Characterization of Flavan-3-ols and Expression of MYB and Late Pathway Genes Involved in Proanthocyanidin Biosynthesis in Foliage of Vitis bellula
Abstract
:1. Introduction
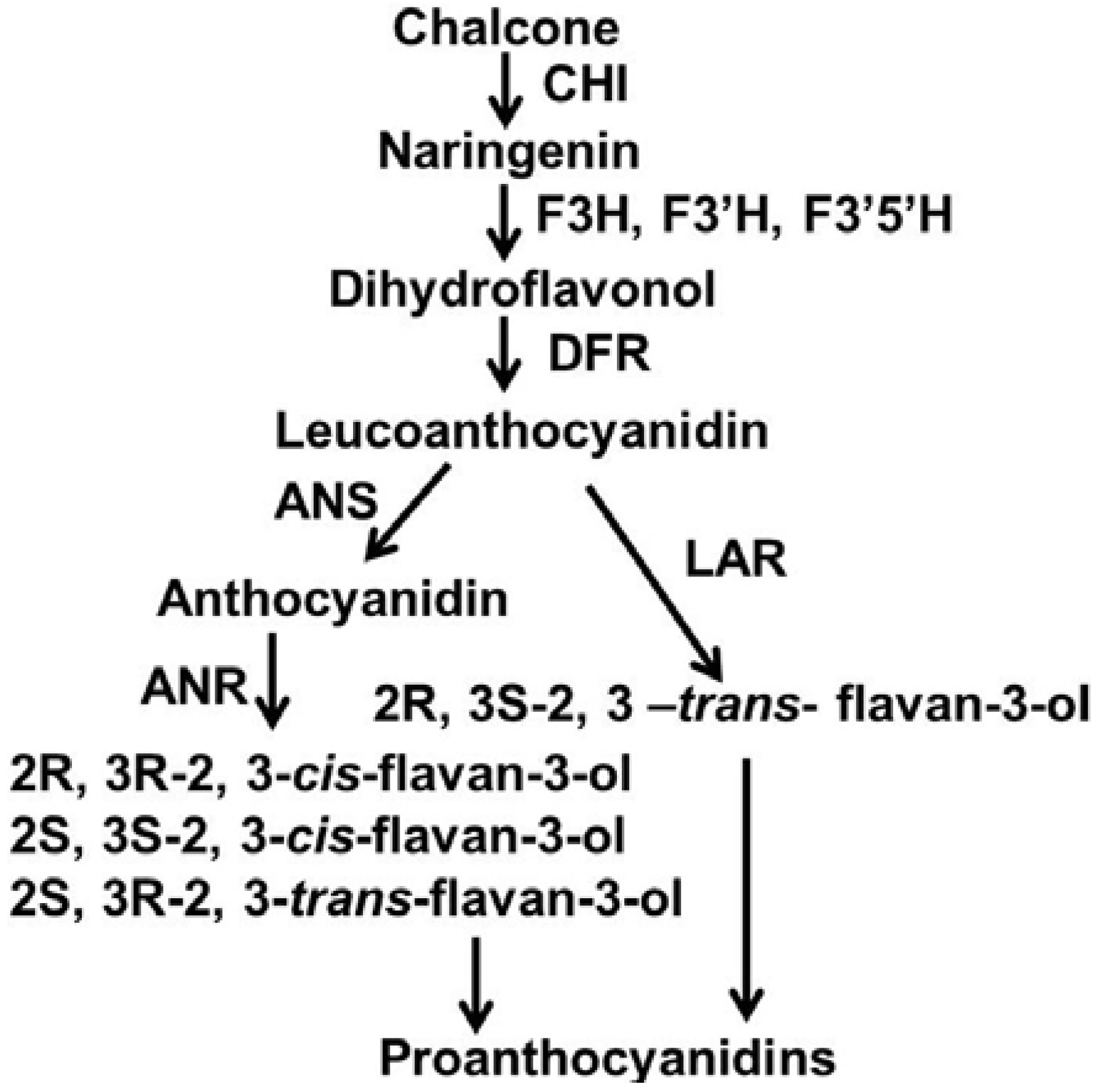
2. Results and Discussion
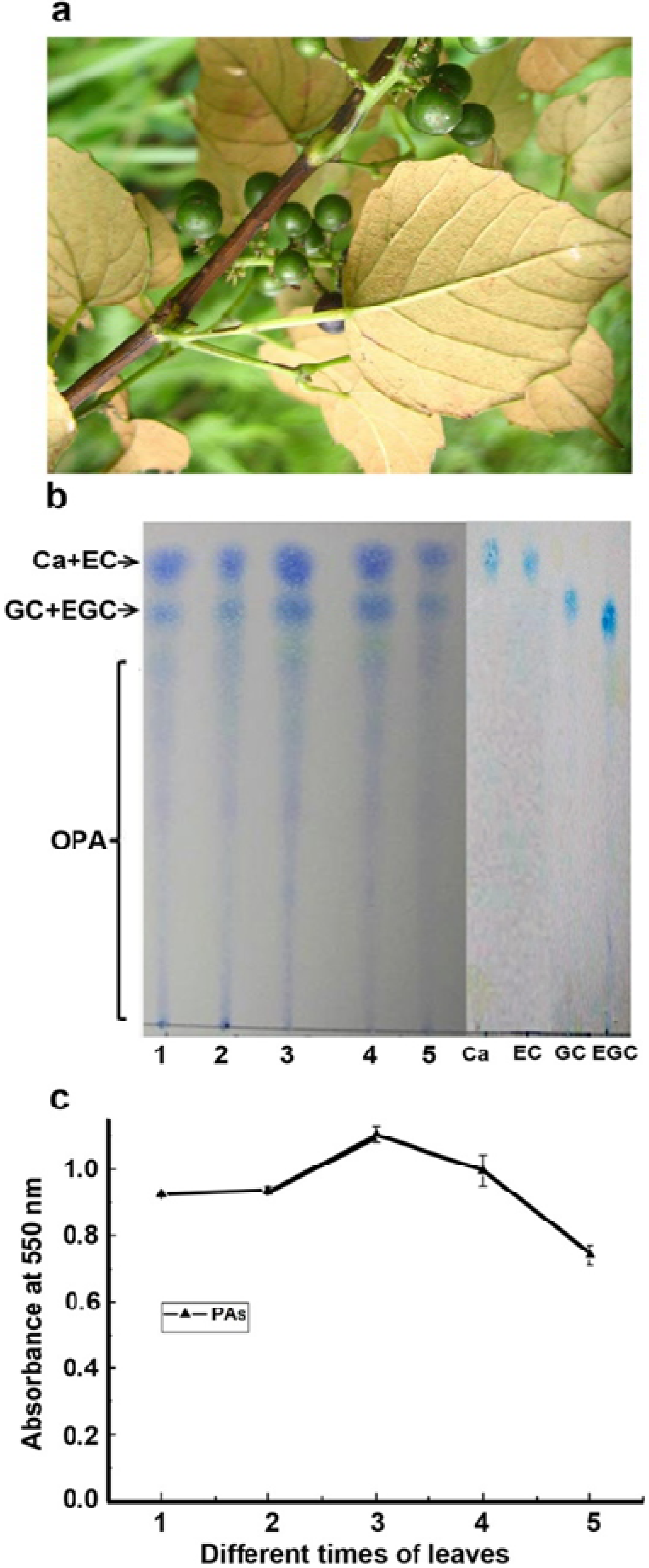
2.1. Characterization of Proanthocyanidin Formation at Different Stages of Leaf Development
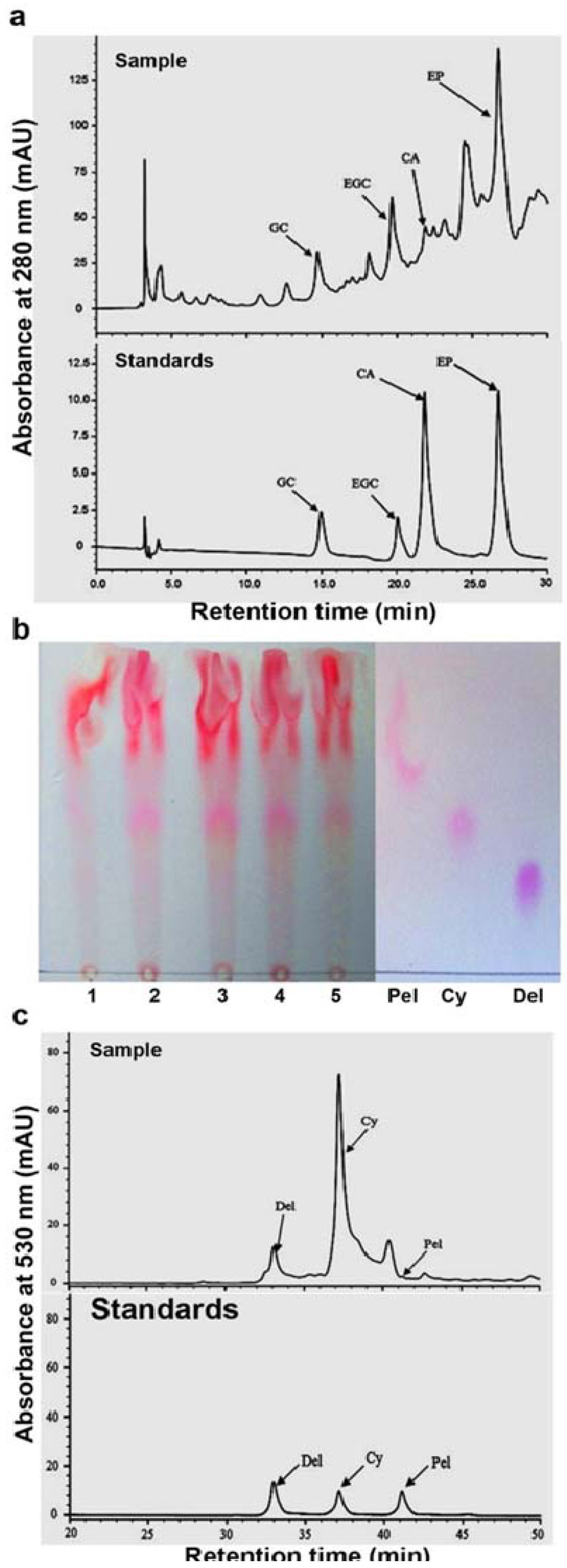
2.2. Characterization of Flavan-3-ols and Extension Units of Proanthocyanidins
2.3. Expression Patterns of ANR, LAR and DFR Homologs During Leaf Development
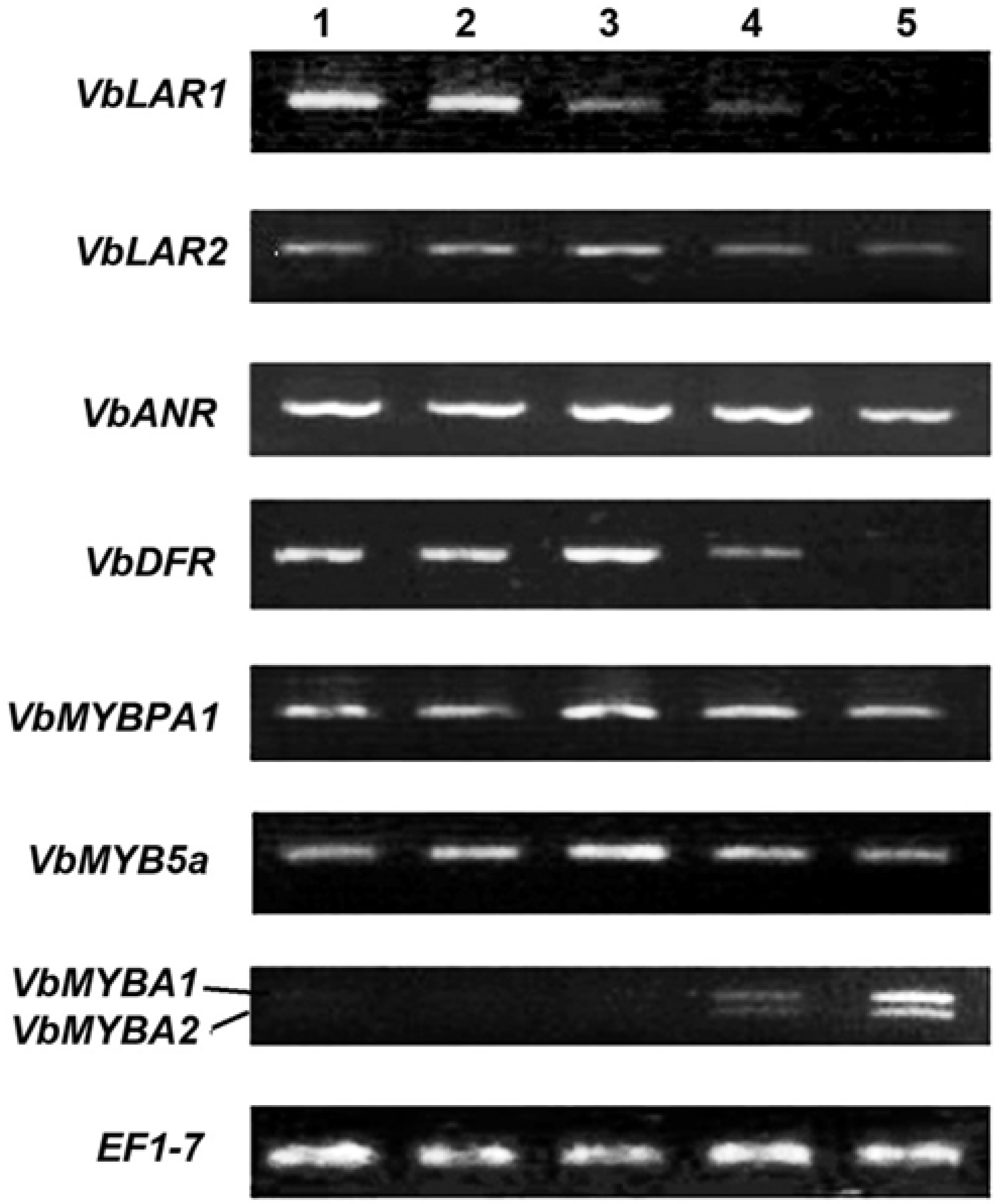
2.4. Expression Patterns of VbMYBPA1, VbMYB5a, VbMYBA1 and VbMYBA2 During Leaf Development
2.5. Discussion
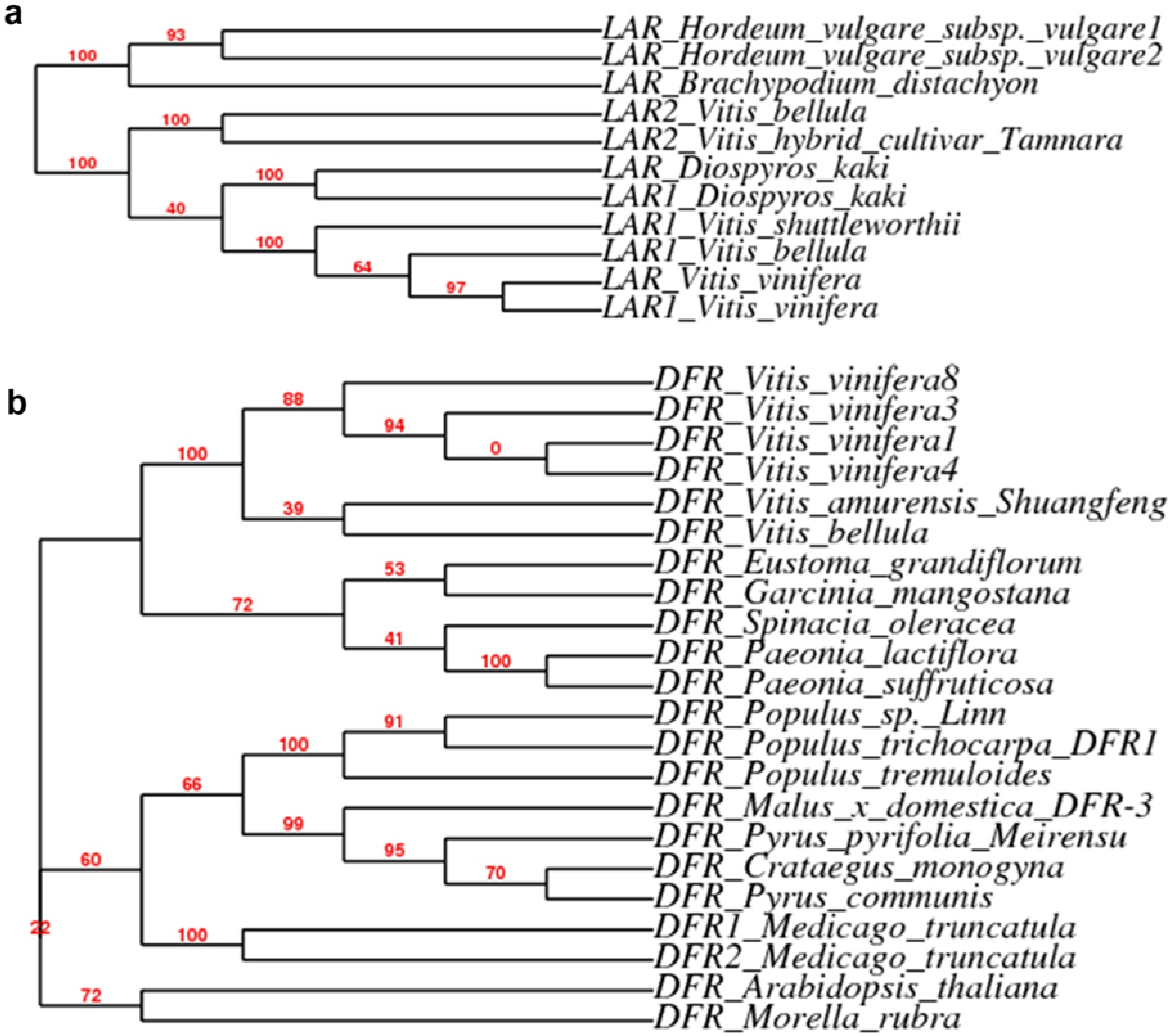
3. Experimental Section
3.1. Plant Materials
3.2. Extraction of Proanthocyanidins And Flavan-3-ols and TLC Assay
3.3. Butanol: HCl Boiling of Proanthocyanidins, Absorbance Measurement and TLC Assay
3.4. HPLC Analysis of Flavan-3-ols and Anthocyanidins
3.5. Total RNA Isolation and Semi-Quantitative RT-PCR Analysis
| cDNAs | Sequence 5'-3' | Thermal Cycle |
|---|---|---|
| VbLAR1 | F: ATGACTGTTTCTCCGGTTCCTTCG | 94 °C 5 min, 30cycles of 94 °C 30 sec, 58 °C 30 sec and 72 °C 45 sec, 72 °C 10 min |
| R: TCAAGCGCAGGTTGCAGTGAC | ||
| VbLAR2 | F: ATGACTGTTTTGTCTGTGAGTACTC | 94 °C 5 min, 30cycles of 94 °C30 sec, 55 °C 30 sec and 72 °C 45 sec, 72 °C 10 min |
| R: TCAGGCGCAGGTAGCAGTGATG | ||
| VbANR | F: ATGGCCACCCAGCACCCCAT | 94 °C 5 min, 30cycles of 94 °C30 sec, 55°C 30 sec and 72 °C 45 sec, 72 °C 10 min |
| R: TCAATTCTGCAATAGCCCCTTGGC | ||
| VbDFR | F: ATGGGTTCACAAAGTGAAACCGTG | 94 °C 5 min, 30cycles of 94 °C30 sec, 50 °C 30 sec and 72 °C 45 sec, 72 °C 10 min |
| R: CTAGGTCTTGCCATCTACAGG | ||
| VbMYBPA1 | F: ATGGGCAGAGCACCTTGTTG | 94 °C 5 min, 30cycles of 94 °C30 sec, 50 °C 30 sec and 72 °C 45 sec, 72 °C 10 min |
| R: TTAAATGAGTAGTGATTCGGCG | ||
| VbMYB5a | F: ATGAGGAATGCATCCTCAGCATCAG | 94 °C 5 min, 30cycles of 94 °C30 sec, 55 °C 30 sec and 2 °C 45 sec, 72 °C 10 min |
| R: TCAGAACCGCTTATCAGGTTGATCG | ||
| VbMYBA1 and 2 | F: ATGGAGAGCTTAGGAGTTAGAAAG | 94 °C 5 min, 30cycles of 94 °C30 sec, 47 °C 30 sec and 72 °C 45 sec, 72 °C 10 min |
| R:TCAGATCAAGTGATTTACTTGTG | ||
| EF1-γ | F: GCGGGCAAGAGATACCTCAA | 94 °C 5 min, 30cycles of 94 °C30 sec, 50 °C 30 sec and 72 °C 30 sec, 72 °C 10 min |
| R:TCAATCTGTCTAGGAAAGGAAG |
3.6. Sequence Analysis
3.7. Statistical Analysis
4. Conclusion
Acknowledgements
Conflict of Interest
References
- Xie, D.-Y.; Dixon, R.A. Proanthocyanidin biosynthesis—Still more questions than answers? Phytochemistry 2005, 66, 2127–2144. [Google Scholar] [CrossRef]
- Dixon, R.A.; Xie, D.Y.; Sharma, S.B. Proanthocyanidins—A final frontier in flavonoid research? New Phytol. 2005, 165:9–165:28. [Google Scholar] [CrossRef]
- Greenspan, P.; Bauer, J.D.; Pollock, S.H.; Gangemi, J.D.; Mayer, E.P.; Ghaffar, A.; Hargrove, J.L.; Hartle, D.K. Antiinflammatory properties of the muscadine grape (Vitis rotundifolia). J. Agric. Food Chem. 2005, 53, 8481–8484. [Google Scholar] [CrossRef]
- Stringer, S.J.; Perkins Veazie, P.M.; Marshall, D.A. Natraceutical profile of selected muscadine (Witis rotundifolia michx.) cultivars and breeding lines. Hortscience 2005, 40, 1068. [Google Scholar]
- Radvanyi, J., Jr.; Stojanovic, B.J.; Drapala, W.J.; Overcash, J.P.; Hegwood, C.P., Jr. Composition and quality of juices and wines of eight Vitis rotundifolia Michx. cultivars. Am. J. Enol. Vitic. 1980, 31, 316–322. [Google Scholar]
- Weinges, K.; Piretti, M.V. Proanthocyanidins. 20. Isolation of C30H26O12-Procyanidin-B1 from grapes. Annalen Der Chemie-Justus Liebig 1971, 748, 218. [Google Scholar] [CrossRef]
- Bagchi, D.; Garg, A.; Krohn, R.L.; Bagchi, M.; Bagchi, D.J.; Balmoori, J.; Stohs, S.J. Protective effects of grape seed proanthocyanidins and selected antioxidants against TPA-induced hepatic and brain lipid peroxidation and DNA fragmentation, and peritoneal macrophage activation in mice. Gen. Pharmacol. 1998, 30, 771–776. [Google Scholar] [CrossRef]
- Kennedy, J.A.; Hayasaka, Y.; Vidal, S.; Waters, E.J.; Jones, G.P. Composition of grape skin proanthocyanidins at different stages of berry development. J. Agric. Food Chem. 2001, 49, 5348–5355. [Google Scholar] [CrossRef]
- Natella, F.; Belelli, F.; Gentili, V.; Ursini, F.; Scaccini, C. Grape seed proanthocyanidins prevent plasma postprandial oxidative stress in humans. J. Agric. Food Chem. 2002, 50, 7720–7725. [Google Scholar] [CrossRef]
- Renaud, S.; de Lorgeril, M. Wine, alcohol, platelets, and the French paradox for coronary heart disease. The Lancet 1992, 339, 1523. [Google Scholar] [CrossRef]
- Somers, T.C. Wine tannins-isolation of condensed flavonoid pigments by gel-filtration. Nature 1966, 209, 368–370. [Google Scholar] [CrossRef]
- Yamakoshi, J.; Kataoka, S.; Koga, T.; Ariga, T. Proanthocyanidin-rich extract from grape seeds attenuates the development of aortic atherosclerosis in cholesterol-fed rabbits. Atherosclerosis 1999, 142, 139–149. [Google Scholar] [CrossRef]
- Sato, M.; Ray, P.S.; Maulik, G.; Maulik, N.; Engelman, R.M.; Bertelli, A.A. E.; Bertelli, A.; Das, D.K. Myocardial protection with red wine extract. J. Cardiovascul. Pharmacol. 2000, 35, 263–268. [Google Scholar] [CrossRef]
- Sanchez-Moreno, C.; Cao, G.; Ou, B.; Prior, R.L. Anthocyanin and proanthocyanidin content in selected white and red Wines. Oxygen radical absorbance capacity comparison with nontraditional wines obtained from Highbush blueberry. J. Agric. Food Chem. 2003, 51, 4889–4896. [Google Scholar] [CrossRef]
- Jordao, A.M.; Goncalves, F.J.; Correia, A.C.; Cantao, J.; Rivero-Perez, M.D.; SanJose, M.L.G. Proanthocyanidin content, antioxidant capacity and scavenger activity of Portuguese sparkling wines (Bairrada Appellation of Origin). J. Sci. Food Agric. 2010, 90, 2144–2152. [Google Scholar]
- Rasmussen, S.E.; Frederiksen, H.; Struntze, K.; Poulsen, K.L. Dietary proanthocyanidins: Occurrence, dietary intake, bioavailability, and protection against cardiovascular disease. Mol. Nutrit. Food Res. 2005, 49, 159–174. [Google Scholar] [CrossRef]
- Sato, M.; Maulik, G.; Ray, P.S.; Bagchi, D.; Das, D.K. Cardioprotective effects of grape seed proanthocyanidin against ischemic reperfusion injury. J. Mol. Cell. Cardiol 1999, 31, 1289–1297. [Google Scholar] [CrossRef]
- Tanner, G.J.; Francki, K.T; Abrahams, S.; Watson, J.M.; Larkin, P.J.; Ashton, A.R. Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J. Biol Chem. 2003, 278, 31647–31656. [Google Scholar]
- Peng, Q.Z.; Zhu, Y.; Liu, Z.; Du, C.; Li, K.G.; Xie, D.Y. An integrated approach to demonstrating the ANR pathway of proanthocyanidin biosynthesis in plants. Planta 2012, 236, 901–918. [Google Scholar] [CrossRef]
- Xie, D-Y.; Sharma, S.B.; Paiva, N.L.; Ferreira, D.; Dixon, R.A. Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 2003, 299, 396–399. [Google Scholar] [CrossRef]
- Gagné, S.; Lacampagne, S.; Claisse, O.; Gény, L. Leucoanthocyanidin reductase and anthocyanidin reductase gene expression and activity in flowers, young berries and skins of Vitis vinifera L. cv. Cabernet-Sauvignon during development. Plant Physiol. Biochem. 2009, 47, 282–290. [Google Scholar] [CrossRef]
- Bogs, J.; Downey, M.O.; Harvey, J.S.; Ashton, A.R.; Tanner, G.J.; Robinson, S.P. Proanthocyanidin synthesis and expression of genes encoding leucoanthocyanidin reductase and anthocyanidin reductase in developing grape berries and grapevine leaves. Plant Physiol. 2005, 139, 652–663. [Google Scholar] [CrossRef]
- Gargouri, M.; Manigand, C.; Mauge, C.; Granier, T.; Langlois d'Estaintot, B.; Cala, O.; Pianet, I.; Bathany, K.; Chaudiere, J.; Gallois, B. Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera. Acta Crystallogr. Sect. D—Biol. Crystallogr. 2009, 65, 989–1000. [Google Scholar] [CrossRef]
- Pfeiffer, J.; Kuhnel, C.; Brandt, J.; Duy, D.; Punyasiri, P.A.N.; Forkmann, G.; Fischer, T.C. Biosynthesis of flavan 3-ols by leucoanthocyanidin 4-reductases and anthocyanidin reductases in leaves of grape (Vitis vinifera L.), apple (Malus x domestica Borkh.) and other crops. Plant Physiol. Biochem. 2006, 44, 323–334. [Google Scholar] [CrossRef]
- Bogs, J.; Jaffe, F.W.; Takos, A.M.; Walker, A.R.; Robinson, S.P. The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 2007, 143, 1347–1361. [Google Scholar] [CrossRef]
- Xie, D.-Y.; Sharma, S.B.; Wright, E.; Wang, Z.-Y.; Dixon, R.A. Metabolic engineering of proanthocyanidins through co-expression of anthocyanidin reductase and the PAP1 MYB transcription factor. Plant J. 2006, 45, 895–907. [Google Scholar] [CrossRef]
- Lacampagne, S.; Gagne, S.; Geny, L. Involvement of abscisic acid in controlling the proanthocyanidin biosynthesis pathway in grape skin: New elements regarding the regulation of tannin composition and leucoanthocyanidin reductase (LAR) and anthocyanidin reductase (ANR) activities and expression. J. Plant Growth Regul. 2010, 29, 81–90. [Google Scholar] [CrossRef]
- Deluc, L.; Barrieu, F.; Marchive, C.; Lauvergeat, V.; Decendit, A.; Richard, T.; Carde, J.-P.; Merillon, J.-M.; Hamdi, S. Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol.200 499–511.
- Kobayashi, S.; Yamamoto, N.G.; Hirochika, H. Association of VvmybA1 gene expression with anthocyanin production in grape (Vitis vinifera) skin - color mutants. J. Jpn. Soc. Hortic. Sci. 2005, 74, 196–203. [Google Scholar] [CrossRef]
- Jeong, S.T.; Goto-Yamamoto, N.; Hashizume, K.; Kobayashi, S.; Esaka, M. Expression of VvmybA1 gene and anthocyanin accumulation in various grape organs. Am. J. Enol. Vitic. 2006, 57, 507–510. [Google Scholar]
- Yakushiji, H.; Kobayashi, S.; Goto-Yamamoto, N.; Jeong, S.T.; Sueta, T.; Mitani, N.; Azuma, A. A skin color mutation of grapevine, from black-skinned Pinot Noir to white-skinned Pinot Blanc, is caused by deletion of the functional VvmybA1 allele. Biosci. Biotechnol. Biochem. 2006, 70, 1506–1508. [Google Scholar] [CrossRef]
- Azuma, A.; Kobayashi, S.; Yakushiji, H.; Yamada, M.; Mitani, N.; Sato, A. VvmybA1 genotype determines grape skin color. Vitis 2007, 46, 154–155. [Google Scholar]
- This, P.; Lacombe, T.; Cadle-Davidson, M.; Owens, C.L. Wine grape (Vitis vinifera L.) color associates with allelic variation in the domestication gene VvmybA1. Theor. Appl. Genet. 2007, 114, 723–730. [Google Scholar] [CrossRef]
- Güner, A.; Gyulai, G.; Tóth, Z.; Başlı1, G.A.; Szabó, Z.; Gyulai, F.; Bittsánszky, A.; Waters, L., Jr.; Heszky, L. Grape (Vitis vinifera) seeds from Antiquity and the Middle Ages Excavated in Hungary - LM and SEM analysis. Anadolu. Univ. J. Sci. Technol. 2009, 10, 205–213. [Google Scholar]
- Gris, E.F.; Mattivi, F.; Ferreira, E.A.; Vrhovsek, U.; Pedrosa, R.C.; Bordignon-Luiz, M.T. Proanthocyanidin profile and antioxidant capacity of Brazilian Vitis vinifera red wines. Food Chem. 2011, 126, 213–220. [Google Scholar]
- Spranger, I.; Sun, B.; Mateus, A. Chemical characterization and antioxidant activities of oligomeric and polymeric procyanidin fractions from grape seeds. Food Chem. 2008, 108, 519–532. [Google Scholar] [CrossRef]
- Vitseva, O.; Varghese, S.; Chakrabarti, S.; Folts, J.; Freedman, J. Grape seed and skin extracts inhibit platelet function and release of reactive oxygen intermediates. J. Cardiovascul. Pharmacol. 2005, 46, 445–451. [Google Scholar] [CrossRef]
- Souquet, J.-M.; Cheynier, V.; Brossaud, F.; Moutounet, M. Polymeric proanthocyanidins from grape skins. Phytochemistry 1996, 43, 509–512. [Google Scholar]
- Fuleki, T.; Ricardo-da-Silva, J.M. Catechin and procyanidin composition of seeds from grape cultivars grown in Ontario. J. Agric. Food Chem. 1997, 45, 156–1160. [Google Scholar]
- Gabetta, B.; Fuzzati, N.; Griffini, A.; Lolla, E.; Pace, R.; Ruffilli, T.; Peterlongo, F. Characterization of proanthocyanidins from grape seeds. Fitoterapia 2000, 71, 162–175. [Google Scholar] [CrossRef]
- Decorde, K.; Teissedre, P.L.; Sutra, T.; Ventura, E.; Cristol, J.P.; Rouanet, J.M. Chardonnay grape seed procyanidin extract supplementation prevents high-fat diet-induced obesity in hamsters by improving adipokine imbalance and oxidative stress markers. Mol. Nutr. Food Res. 2009, 53, 659–666. [Google Scholar] [CrossRef]
- Gonzalez-Paramas, A.M.; da Silva, F.L.; Martin-Lopez, P.; Macz-Pop, G.; Gonzalez-Manzano, S.; Alcalde-Eon, C.; Perez-Alonso, J.J.; Escribano-Bailon, M.T.; Rivas-Gonzalo, J.C.; Santos-Buelga, C. Flavanol-anthocyanin condensed pigments in plant extracts. Food Chem. 2006, 94, 428–436. [Google Scholar] [CrossRef]
- Castillo-Munoz, N.; Fernandez-Gonzalez, M.; Gomez-Alonso, S.; Garcia-Romero, E.; Hermosin-Gutierrez, I. Red-color related phenolic composition of Garnacha Tintorera (Vitis vinifera L.) grapes and red wines. J. Agric. Food Chem. 2009, 57, 7883–7891. [Google Scholar] [CrossRef]
- He, F.; He, J.J.; Pan, Q.H.; Duan, C.Q. Mass-spectrometry evidence confirming the presence of pelargonidin-3-O-glucoside in the berry skins of Cabernet Sauvignon and Pinot Noir (Vitis vinifera L.). Aust. J. Grape Wine Res. 2010, 16, 464–468. [Google Scholar] [CrossRef]
- He, J.J.; Liu, Y.X.; Pan, Q.H.; Cui, X.Y.; Duan, C.Q. Different anthocyanin profiles of the skin and the pulp of Yan73 (Muscat Hamburg x Alicante Bouschet) grape berries. Molecules 2010, 15, 1141–1153. [Google Scholar] [CrossRef]
- Zhao, Q.; Duan, C.Q.; Wang, J. Anthocyanins profile of grape berries of Vitis amurensis, its hybrids and their wines. Int J. Mol. Sci. 2010, 11, 2212–2228. [Google Scholar] [CrossRef]
- Punyasiri, P.A.N.; Abeysinghe, I.S.B.; Kumar, V.; Treutter, D.; Duy, D.; Gosch, C.; Martens, S.; Forkmann, G.; Fischer, T.C. Flavonoid biosynthesis in the tea plant Camellia sinensis: Properties of enzymes of the prominent epicatechin and catechin pathways. Arch. Biochem. Biophys. 2004, 431, 22–30. [Google Scholar] [CrossRef]
- Shen, G.A.; Pang, Y.Z.; Wu, W.S.; Liu, X.F.; Zhao, L.X.; Sun, X.F.; Tang, K.X. Isolation and characterization of a putative anthocyanidin reductase gene from Ginkgo biloba. J. Plant Physiol. 2006, 163, 224–227. [Google Scholar] [CrossRef]
- Terrier, N.; Torregrosa, L.; Ageorges, A.; Vialet, S.; Verries, C.; Cheynier, V.; Romieu, C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009, 149, 1028–1041. [Google Scholar]
- Deluc, L.; Bogs, J.; Walker, A.R.; Ferrier, T.; Decendit, A.; Merillon, J.-M.; Robinson, S.P.; Barrieu, F. The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 2008, 147, 2041–2053. [Google Scholar] [CrossRef]
- Walker, A.R.; Lee, E.; Bogs, J.; McDavid, D.A.J.; Thomas, M.R.; Robinson, S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007, 49, 772–785. [Google Scholar] [CrossRef]
- Azuma, A.; Kobayashi, S.; Goto-Yamamoto, N.; Shiraishi, M.; Mitani, N.; Yakushiji, H.; Koshita, Y. Color recovery in berries of grape (Vitis vinifera L.) 'Benitaka', a bud sport of 'Italia', is caused by a novel allele at the VvmybA1 locus. Plant Sci. 2009, 176, 470–478. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36 (Suppl 2), W465–W469. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Zhu, Y.; Peng, Q.-Z.; Du, C.; Li, K.-G.; Xie, D.-Y. Characterization of Flavan-3-ols and Expression of MYB and Late Pathway Genes Involved in Proanthocyanidin Biosynthesis in Foliage of Vitis bellula. Metabolites 2013, 3, 185-203. https://doi.org/10.3390/metabo3010185
Zhu Y, Peng Q-Z, Du C, Li K-G, Xie D-Y. Characterization of Flavan-3-ols and Expression of MYB and Late Pathway Genes Involved in Proanthocyanidin Biosynthesis in Foliage of Vitis bellula. Metabolites. 2013; 3(1):185-203. https://doi.org/10.3390/metabo3010185
Chicago/Turabian StyleZhu, Yue, Qing-Zhong Peng, Ci Du, Ke-Gang Li, and De-Yu Xie. 2013. "Characterization of Flavan-3-ols and Expression of MYB and Late Pathway Genes Involved in Proanthocyanidin Biosynthesis in Foliage of Vitis bellula" Metabolites 3, no. 1: 185-203. https://doi.org/10.3390/metabo3010185
APA StyleZhu, Y., Peng, Q.-Z., Du, C., Li, K.-G., & Xie, D.-Y. (2013). Characterization of Flavan-3-ols and Expression of MYB and Late Pathway Genes Involved in Proanthocyanidin Biosynthesis in Foliage of Vitis bellula. Metabolites, 3(1), 185-203. https://doi.org/10.3390/metabo3010185





