Advances in Metabolic Engineering of Cyanobacteria for Photosynthetic Biochemical Production
Abstract
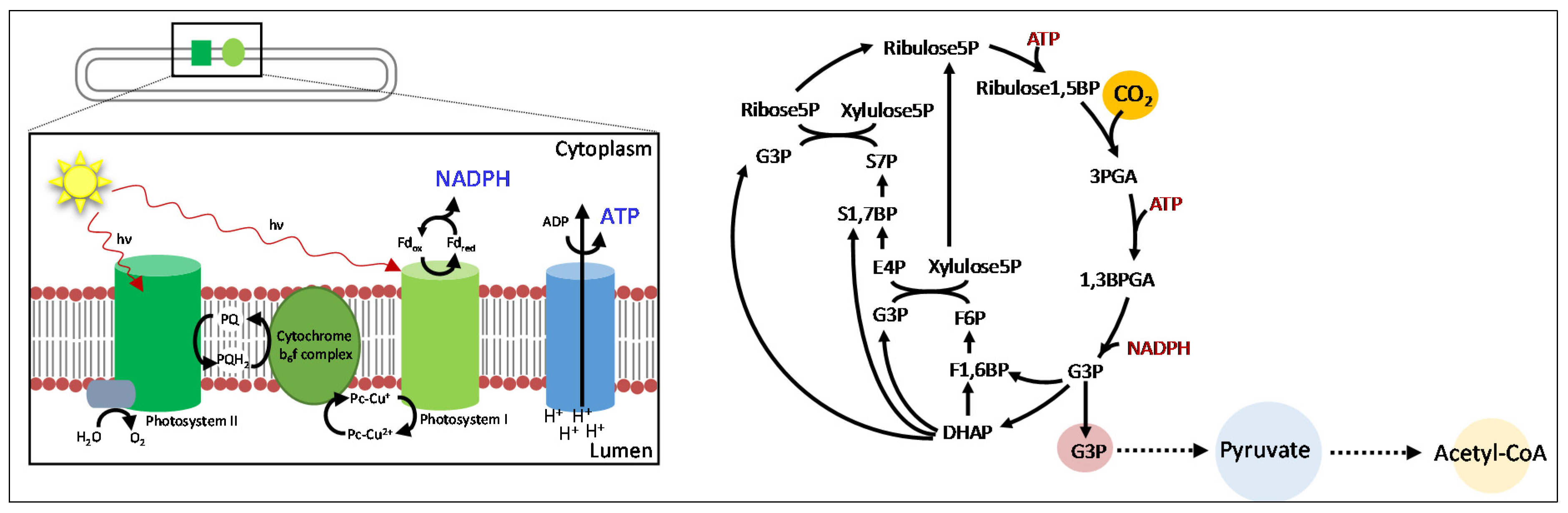
:1. Introduction
| Strain | Synechocystis PCC 6803 | Synechococcus sp.PCC 7002 | Synechococcus elongatus PCC 7942 | Anabaena sp. 7120 |
|---|---|---|---|---|
| Genome | 3.6 Mb Chromosome + 7 plasmids size ranging from 2.3 to 120 kb | 3.0 Mb chromosome + 6 plasmids size ranging from 4.8 to 186 kb | 2.7 Mb Chromosome + 46 kb plasmid | 6.4 Mb chromosome + 6 plasmids size ranging from 5.6 to 408 kb |
| Description | Freshwater | Salt tolerant | Freshwater | Diazotrophic |
| Transformation | Naturally competent | Naturally competent | Naturally competent | Conjugation |
| Chemical Target | Strain | Promoter(s) Used | Gene(s) Expressed | Gene Knockout(s) | Titer (mg/L) | Days of Cultivation | Relevant Central Metabolite | Comments | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Ethanol | PCC 6803 | Prbc | pdc, adh2 | phb | 5500 | 26 | Pyruvate | Two copies of pdc, optimized cultivation | [11] |
| Ethanol | PCC 6803 | PpsbA2 | pdc, adh | 550 | 6 | Pyruvate | Decarboxylation of pyruvate serves as efficient driving force | [12] | |
| Ethanol | PCC 7942 | Ptrc | pduP, yqhD | 182 | 10 | Acetyl-CoA | Oxygen tolerant aldehyde dehydrogenase | [13] | |
| Ethanol | PCC 7942 | PrbcLS | pdc, adh | 0.02 | 7 | pyruvate | expression of pdc and adh on plasmid | [14] | |
| Isopropanol | PCC 7942 | Ptrc | thl-atoAD-adc-adh | 146 | 15 | Acetyl-CoA | Medium optimization, 2 phase cultivation | [15] | |
| Isopropanol | PCC 7942 | Ptrc | thl-atoAD-adc-adh | 26.5 | 9 | Acetyl-CoA | decarboxylation of acetoacetate traps carbon to acetone | [16] | |
| 1-Butanol | PCC 7942 | Ptrc/PLlacO1 | ter/nphT7, pduP, yqhD, crt, hbd | 317 | 12 | Acetyl-CoA | Oxygen tolerant aldehyde dehydrogenase | [13] | |
| 1-Butanol | PCC 7942 | Ptrc/PLlacO1 | ter/nphT7, bldh, yqhD, crt, hbd | 30 | 17 | Acetyl-CoA | ATP driving force through acetoacetyl-CoA synthase | [17] | |
| 1-Butanol | PCC 7942 | Ptrc/PLlacO1 | ter/atoB, adhE2, crt, hbd | 14.5 | 7 | Acetyl-CoA | Dark anaerobic incubation | [18] | |
| Isobutyraldehyde | PCC 7942 | Ptrc/PlacO1 | kivd/alsS-ilvC-ilvD | 1100 | 8 | Pyruvate | Decarboxylation of KIV serves as effective driving force. In situ product removal | [19] | |
| Isobutanol | PCC 6803 | Ptac | kivD, adhA | 240 | 21 | Pyruvate | Oleyol alcohol trap | [20] | |
| Isobutanol | PCC 7942 | Ptrc | kvid, YqhD | 450 | 6 | Pyruvate | Decarboxylation of KIV serves as effective driving force. In situ product removal | [19] | |
| 2-Methylbutanol | PCC 7942 | Ptrc | kivD, yqhD, cimA, leuBCD | 178 | 12 | Pyruvate/acetyl-CoA | decarboxylation, native highly active AHAS | [21] | |
| Fatty alcohol | PCC 6803 | Prbc | jojoba FAR | 0.2 | 18 | Acetyl-CoA | using native fatty-acyl-ACP synthesis & expression of jojoba FAR | [22] | |
| Fatty alcohol | PCC 6803 | Prbc/PpsbA2 | jojoba FAR/aas | 0.17 | 10 | Acetyl-CoA | expression of jojoba FAR & overexpression of acyl-ACP synthetase | [23] | |
| 1,2-Propanediol | PCC 7942 | Ptrc | sADH (C.beijerinkii), yqhD, mgsA | 150 | 10 | Pyruvate | NADPH utilization | [24] | |
| 2,3-Butanediol | PCC 6803 | Ptrc | als, aldc, ar | 585 | 29 | Pyruvate | Codon optimization | [25] | |
| 2,3-Butanediol | PCC 7942 | PLlacO1 | alsS, alsD, adh | 2380 | 20 | Pyruvate | pyruvate pool coupled to decarboxylation and product low toxicity | [26] | |
| Glycerol | PCC 6803 | Ptrc | gpp2 | 1068 | 17 | DHAP | Salt stress can stimulate glycerol production even in wildtype for about 0.7 mM | [27] | |
| Glycerol | PCC 7942 | Ptrc | gpp1 | 1170 | 20 | DHAP | aeration, thermodynamically favorable glycerol phosphatase | [28] | |
| D-Lactate | PCC 6803 | Ptrc | gldA101, sth | 1140 | 24 | Pyruvate | expression of transhydrogenase, codon optmized mutated glycerol dehydrogenase, addition of acetate helped production | [29] | |
| D-Lactate | PCC 6803 | Pcpc560 | Dldh | pta,phaCE | 1060 | 4 | Pyruvate | knockout of PHB synthesis & acetate formation, expression codon optimized ldh from Lactobacillus delbrueckii | [30] |
| D-Lactate | PCC 7942 | Ptrc | ldhD, lldP | 829 | 10 | Pyruvate | expression of lactate transporter, engineered Ldh to use NADPH | [31] | |
| D-Lactate | PCC 7942 | Plac | ldhA, lldP, udhA | 55 | 4 | Pyruvate | Expression of LldP protein from E. coli (transporter) | [32] | |
| L-Lactate | PCC 6803 | Ptrc | ldh | 1800 | 40 | Pyruvate | Long term production | [33] | |
| L-Lactate | PCC 6803 | Ptrc2/Ptrc2 | pk/ldh | knockdown PPC | 837 | 14 | Pyruvate | codon optimization & natural copy | [34] |
| L-Lactate | PCC 6803 | Ptrc/Ptrc | ldh/sth | 288 | 14 | Pyruvate | expression of transhydrogenase, Bacillus subtilis Ldh | [35] | |
| L-Lactate | PCC 6803 | PpsbA2 | ldh, ldhP | 15.3 | 18 | Pyruvate | Tested various ldh genes. | [36] | |
| 3-Hydroxypropionate | PCC 7942 | Ptrc | mcr, msr | 659 | 16 | Acetyl-CoA | selection of best performing malonate semialdehyde reductase (Msr), two NADPH utilizing steps | [37] | |
| 3-Hydroxypropionate | PCC 7942 | Ptrc | gpp1, dhaB, puuC | 31.7 | 10 | DHAP | oxygen sensitive, Dark anaerobic with nutrient limitation | [28] | |
| 3-Hydroxybutyrate | PCC 6803 | Ptca/Ptac | tesB/phaA-phaB | 533 | 21 | Acetyl-CoA | Nutrient limitation, NADPH | [38] | |
| Itaconic acid | PCC 6803 | Ptac | cad | 14.5 | 16 | Isocitrate | Expression of cad | [39] | |
| p-Coumaric acid | PCC 6803 | PpsbA2 | sam8 | slr1573 (laccase) | 82.6 | 4 | Tyrosine | knockout of competing pathway for phenolic compound degradation | [40] |
| Fatty acids | PCC 6803 | Ptrc/Pcpc/Prbc | tesA, fatB1, fatB2/accBC/accDA, fat B2 | aas, pta, phb genes,(see comments) | 197 | 2 | Acetyl-CoA | construct six generation strain: extensive knock outs of PHB synthesis, peptidoglycan layer protein, hemolysin-like surface layer protein, cyanophycin synthesis | [41] |
| Fatty acids | PCC 7002 | Ptrc/PpsbA1 | tesA/rbcLS | fadD | 131 | 20 | Acetyl-CoA | Overexpression of rubisco | [42] |
| Fatty acids | PCC 7942 | Ptrc | tesA | aas | 45 | 20 | Acetyl-CoA | knockout of acylACP synthetase blocks utilization of fatty acids | [43] |
| Fatty acids | PCC 7942 | Ptrc/PpsbA1 | fat1/rbcLS | aas | 35 | 20 | Acetyl-CoA | overexpression of ACCase hurts production | [44] |
| D-Mannitol | PCC 7002 | PpsbA | mtlD, mlp | glgA1, glgA2 | 1100 | 12 | F6P | codon optimization, artificial carbon sink | [45] |
| Hexose | PCC 7942 | Ptrc | glf, invA, galU | 45 | 5 | Glc6P | expression of sugar transporter | [32] | |
| Sucrose | PCC 7942 | Ptrc | cscB | invA, glgC | 2700 | 7 | Glc6P | Salt stress, knockout of natural carbon/electron sink | [46] |
| Sucrose | PCC 6803 | PpetE | cscB, sps, spp, ugp | ggpS, ggtCD | 140 | 10 | Glc6P | Salt stress, knockout of competing pathways, expression of sucrose synthesis genes | [47] |
| Glucosylglycerol | PCC 6803 | -- | -- | ggtCD, ggpR | 981 | 24 | G3P/Glc6P | salt shock, hypoosmotic shock | [48] |
| Ethylene | PCC 6803 | Ptrc | efe | 240 nL/mL/d | α-Ketoglutarate | compared various promoters, used plasmid based expression | [49] | ||
| Ethylene | PCC 6803 | PpsbA | efe | 171 mg/L/d | α-Ketoglutarate | Multiple copies of EFE | [50] | ||
| Ethylene | PCC 7942 | Ptrc | ACS-Ctdoc-ACO-Acdoc-Cip2 | 81.6 nL/mL/d/OD | SAM | Chimeric protein fusion | [51] | ||
| Ethylene | PCC 7942 | PpsbA1 | efe | 10.82 μL/mL/D/OD | α-Ketoglutarate | choose a strong promoter site in 7942 and rpsl2-mediated gene replacement | [52] | ||
| Isoprene | PCC 6803 | PpsbA2 | IspS (Pueraria montana) | 0.35 | 8 | G3P/Pyruvate | Gaseous/aqueous two-phase photobioreactor | [53] | |
| Isoprene | PCC 6803 | PpsbA2 | IspS, hmgS, hmgR, fni, mk, pmd, pmk | 0.3 | 8 | Acetyl-CoA | expression of both pathways to IPP increases isoprene production | [54] | |
| Isoprene | PCC 6803 | PpsbA2 | IspS (Pueraria montana) | 50 μg/gDCW/d | G3P/Pyruvate | Expression of isoprene synthase | [55] | ||
| Limonene | PCC 6803 | Ptrc | limS (Schizonepeta tenuifolia), dxs, crtE, ipi | 1 | 30 | G3P/Pyruvate | codon optimization, enhancing flux through MEP pathway | [56] | |
| Limonene | PCC 7002 | PcpcBA | limS (Mentha spicata) | 4 | 4 | G3P/Pyruvate | product trap by dodecane overlayLan | [57] | |
| Limonene | PCC 7120 | Pnir::PpsbA1 | limS (Picea sitchensis), dxs, ipphp, gpps | 0.52 | 12 | G3P/Pyruvate | enhancing flux through MEP pathway by gene overexpression, high light density | [58] | |
| Farnesene | PCC 7120 | Pnir, PpsbA1 | faS (Picea abies) | 0.31 | 15 | G3P/Pyruvate | codon optimization | [59] | |
| Bisabolene | PCC 7002 | PcpcBA | bis (Abies grandis) | 0.6 | 4 | G3P/Pyruvate | product trap by dodecane overlay | [57] | |
| Tocopherols | PCC 6803 | PnirA | hpd (Arabidopsis thaliana) | 0.250 mg/gDCW | 12 | G3P/Pyruvate | Nitrate inducible promoter | [60] | |
| β-Caryophyllene | PCC 6803 | PpsbA2 | QHS1 | 0.046 | 7 | G3P/Pyruvate | use similar pathway in 6803 to produce plant's second metabolite, only need few key enzyme | [61] | |
| β-Phellandrene | PCC 6803 | PpsbA2-trc-T7 | PLHS (Lavandula angustifolia) | 0.9 | 2 | G3P/Pyruvate | codon optimization, High light with psba2-trc-T7 fused promoter | [62] | |
| β-Phellandrene | PCC 6803 | PpsbA2 | PHLS (Lavandula angustifolia) | 0.2 | 8 | G3P/Pyruvate | codon optimization | [63] | |
| Dihydroxyacetone | PCC 7942 | Ptrc | gpp1, dhaD | 78.6 | 16 | DHAP | NAD-dependent DhaD could not efficiently reduce glycerol | [28] | |
| Acetone | PCC 6803 | Prbc/Pcpc | cftAB/adc | phaCE,pta | 36 | 4 | Acetyl-CoA | Increasing acetyl-CoA pool | [64] |
| Alkanes | PCC 7120 | Pado | aar, ado (A. halophytica) | 1.25 mg/gDCW | 5 | Acetyl-CoA | Salt stress | [65] | |
2. Short Chain Alcohols
2.1. Ethanol
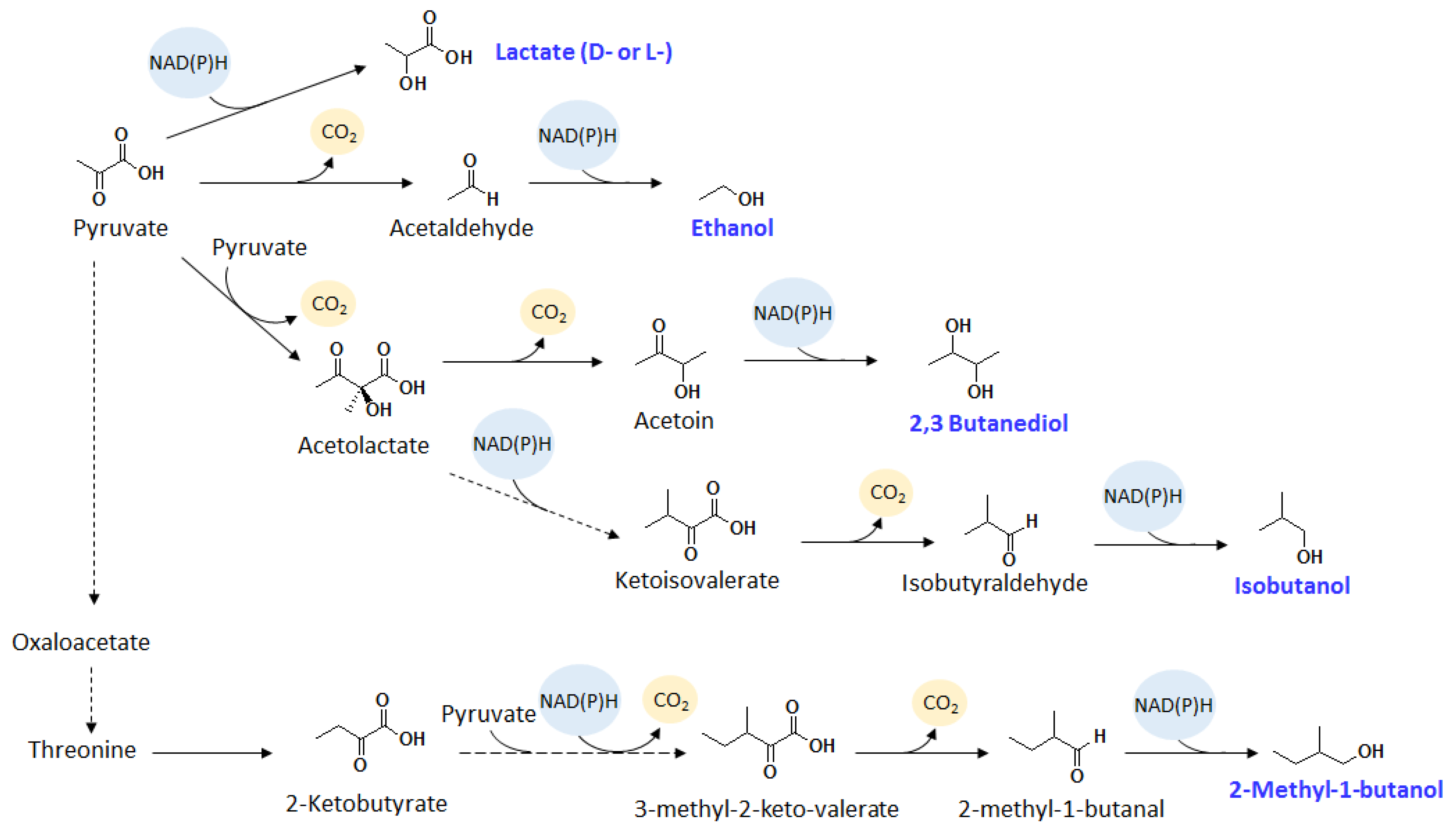
2.2. Isopropanol
2.3. Isobutanol
2.4. 1-Butanol
2.5. 2-Methyl-1-Butanol
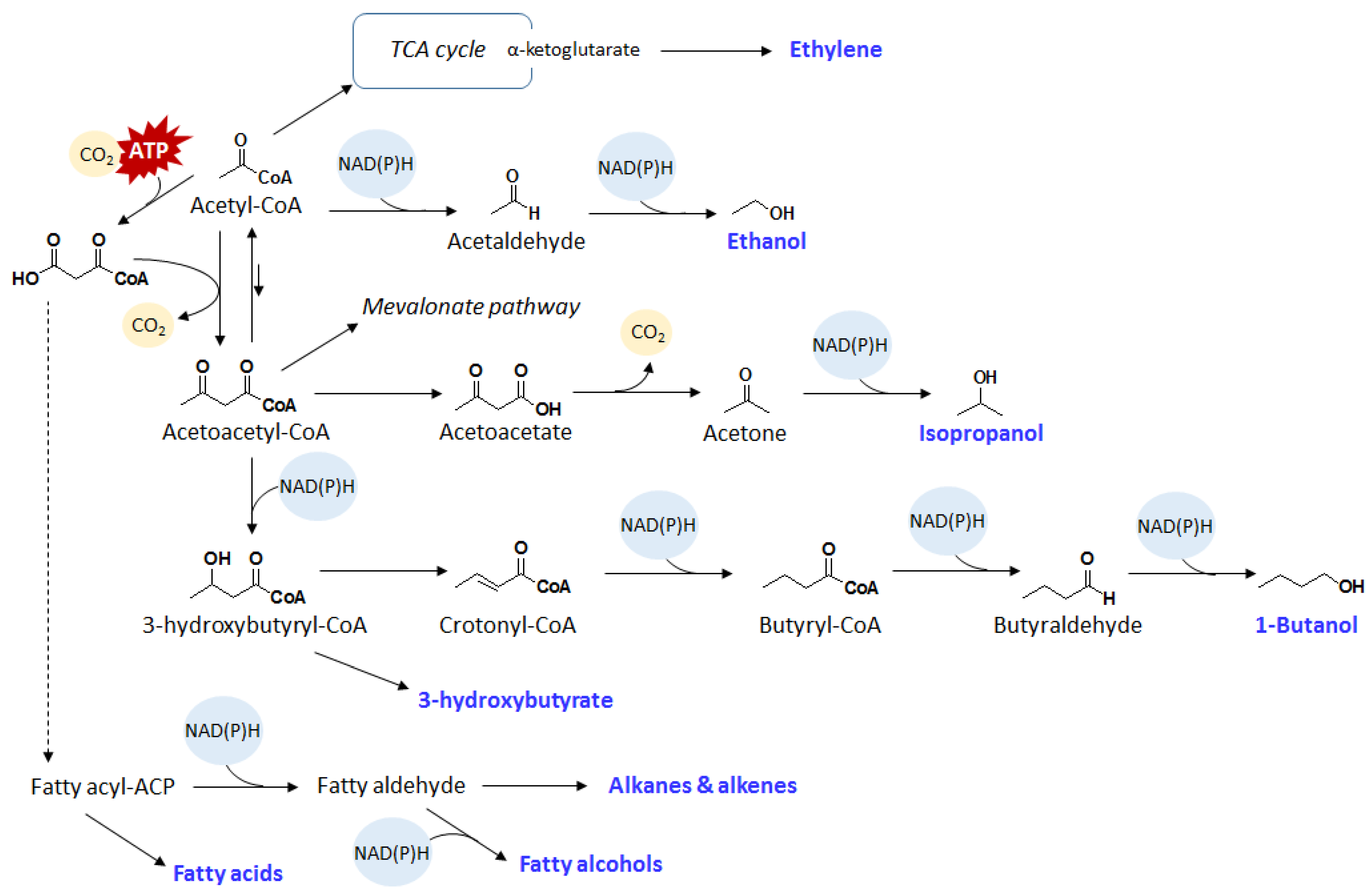
3. Fatty Acids and Hydrocarbons
3.1. Fatty Acids
3.2. Fatty Alcohols and Hydrocarbons
4. Olefins
4.1. Ethylene
4.2. Isoprene
4.3. Terpenoids
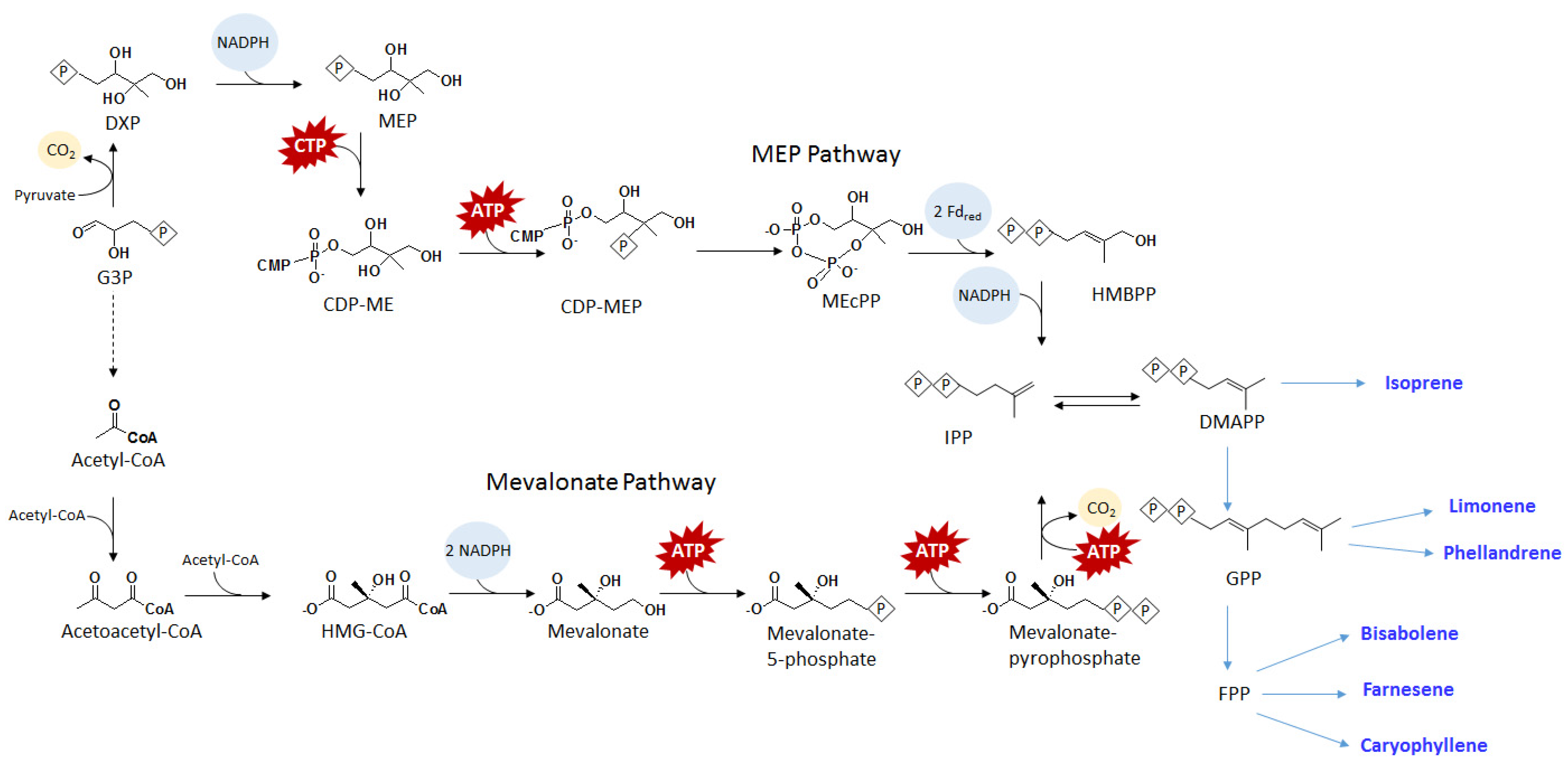
5. Organic Acids
5.1. Lactate
5.2. 3-Hydroxybutyrate
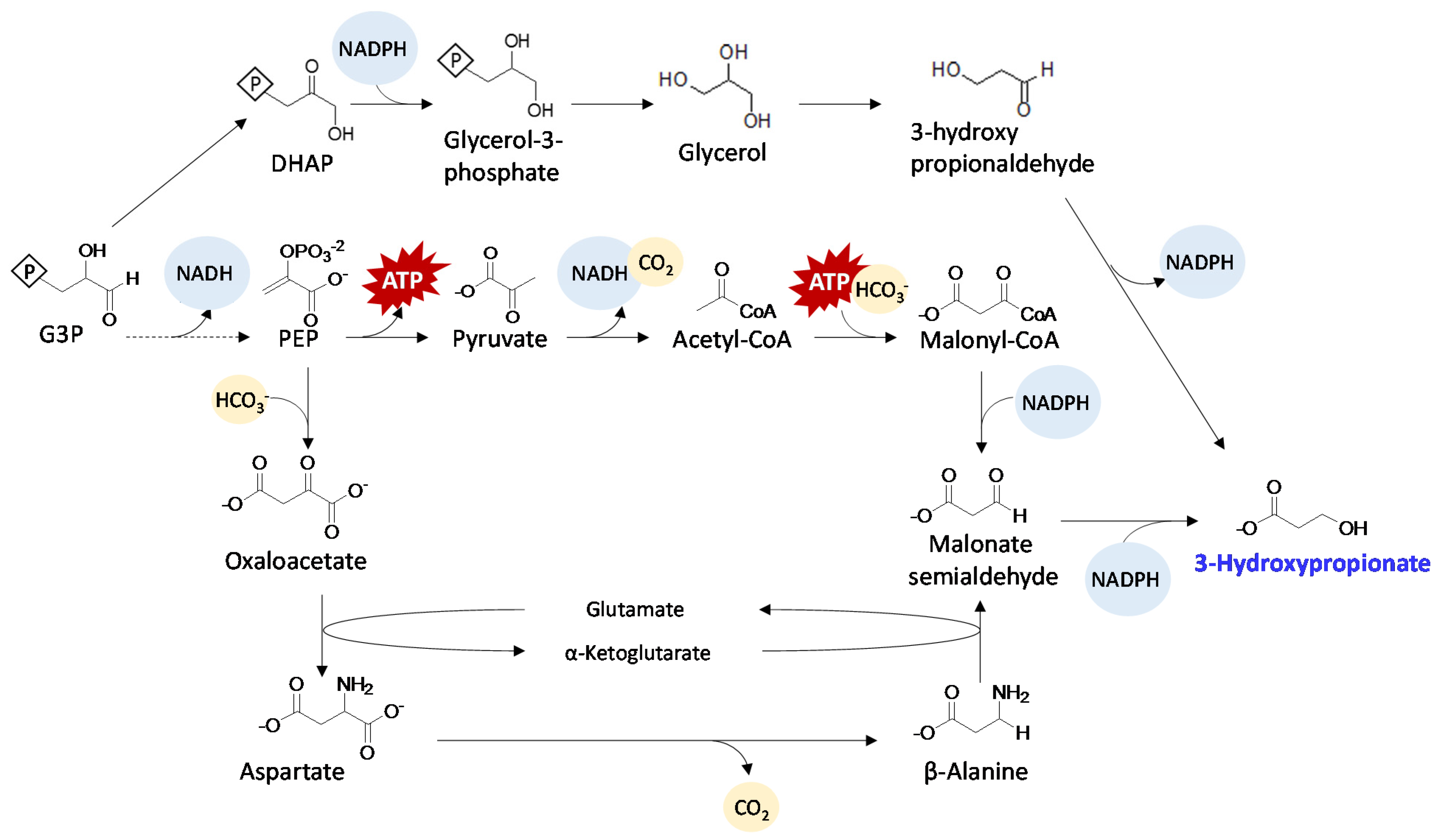
5.3. 3-Hydroxpropionate
5.4. p-Coumaric Acid
6. Sugars
7. Diols and Polyol
7.1. 2,3-Butanediol
7.2. 1,2-Propanediol
7.3. Glycerol
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| G3P | glyceraldehyde-3-phosphate |
| DXP | 1-Deoxy-D-xylulose 5-phosphate |
| MEP | methylerythritol 4-phosphate |
| CDP-ME | 4-diphosphocytidyl-2-C-methylerythritol |
| CDP-MEP | 4-diphosphocytidyl-2-C-methyl-D-erythritol 2-phosphate |
| MEcPP | 2-C-methyl-D-erythritol 2,4-cyclopyrophosphate |
| HMBPP | (E)-4-Hydroxy-3-methyl-but-2-enyl pyrophosphate |
| IPP | Isopentenyl pyrophosphate |
| DMAPP | Dimethylallyl pyrophosphate |
| GPP | Geranyl pyrophosphate |
| FPP | Farnesyl pyrophosphate |
| HMG-CoA | 3-hydroxy-3-methylglutaryl-CoA |
| F6P | fructose-6-phosphate |
| Glc6P | glucose-6-phosphate |
| DHAP | dihydroxyacetone phosphate |
| SAM | S-adenosylmethionine |
References
- Nakao, M.; Okamoto, S.; Kohara, M.; Fujishiro, T.; Fujisawa, T.; Sato, S.; Tabata, S.; Kaneko, T.; Nakamura, Y. Cyanobase: The cyanobacteria genome database update 2010. Nucleic Acids Res. 2010, 38, D379–D381. [Google Scholar] [PubMed]
- Dempo, Y.; Ohta, E.; Nakayama, Y.; Bamba, T.; Fukusaki, E. Molar-based targeted metabolic profiling of cyanobacterial strains with potential for biological production. Metabolites 2014, 4, 499–516. [Google Scholar] [CrossRef] [PubMed]
- Hasunuma, T.; Kikuyama, F.; Matsuda, M.; Aikawa, S.; Izumi, Y.; Kondo, A. Dynamic metabolic profiling of cyanobacterial glycogen biosynthesis under conditions of nitrate depletion. J. Exp. Bot. 2013, 64, 2943–2954. [Google Scholar] [CrossRef] [PubMed]
- Nakajima, T.; Kajihata, S.; Yoshikawa, K.; Matsuda, F.; Furusawa, C.; Hirasawa, T.; Shimizu, H. Integrated metabolic flux and omics analysis of Synechocystis sp. PCC 6803 under mixotrophic and photoheterotrophic conditions. Plant Cell Physiol. 2014, 55, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Triana, J.; Montagud, A.; Siurana, M.; Fuente, D.; Urchueguia, A.; Gamermann, D.; Torres, J.; Tena, J.; de Cordoba, P.F.; Urchueguia, J.F. Generation and evaluation of a genome-scale metabolic network model of Synechococcus elongatus PCC7942. Metabolites 2014, 4, 680–698. [Google Scholar] [CrossRef] [PubMed]
- Knoop, H.; Grundel, M.; Zilliges, Y.; Lehmann, R.; Hoffmann, S.; Lockau, W.; Steuer, R. Flux balance analysis of cyanobacterial metabolism: The metabolic network of Synechocystis sp. PCC 6803. PLoS Comput. Biol. 2013, 9, e1003081. [Google Scholar] [CrossRef] [PubMed]
- Mueller, T.J.; Berla, B.M.; Pakrasi, H.B.; Maranas, C.D. Rapid construction of metabolic models for a family of cyanobacteria using a multiple source annotation workflow. BMC Syst. Biol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Kufryk, G.I.; Sachet, M.; Schmetterer, G.; Vermaas, W.F.J. Transformation of the cyanobacterium Synechocystis sp. PCC 6803 as a tool for genetic mapping: Optimization of efficiency. FEMS Microbiol. Lett. 2002, 206, 215–219. [Google Scholar] [CrossRef] [PubMed]
- Bustos, S.A.; Golden, S.S. Expression of the psbDII gene in Synechococcus sp. Strain PCC 7942 requires sequences downstream of the transcription start site. J. Bacteriol. 1991, 173, 7525–7533. [Google Scholar] [PubMed]
- ElhaI, J.; Vepritskiy, A.; MuroPastor, A.M.; Flores, E.; Wolk, C.P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. Strain PCC 7120. J. Bacteriol. 1997, 179, 1998–2005. [Google Scholar] [PubMed]
- Gao, Z.X.; Zhao, H.; Li, Z.M.; Tan, X.M.; Lu, X.F. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy Environ. Sci. 2012, 5, 9857–9865. [Google Scholar] [CrossRef]
- Dexter, J.; Fu, P.C. Metabolic engineering of cyanobacteria for ethanol production. Energy Environ. Sci. 2009, 2, 857–864. [Google Scholar] [CrossRef]
- Lan, E.I.; Ro, S.Y.; Liao, J.C. Oxygen-tolerant coenzyme a-acylating aldehyde dehydrogenase facilitates efficient photosynthetic n-butanol biosynthesis in cyanobacteria. Energy Environ. Sci. 2013, 6, 2672–2681. [Google Scholar] [CrossRef]
- Deng, M.D.; Coleman, J.R. Ethanol synthesis by genetic engineering in cyanobacteria. Appl. Environ. Microbiol. 1999, 65, 523–528. [Google Scholar] [PubMed]
- Hirokawa, Y.; Suzuki, I.; Hanai, T. Optimization of isopropanol production by engineered cyanobacteria with a synthetic metabolic pathway. J. Biosci. Bioeng. 2015, 119, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Kusakabe, T.; Tatsuke, T.; Tsuruno, K.; Hirokawa, Y.; Atsumi, S.; Liao, J.C.; Hanai, T. Engineering a synthetic pathway in cyanobacteria for isopropanol production directly from carbon dioxide and light. Metab. Eng. 2013, 20, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.I.; Liao, J.C. Atp drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 6018–6023. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.I.; Liao, J.C. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 2011, 13, 353–363. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Higashide, W.; Liao, J.C. Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol. 2009, 27, 1177–1180. [Google Scholar] [CrossRef] [PubMed]
- Varman, A.M.; Xiao, Y.; Pakrasi, H.B.; Tang, Y.J.J. Metabolic engineering of Synechocystis sp. strain PCC 6803 for isobutanol production. Appl. Environ. Microbiol. 2013, 79, 908–914. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.R.; Liao, J.C. Photosynthetic production of 2-methyl-1-butanol from CO2 in cyanobacterium Synechococcus elongatus PCC7942 and characterization of the native acetohydroxyacid synthase. Energy Environ. Sci. 2012, 5, 9574–9583. [Google Scholar] [CrossRef]
- Tan, X.M.; Yao, L.; Gao, Q.Q.; Wang, W.H.; Qi, F.X.; Lu, X.F. Photosynthesis driven conversion of carbon dioxide to fatty alcohols and hydrocarbons in cyanobacteria. Metab. Eng. 2011, 13, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.Q.; Wang, W.H.; Zhao, H.; Lu, X.F. Effects of fatty acid activation on photosynthetic production of fatty acid-based biofuels in Synechocystis sp. PCC6803. Biotechnol. Biofuels 2012, 5, 17. [Google Scholar] [PubMed]
- Li, H.; Liao, J.C. Engineering a cyanobacterium as the catalyst for the photosynthetic conversion of CO2 to 1,2-propanediol. Microb. Cell Fact. 2013. [Google Scholar] [CrossRef] [PubMed]
- Savakis, P.E.; Angermayr, S.A.; Hellingwerf, K.J. Synthesis of 2,3-butanediol by Synechocystis sp. PCC6803 via heterologous expression of a catabolic pathway from lactic acid- and enterobacteria. Metab. Eng. 2013, 20, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.W.; Machado, I.M.; Yoneda, H.; Atsumi, S. Cyanobacterial conversion of carbon dioxide to 2,3-butanediol. Proc. Natl. Acad. Sci. USA 2013, 110, 1249–1254. [Google Scholar] [CrossRef] [PubMed]
- Savakis, P.; Tan, X.M.; Du, W.; dos Santos, F.B.; Lu, X.F.; Hellingwerf, K.J. Photosynthetic production of glycerol by a recombinant cyanobacterium. J. Biotechnol. 2015, 195, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tao, F.; Ni, J.; Li, C.; Xu, P. Production of C3 platform chemicals from CO2 by genetically engineered cyanobacteria. Green Chem. 2015, 17, 3100–3110. [Google Scholar] [CrossRef]
- Varman, A.M.; Yu, Y.; You, L.; Tang, Y.J.J. Photoautotrophic production of d-lactic acid in an engineered cyanobacterium. Microb. Cell Fact. 2013. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, H.F.; Meng, H.K.; Zhang, Y.P.; Li, Y. Production of optically pure d-lactate from CO2 by blocking the phb and acetate pathways and expressing d-lactate dehydrogenase in cyanobacterium Synechocystis sp. PCC 6803. Process Biochem. 2014, 49, 2071–2077. [Google Scholar] [CrossRef]
- Li, C.; Tao, F.; Ni, J.; Wang, Y.; Yao, F.; Xu, P. Enhancing the light-driven production of d-lactate by engineering cyanobacterium using a combinational strategy. Sci. Rep. 2015, 5, 9777. [Google Scholar] [CrossRef] [PubMed]
- Niederholtmeyer, H.; Wolfstadter, B.T.; Savage, D.F.; Silver, P.A.; Way, J.C. Engineering cyanobacteria to synthesize and export hydrophilic products. Appl. Environ. Microbiol. 2010, 76, 3462–3466. [Google Scholar] [CrossRef] [PubMed]
- Angermayr, S.A.; Hellingwerf, K.J. On the use of metabolic control analysis in the optimization of cyanobacterial biosolar cell factories. J. Phys. Chem. B 2013, 117, 11169–11175. [Google Scholar] [CrossRef] [PubMed]
- Angermayr, S.A.; van der Woude, A.D.; Correddu, D.; Vreugdenhil, A.; Verrone, V.; Hellingwerf, K.J. Exploring metabolic engineering design principles for the photosynthetic production of lactic acid by Synechocystis sp. PCC6803. Biotechnol. Biofuels 2014, 7, 99. [Google Scholar] [CrossRef] [PubMed]
- Angermayr, S.A.; Paszota, M.; Hellingwerf, K.J. Engineering a cyanobacterial cell factory for production of lactic acid. Appl. Environ. Microbiol. 2012, 78, 7098–7106. [Google Scholar] [CrossRef] [PubMed]
- Joseph, A.; Aikawa, S.; Sasaki, K.; Tsuge, Y.; Matsuda, F.; Tanaka, T.; Kondo, A. Utilization of lactic acid bacterial genes in Synechocystis sp. PCC 6803 in the production of lactic acid. Biosci. Biotech. Biochem. 2013, 77, 966–970. [Google Scholar] [CrossRef] [PubMed]
- Lan, E.I.; Chuang, D.S.; Shen, C.R.; Lee, A.M.; Ro, S.Y.; Liao, J.C. Metabolic engineering of cyanobacteria for photosynthetic 3-hydroxypropionic acid production from CO2 using Synechococcus elongatus PCC 7942. Metab. Eng. 2015, 31, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Pugh, S.; Nielsen, D.R.; Zhang, W.; Meldrum, D.R. Engineering cyanobacteria for photosynthetic production of 3-hydroxybutyrate directly from CO2. Metab. Eng. 2013, 16C, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Chin, T.; Sano, M.; Takahashi, T.; Ohara, H.; Aso, Y. Photosynthetic production of itaconic acid in Synechocystis sp. PCC6803. J. Biotechnol. 2015, 195, 43–45. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, Y.; Cheng, D.; Daddy, S.; He, Q.F. Genetically engineering Synechocystis sp. pasteur culture collection 6803 for the sustainable production of the plant secondary metabolite p-coumaric acid. Proc. Natl. Acad. Sci. USA 2014, 111, 9449–9454. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Sheng, J.; Curtiss, R. Fatty acid production in genetically modified cyanobacteria. Proc. Natl. Acad. Sci. USA 2011, 108, 6899–6904. [Google Scholar] [CrossRef] [PubMed]
- Ruffing, A.M. Improved free fatty acid production in cyanobacteria with Synechococcus sp. PCC 7002 as host. Front. Bioeng. Biotechnol. 2014, 2, 17. [Google Scholar] [CrossRef] [PubMed]
- Ruffing, A.M.; Jones, H.D. Physiological effects of free fatty acid production in genetically engineered Synechococcus elongatus PCC 7942. Biotechnol. Bioeng. 2012, 109, 2190–2199. [Google Scholar] [CrossRef] [PubMed]
- Ruffing, A.M. Borrowing genes from chlamydomonas reinhardtii for free fatty acid production in engineered cyanobacteria. J. Appl. Phycol. 2013, 25, 1495–1507. [Google Scholar] [CrossRef]
- Jacobsen, J.H.; Frigaard, N.U. Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab. Eng. 2014, 21, 60–70. [Google Scholar] [PubMed]
- Ducat, D.C.; Avelar-Rivas, J.A.; Way, J.C.; Silver, P.A. Rerouting carbon flux to enhance photosynthetic productivity. Appl. Environ. Microbiol. 2012, 78, 2660–2668. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Liang, F.Y.; Duan, Y.K.; Tan, X.M.; Lu, X.F. Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab. Eng. 2013, 19, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.M.; Du, W.; Lu, X.F. Photosynthetic and extracellular production of glucosylglycerol by genetically engineered and gel-encapsulated cyanobacteria. Appl. Microbiol. Biotechnol. 2015, 99, 2147–2154. [Google Scholar] [CrossRef] [PubMed]
- Guerrero, F.; Carbonell, V.; Cossu, M.; Correddu, D.; Jones, P.R. Ethylene synthesis and regulated expression of recombinant protein in Synechocystis sp. PCC 6803. PLoS ONE 2012, 7, e50470. [Google Scholar] [PubMed]
- Ungerer, J.; Tao, L.; Davis, M.; Ghirardi, M.; Maness, P.-C.; Yu, J. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy Environ. Sci. 2012, 5, 8998–9006. [Google Scholar] [CrossRef]
- Jindou, S.; Ito, Y.; Mito, N.; Uematsu, K.; Hosoda, A.; Tamura, H. Engineered platform for bioethylene production by a cyanobacterium expressing a chimeric complex of plant enzymes. ACS Synth. Biol. 2014, 3, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Takahama, K.; Matsuoka, M.; Nagahama, K.; Ogawa, T. Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbai locus. J. Biosci. Bioeng. 2003, 95, 302–305. [Google Scholar] [CrossRef]
- Bentley, F.K.; Melis, A. Diffusion-based process for carbon dioxide uptake and isoprene emission in gaseous/aqueous two-phase photobioreactors by photosynthetic microorganisms. Biotechnol. Bioeng. 2012, 109, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Bentley, F.K.; Zurbriggen, A.; Melis, A. Heterologous expression of the mevalonic acid pathway in cyanobacteria enhances endogenous carbon partitioning to isoprene. Mol. Plant 2014, 7, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, P.; Park, S.; Melis, A. Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 2010, 12, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Kiyota, H.; Okuda, Y.; Ito, M.; Hirai, M.Y.; Ikeuchi, M. Engineering of cyanobacteria for the photosynthetic production of limonene from CO2. J. Biotechnol. 2014, 185, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Davies, F.K.; Work, V.H.; Beliaev, A.S.; Posewitz, M.C. Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2014, 2, 21. [Google Scholar] [CrossRef] [PubMed]
- Halfmann, C.; Gu, L.P.; Zhou, R.B. Engineering cyanobacteria for the production of a cyclic hydrocarbon fuel from CO2 and H2O. Green Chem. 2014, 16, 3175–3185. [Google Scholar] [CrossRef]
- Halfmann, C.; Gu, L.P.; Gibbons, W.; Zhou, R.B. Genetically engineering cyanobacteria to convert CO2, water, and light into the long-chain hydrocarbon farnesene. Appl. Microbiol. Biotechnol. 2014, 98, 9869–9877. [Google Scholar] [CrossRef] [PubMed]
- Qi, Q.G.; Hao, M.; Ng, W.O.; Slater, S.C.; Baszis, S.R.; Weiss, J.D.; Valentin, H.E. Application of the Synechococcus nirA promoter to establish an inducible expression system for engineering the Synechocystis tocopherol pathway. Appl. Environ. Microbiol. 2005, 71, 5678–5684. [Google Scholar] [CrossRef] [PubMed]
- Reinsvold, R.E.; Jinkerson, R.E.; Radakovits, R.; Posewitz, M.C.; Basu, C. The production of the sesquiterpene beta-caryophyllene in a transgenic strain of the cyanobacterium Synechocystis. J. Plant. Physiol. 2011, 168, 848–852. [Google Scholar] [CrossRef] [PubMed]
- Formighieri, C.; Melis, A. Regulation of beta-phellandrene synthase gene expression, recombinant protein accumulation, and monoterpene hydrocarbons production in Synechocystis transformants. Planta 2014, 240, 309–324. [Google Scholar] [CrossRef] [PubMed]
- Bentley, F.; García-Cerdán, J.; Chen, H.-C.; Melis, A. Paradigm of monoterpene (β-phellandrene) hydrocarbons production via photosynthesis in cyanobacteria. Bioenergy Res. 2013, 6, 917–929. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, H.; Zhang, Y.; Li, Y.; Ma, Y. Designing and creating a modularized synthetic pathway in cyanobacterium Synechocystis enables production of acetone from carbon dioxide. Metab. Eng. 2012, 14, 394–400. [Google Scholar] [PubMed]
- Kageyama, H.; Waditee-Sirisattha, R.; Sirisattha, S.; Tanaka, Y.; Mahakhant, A.; Takabe, T. Improved alkane production in nitrogen-fixing and halotolerant cyanobacteria via abiotic stresses and genetic manipulation of alkane synthetic genes. Curr. Microbiol. 2015, 71, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Stal, L.J.; Moezelaar, R. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 1997, 21, 179–211. [Google Scholar] [CrossRef]
- Shen, C.R.; Lan, E.I.; Dekishima, Y.; Baez, A.; Cho, K.M.; Liao, J.C. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2905–2915. [Google Scholar] [CrossRef] [PubMed]
- Bond-Watts, B.B.; Bellerose, R.J.; Chang, M.C.Y. Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 2011, 7, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Atsumi, S.; Liao, J.C. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 7802–7808. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fallon, S.; Sheng, J.; Curtiss, R., 3rd. CO2-limitation-inducible green recovery of fatty acids from cyanobacterial biomass. Proc. Natl. Acad. Sci. USA 2011, 108, 6905–6908. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; McCarthy, E.D.; Hoeven, W.V.; Calvin, M.; Bradley, W.H. Organic geochemical studies, ii. A preliminary report on the distribution of aliphatic hydrocarbons in algae, in bacteria, and in a recent lake sediment. Proc. Natl. Acad. Sci. USA 1968, 59, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Schirmer, A.; Rude, M.A.; Li, X.; Popova, E.; del Cardayre, S.B. Microbial biosynthesis of alkanes. Science 2010, 329, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Perez, D.; Herman, N.A.; Pfleger, B.F. A desaturase gene involved in the formation of 1,14-nonadecadiene in Synechococcus sp. Strain PCC 7002. Appl. Environ. Microbiol. 2014, 80, 6073–6079. [Google Scholar] [CrossRef] [PubMed]
- Peramuna, A.; Morton, R.; Summers, M.L. Enhancing alkane production in cyanobacterial lipid droplets: A model platform for industrially relevant compound production. Life 2015, 5, 1111–1126. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.H.; Liu, X.F.; Lu, X.F. Engineering cyanobacteria to improve photosynthetic production of alka(e)nes. Biotechnol. Biofuels 2013, 6. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lee, S.Y. Microbial production of short-chain alkanes. Nature 2013, 502, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, H.; Sakai, M.; Nagahama, K.; Fujii, T.; Matsuoka, M.; Inoue, Y.; Ogawa, T. Heterologous expression of the gene for the ethylene-forming enzyme from Pseudomonas syringae in the cyanobacterium Synechococcus. Biotechnol. Lett. 1994, 16, 1–6. [Google Scholar] [CrossRef]
- Sakai, M.; Ogawa, T.; Matsuoka, M.; Fukuda, H. Photosynthetic conversion of carbon dioxide to ethylene by the recombinant cyanobacterium, Synechococcus sp. PCC 7942, which harbors a gene for the ethylene-forming enzyme of Pseudomonas syringae. J. Ferment. Bioeng. 1997, 84, 434–443. [Google Scholar] [CrossRef]
- Lee, T.C.; Xiong, W.; Paddock, T.; Carrieri, D.; Chang, I.F.; Chiu, H.F.; Ungerer, J.; Hank Juo, S.H.; Maness, P.C.; Yu, J. Engineered xylose utilization enhances bio-products productivity in the cyanobacterium Synechocystis sp. PCC 6803. Metab. Eng. 2015, 30, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Englund, E.; Andersen-Ranberg, J.; Miao, R.; Hamberger, B.; Lindberg, P. Metabolic engineering of Synechocystis sp. PCC 6803 for production of the plant diterpenoid manoyl oxide. ACS Synth. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hollinshead, W.D.; Varman, A.M.; You, L.; Hembree, Z.; Tang, Y.J.J. Boosting d-lactate production in engineered cyanobacteria using sterilized anaerobic digestion effluents. Bioresour. Technol. 2014, 169, 462–467. [Google Scholar] [CrossRef] [PubMed]
- Begemann, M.B.; Zess, E.K.; Walters, E.M.; Schmitt, E.F.; Markley, A.L.; Pfleger, B.F. An organic acid based counter selection system for cyanobacteria. PLoS ONE 2013, 8, e76594. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.X.; Cho, K.M.; Rivera, J.G.L.; Monte, E.; Shen, C.R.; Yan, Y.J.; Liao, J.C. Conversion of proteins into biofuels by engineering nitrogen flux. Nat. Biotechnol. 2011, 29, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, Y.; Grace, S.; He, Q. Functional expression of an Arabidopsis p450 enzyme, p-coumarate-3-hydroxylase, in the cyanobacterium Synechocystis PCC 6803 for the biosynthesis of caffeic acid. J. Appl. Phycol. 2014, 26, 219–226. [Google Scholar] [CrossRef]
- Klahn, S.; Hagemann, M. Compatible solute biosynthesis in cyanobacteria. Environ. Microbiol. 2011, 13, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, E.; Ohkawa, H.; Moriya, K.; Matsubara, T.; Nagaike, Y.; Iwasaki, I.; Fujiwara, S.; Tsuzuki, M.; Nakamura, Y. Carbohydrate metabolism in mutants of the cyanobacterium Synechococcus elongatus PCC 7942 defective in glycogen synthesis. Appl. Environ. Microbiol. 2010, 76, 3153–3159. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.W.; Machado, I.M.; Yoneda, H.; Atsumi, S. Combinatorial optimization of cyanobacterial 2,3-butanediol production. Metab. Eng. 2014, 22, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Oliver, J.W.; Atsumi, S. A carbon sink pathway increases carbon productivity in cyanobacteria. Metab. Eng. 2015, 29, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Nozzi, N.E.; Atsumi, S. Genome engineering of the 2,3-butanediol biosynthetic pathway for tight regulation in cyanobacteria. ACS Synth. Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Clomburg, J.M.; Gonzalez, R. Anaerobic fermentation of glycerol: A platform for renewable fuels and chemicals. Trends Biotechnol. 2013, 31, 20–28. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, M.C.; Lan, E.I. Advances in Metabolic Engineering of Cyanobacteria for Photosynthetic Biochemical Production. Metabolites 2015, 5, 636-658. https://doi.org/10.3390/metabo5040636
Lai MC, Lan EI. Advances in Metabolic Engineering of Cyanobacteria for Photosynthetic Biochemical Production. Metabolites. 2015; 5(4):636-658. https://doi.org/10.3390/metabo5040636
Chicago/Turabian StyleLai, Martin C., and Ethan I. Lan. 2015. "Advances in Metabolic Engineering of Cyanobacteria for Photosynthetic Biochemical Production" Metabolites 5, no. 4: 636-658. https://doi.org/10.3390/metabo5040636
APA StyleLai, M. C., & Lan, E. I. (2015). Advances in Metabolic Engineering of Cyanobacteria for Photosynthetic Biochemical Production. Metabolites, 5(4), 636-658. https://doi.org/10.3390/metabo5040636






