enhanced wastewater treatment and electricity harvest: A review. Chemosphere 2015, 140, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Bustard, M.T.; Whiting, S.; Cowan, D.A.; Wright, P.C. Biodegradation of high-concentration isopropanol by a solvent-tolerant thermophile, Bacillus pallidus. Extremophiles 2002, 6, 319–323. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Poh, C.L. Insights into environmental bioremediation by microorganisms through functional genomics and proteomics. Proteomics 2008, 8, 874–881. [Google Scholar] [CrossRef] [PubMed]
- Heipieper, H.J.; Neumann, G.; Cornelissen, S.; Meinhardt, F. Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 2007, 74, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Kabelitz, N.; Zehnsdorf, A.; Miltner, A.; Lippold, H.; Meyer, D.; Schmid, A.; Heipieper, H.J. Prediction of the adaptability of Pseudomonas putida DOT-T1E to a second phase of a solvent for economically sound two-phase biotransformations. Appl. Environ. Microbiol. 2005, 71, 6606–6612. [Google Scholar] [CrossRef] [PubMed]
- Bruce, L.J.; Daugulis, A.J. Solvent selection-strategies for extractive biocatalysis. Biotechnol. Prog. 1991, 7, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Leon, R.; Fernandes, P.; Pinheiro, H.M.; Cabral, J.M.S. Whole-cell biocatalysis in organic media. Enzyme Microb. Technol. 1998, 23, 483–500. [Google Scholar] [CrossRef]
- Ramos, J.L.; Duque, E.; Godoy, P.; Segura, A. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 1998, 180, 3323–3329. [Google Scholar] [PubMed]
- Horikoshi, K.; Antranikaian, G.; Bull, A.T.; Robb, F.T.; Stetter, K.O. Extremophiles Handbook; Springer Verlag: Tokyo, Japan, 2011; Volume 1. [Google Scholar]
- Goodacre, R.; Vaidyanathan, S.; Dunn, W.B.; Harrigan, G.G.; Kell, D.B. Metabolomics by numbers: Acquiring and understanding global metabolite data. Trends Biotechnol. 2004, 22, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef]
- Dunn, W.B.; Bailey, N.J.C.; Johnson, H.E. Measuring the metabolome: Current analytical technologies. Analyst 2005, 130, 606–625. [Google Scholar] [CrossRef] [PubMed]
- Winder, C.L.; Dunn, W.B.; Goodacre, R. Tardis-based microbial metabolomics: Time and relative differences in systems. Trends Microbiol. 2011, 19, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.I.; Dunn, W.B.; Griffin, J.L.; Allwood, J.W.; Goodacre, R. Metabolic fingerprinting as a diagnostic tool. Pharmacogenomics 2007, 8, 1243–1266. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.I.; Goodacre, R. Metabolic fingerprinting in disease diagnosis: Biomedical applications of infrared and Raman spectroscopy. Analyst 2006, 131, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R.; Timmins, E.M.; Burton, R.; Kaderbhai, N.; Woodward, A.M.; Kell, D.B.; Rooney, P.J. Rapid identification of urinary tract infection bacteria using hyperspectral whole-organism fingerprinting and artificial neural networks. Microbiology 1998, 144, 1157–1170. [Google Scholar] [CrossRef] [PubMed]
- Lang, P.L.; Sang, S.C. The in situ infrared microspectroscopy of bacterial colonies on agar plates. Cell. Mol. Biol. 1998, 44, 231–238. [Google Scholar] [PubMed]
- Naumann, D.; Helm, D.; Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 1991, 351, 81–82. [Google Scholar] [CrossRef] [PubMed]
- Naumann, D. Infrared Spectroscopy in Microbiology; Encyclopedia of Analytical Chemistry: Chichester, UK, 2006. [Google Scholar]
- Gillie, J.K.; Hochlowski, J.; Arbuckle-Keil, G.A. Infrared spectroscopy. Anal. Chem. 2000, 72, 71–79. [Google Scholar] [CrossRef]
- Ellis, D.I.; Broadhurst, D.; Kell, D.B.; Rowland, J.J.; Goodacre, R. Rapid and quantitative detection of the microbial spoilage of meat by Fourier transform infrared spectroscopy and machine learning. Appl. Environ. Microbiol. 2002, 68, 2822–2828. [Google Scholar] [CrossRef] [PubMed]
- Winder, C.L.; Gordon, S.V.; Dale, J.; Hewinson, R.G.; Goodacre, R. Metabolic fingerprints of Mycobacterium bovis cluster with molecular type: Implications for genotype-phenotype links. Microbiology 2006, 152, 2757–2765. [Google Scholar] [CrossRef] [PubMed]
- Martens, H.; Nielsen, J.P.; Engelsen, S.B. Light scattering and light absorbance separated by extended multiplicative signal correction. Application to near-infrared transmission analysis of powder mixtures. Anal. Chem. 2003, 75, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Joliffe, I.T. Principal Component Analysis; Springer: New York, NY, USA, 1986. [Google Scholar]
- Johnson, H.E.; Broadhurst, D.; Goodacre, R.; Smith, A.R. Metabolic fingerprinting of salt-stressed tomatoes. Phytochemistry 2003, 62, 919–928. [Google Scholar] [CrossRef]
- Macfie, H.J.H.; Gutteridge, C.S.; Norris, J.R. Use of canonical variates analysis in differentiation of bacteria by pyrolysis gas-liquid-chromatography. J. Gen. Microbiol. 1978, 104, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Abe, A.; Inoue, A.; Usami, R.; Moriya, K.; Horikoshi, K. Degradation of polyaromatic hydrocarbons by organic solvent-tolerant bacteria from deep-sea. Biosci. Biotechnol. Biochem. 1995, 59, 1154–1156. [Google Scholar] [CrossRef] [PubMed]
- Aono, R.; Ito, M.; Inoue, A.; Horikoshi, K. Isolation of novel toluene-tolerant strain of Pseudomonas-aeruginosa. Biosci. Biotechnol. Biochem. 1992, 56, 145–146. [Google Scholar] [CrossRef]
- Ramos, J.L.; Duque, E.; RodriguezHerva, J.J.; Godoy, P.; Haidour, A.; Reyes, F.; FernandezBarrero, A. Mechanisms for solvent tolerance in bacteria. J. Biol. Chem. 1997, 272, 3887–3890. [Google Scholar] [CrossRef] [PubMed]
- Isken, S.; de Bont, J.A.M. Active efflux of toluene in a solvent-resistant bacterium. J. Bacteriol. 1996, 178, 6056–6058. [Google Scholar] [PubMed]
- Kieboom, J.; Dennis, J.J.; de Bont, J.A.M.; Zylstra, G.J. Identification and molecular characterization of an efflux pump involved in Pseudomonas putida S12 solvent tolerance. J. Biol. Chem. 1998, 273, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Rojo, F. Carbon catabolite repression in Pseudomonas: Optimizing metabolic versatility and interactions with the environment. FEMS Microbiol. Rev. 2010, 34, 658–684. [Google Scholar] [CrossRef] [PubMed]
- Isken, S.; Derks, A.; Wolffs, P.F.G.; de Bont, J.A.M. Effect of organic solvents on the yield of solvent-tolerant Pseudomonas putida S12. Appl. Environ. Microbiol. 1999, 65, 2631–2635. [Google Scholar] [PubMed]
- Stanlotter, H.; Gupta, M.; Sanderson, K.E. Influence of cations on the permeability of the outer-membrane of Salmonella-typhimurium and other Gram-negative bacteria. Can. J. Microbiol. 1979, 25, 475–485. [Google Scholar] [CrossRef]
- Brito-Echeverria, J.; Lucio, M.; Lopez-Lopez, A.; Anton, J.; Schmitt-Kopplin, P.; Rossello-Mora, R. Response to adverse conditions in two strains of the extremely halophilic species Salinibacter ruber. Extremophiles 2011, 15, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Kol, S.; Merlo, M.E.; Scheltema, R.A.; de Vries, M.; Vonk, R.J.; Kikkert, N.A.; Dijkhuizen, L.; Breitling, R.; Takano, E. Metabolomic characterization of the salt stress response in Streptomyces coelicolor. Appl. Environ. Microbiol. 2010, 76, 2574–2581. [Google Scholar] [CrossRef] [PubMed]
- Allwood, J.W.; AlRabiah, H.; Correa, E.; Vaughan, A.; Xu, Y.; Upton, M.; Goodacre, R. A workflow for bacterial metabolic fingerprinting and lipid profiling: Application to ciprofloxacin challenged Escherichia coli. Metabolomics 2015, 11, 438–453. [Google Scholar] [CrossRef]
- Anton, J.; Lucio, M.; Pena, A.; Cifuentes, A.; Brito-Echeverria, J.; Moritz, F.; Tziotis, D.; Lopez, C.; Urdiain, M.; Schmitt-Kopplin, P.; et al. High metabolomic microdiversity within co-occurring isolates of the extremely halophilic bacterium Salinibacter ruber. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Allwood, J.W.; Moore, R.E.; Marsden-Edwards, E.; Dunn, W.B.; Xu, Y.; Hampson, L.; Hampson, I.N.; Goodacre, R. A metabolomics investigation into the effects of HIV protease inhibitors on HPV16 E6 expressing cervical carcinoma cells. Mol. Biosyst. 2014, 10, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Lin, W.; Broadhurst, D.; Begley, P.; Brown, M.; Zelena, E.; Vaughan, A.A.; Halsall, A.; Harding, N.; Knowles, J.D.; et al. Molecular phenotyping of a UK population: Defining the human serum metabolome. Metabolomics 2015, 11, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.I.; Goodacre, R. Metabolomics-assisted synthetic biology. Curr. Opin. Biotechnol. 2012, 23, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Ellis, D.I.; Brewster, V.L.; Dunn, W.B.; Allwood, J.W.; Golovanov, A.P.; Goodacre, R. Fingerprinting food: Current technologies for the detection of food adulteration and contamination. Chem. Soc. Rev. 2012, 41, 5706–5727. [Google Scholar] [CrossRef] [PubMed]
- Goodacre, R. Explanatory analysis of spectroscopic data using machine learning of simple, interpretable rules. Vib. Spectrosc. 2003, 32, 33–45. [Google Scholar] [CrossRef]
- Teran, W.; Felipe, A.; Segura, A.; Rojas, A.; Ramos, J.L.; Gallegos, M.T. Antibiotic-dependent induction of Pseudomonas putida DOT-T1E ttgABC efflux pump is mediated by the drug binding repressor ttgR. Antimicrob. Agents Chemother. 2003, 47, 3067–3072. [Google Scholar] [CrossRef] [PubMed]
- Duque, E.; Rodriguez-Herva, J.-J.; de la Torre, J.; Dominguez-Cuevas, P.; Munoz-Rojas, J.; Ramos, J.-L. The RpoT regulon of Pseudomonas putida DOT-T1E and its role in stress endurance against solvents. J. Bacteriol. 2007, 189, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Roca, A.; Rodriguez-Herva, J.-J.; Duque, E.; Ramos, J.L. Physiological responses of Pseudomonas putida to formaldehyde during detoxification. Microb. Biotechnol. 2008, 1, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-H.; Jarvis, R.M.; Xu, Y.; Oliver, A.W.; Allwood, J.W.; Hampson, L.; Hampson, I.N.; Goodacre, R. Combining metabolic fingerprinting and footprinting to understand the phenotypic response of HPV16 E6 expressing cervical carcinoma cells exposed to the HIV anti-viral drug lopinavir. Analyst 2010, 135, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H.; Vaara, M. Molecular basis of bacterial outer membrane permeability. Microbiol. Rev. 1985, 49, 1. [Google Scholar] [CrossRef] [PubMed]
- Schneck, E.; Schubert, T.; Konovalov, O.V.; Quinn, B.E.; Gutsmann, T.; Brandenburg, K.; Oliveira, R.G.; Pink, D.A.; Tanaka, M. Quantitative determination of ion distributions in bacterial lipopolysaccharide membranes by grazing-incidence X-ray fluorescence. Proc. Natl. Acad. Sci. USA 2010, 107, 9147–9151. [Google Scholar] [CrossRef] [PubMed]
- Labischinski, H.; Barnickel, G.; Bradaczek, H.; Naumann, D.; Rietschel, E.T.; Giesbrecht, P. High state of order of isolated bacterial lipopolysaccharide and its possible contribution to the permeation barrier property of the outer-membrane. J. Bacteriol. 1985, 162, 9–20. [Google Scholar] [PubMed]
- Sheu, C.W.; Freese, E. Lipopolysaccharide layer protection of Gram-negative bacteria against inhibition by long-chain fatty-acids. J. Bacteriol. 1973, 115, 869–875. [Google Scholar]
- Hancock, R.E.W. Alterations in outer-membrane permeability. Annu. Rev. Microbiol. 1984, 38, 237–264. [Google Scholar] [CrossRef] [PubMed]
- Vaara, M. Antibiotic-supersusceptible mutants of Escherichia-coli and Salmonella-typhimurium. Antimicrob. Agents Chemother. 1993, 37, 2255–2260. [Google Scholar] [CrossRef] [PubMed]
- Junker, F.; Rodriguez-Herva, J.J.; Duque, E.; Ramos-Gonzalez, M.I.; Llamas, M.; Ramos, J.L. A wbpl mutant of Pseudomonas putida DOT-T1E strain, which lacks the O-antigenic side chain of lipopolysaccharides, is tolerant to organic solvent shocks. Extremophiles 2001, 5, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.J.; Dhillon, P.; Yan, H.; Farmer, S.; Hancock, R.E.W. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 3317–3321. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Clifton, L.A.; Skoda, M.W.A.; Le Brun, A.P.; Ciesielski, F.; Kuzmenko, I.; Holt, S.A.; Lakey, J.H. Effect of divalent cation removal on the structure of Gram-negative bacterial outer membrane models. Langmuir 2015, 31, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Pink, D.A.; Hansen, L.T.; Gill, T.A.; Quinn, B.E.; Jericho, M.H.; Beveridge, T.J. Divalent calcium ions inhibit the penetration of protamine through the polysaccharide brush of the outer membrane of Gram-negative bacteria. Langmuir 2003, 19, 8852–8858. [Google Scholar] [CrossRef]
- Lam, N.H.; Ma, Z.; Ha, B.-Y. Electrostatic modification of the lipopolysaccharide layer: Competing effects of divalent cations and polycationic or polyanionic molecules. Soft Matter 2014, 10, 7528–7544. [Google Scholar] [CrossRef] [PubMed]
- Wijte, D.; van Baar, B.L.M.; Heck, A.J.R.; Altelaar, A.F.M. Probing the proteome response to toluene exposure in the solvent tolerant Pseudomonas putida S12. J. Proteome Res. 2011, 10, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Schneck, E.; Papp-Szabo, E.; Quinn, B.E.; Konovalov, O.V.; Beveridge, T.J.; Pink, D.A.; Tanaka, M. Calcium ions induce collapse of charged O-side chains of lipopolysaccharides from Pseudomonas aeruginosa. J. R. Soc. Interface 2009, 6, S671–S678. [Google Scholar] [CrossRef] [PubMed]
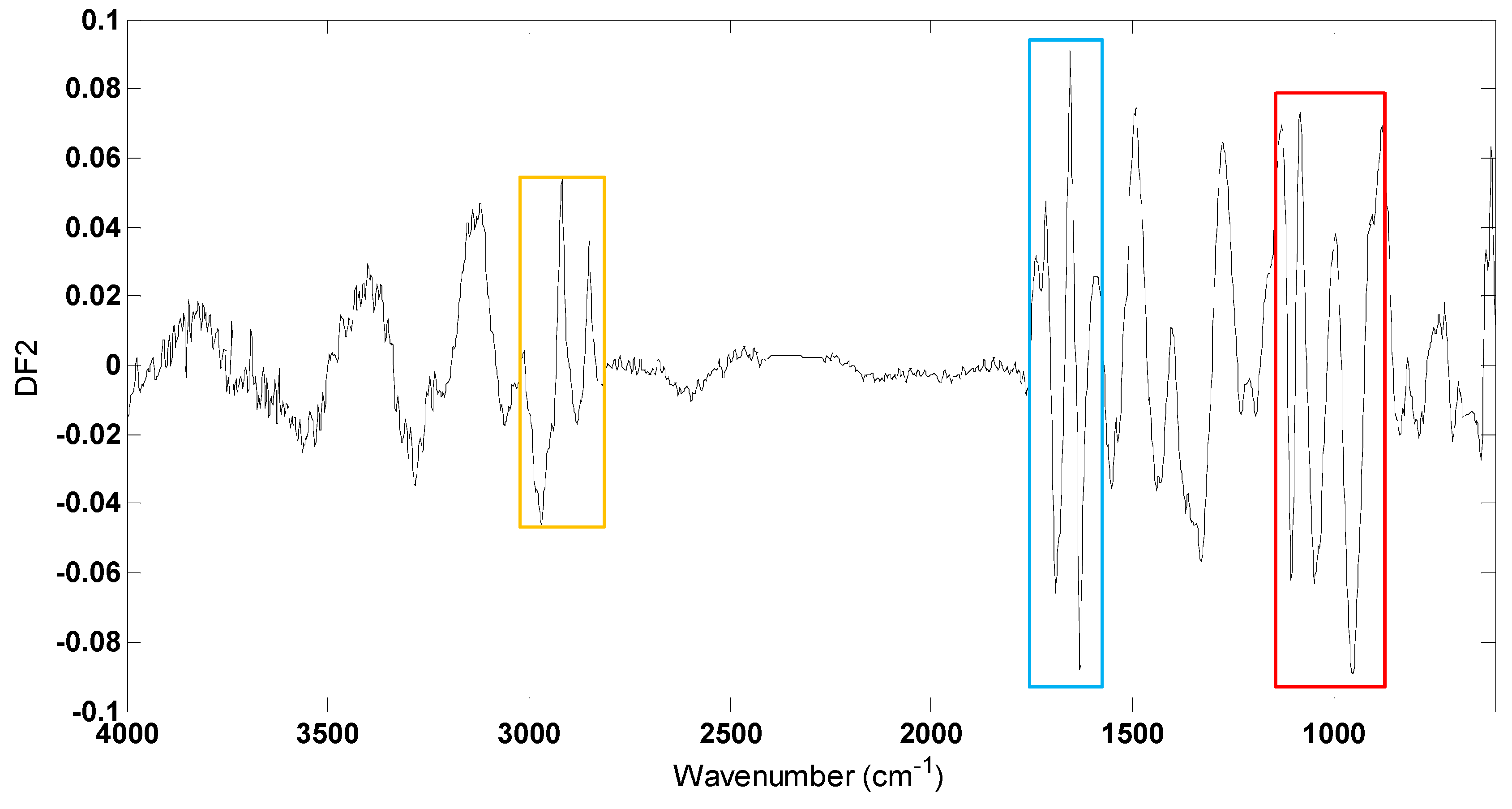
| Wavenumbers (cm−1) | Assignment | FT-IR Vibrational Modes |
|---|---|---|
| Lipid | ||
| (2958–2873) | Membrane lipid | Asymmetric CH3 stretches mode of CH3 end groups from membrane lipid |
| (2924–2850) | Membrane lipid | Symmetric CH2 stretches mode of CH2 chain from membrane lipid |
| Protein | ||
| (3400–3300) | Amide A | N-H stretching |
| (1690–1620) | Amide I | C=O stretching |
| (1590–1530) | Amide II | C-N stretching and N-H bending |
| (1450–1200) | COOH of proteins, free amino acids, polysaccharides | |
| Carbohydrate | ||
| (1200–900) | Polysaccharides | C-O or O-H stretching from polysaccharides |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sayqal, A.; Xu, Y.; Trivedi, D.K.; AlMasoud, N.; Ellis, D.I.; Goodacre, R. Metabolic Fingerprinting of Pseudomonas putida DOT-T1E Strains: Understanding the Influence of Divalent Cations in Adaptation Mechanisms Following Exposure to Toluene. Metabolites 2016, 6, 14. https://doi.org/10.3390/metabo6020014
Sayqal A, Xu Y, Trivedi DK, AlMasoud N, Ellis DI, Goodacre R. Metabolic Fingerprinting of Pseudomonas putida DOT-T1E Strains: Understanding the Influence of Divalent Cations in Adaptation Mechanisms Following Exposure to Toluene. Metabolites. 2016; 6(2):14. https://doi.org/10.3390/metabo6020014
Chicago/Turabian StyleSayqal, Ali, Yun Xu, Drupad K. Trivedi, Najla AlMasoud, David I. Ellis, and Royston Goodacre. 2016. "Metabolic Fingerprinting of Pseudomonas putida DOT-T1E Strains: Understanding the Influence of Divalent Cations in Adaptation Mechanisms Following Exposure to Toluene" Metabolites 6, no. 2: 14. https://doi.org/10.3390/metabo6020014
APA StyleSayqal, A., Xu, Y., Trivedi, D. K., AlMasoud, N., Ellis, D. I., & Goodacre, R. (2016). Metabolic Fingerprinting of Pseudomonas putida DOT-T1E Strains: Understanding the Influence of Divalent Cations in Adaptation Mechanisms Following Exposure to Toluene. Metabolites, 6(2), 14. https://doi.org/10.3390/metabo6020014








