Separation Technique for the Determination of Highly Polar Metabolites in Biological Samples
Abstract
:1. Introduction
2. Non-Target and Target Metabolomics
3. Separation Technique of Highly Polar Metabolites
3.1. Capillary Electrophoresis
| Sample | Analyte | Separation method | Sample preparation | Separation condition | Reference |
|---|---|---|---|---|---|
| Rat urine | Non-targeted | CD-MERK | Filtered urine | 25 mM sodium tetraborate decahydrate, 75 mM SDS and 6.25 mM sulfated β-cyclodextrin. The pH was adjusted to pH 9.50 (apparent) with 2 M NaOH (after addition of SDS and cyclodextrin) with 2.25% v/v hexafluoroisopropanol. | [50] |
| CZE | |||||
| Red blood cell | Amino acid | CE-ESI-MS | Prepared in 200 mM ammonium acetate | 1.4 M formic acid (pH 1.8) | [51] |
| Tumor tissues (colon and stomach) | Anionic, cationic metabolites and nucleotide | CE-ESI-time-of-flight mass spectrometry (TOFMS) | Homogenized in water. Liquid-liquid extraction with water and chloroform. Filtered through a 5-kDa cutoff filter. | Anionic metabolites: 50 mM ammonium acetate | [52] |
| Cationic metabolites: 1 M formic acid | |||||
| Neurons ( Aplysia californica) | Neurotransmitters | CE-ESI-TOFMS | After the protease treatment, ganglia were washed in artificial seawater. | 1% formic acid | [53] |
| Plant extract | Anionic metabolite | CE-ESI-TOFMS | The frozen leaf disc was homogenized in methanol. Liquid-liquid extraction with water and chloroform. | 50 mM ammonium acetate adjusted to pH 9.0 by the addition of ammonium hydroxide. | [54] |
| Mouse blood and tissue | Anionic and cationic metabolites | CE-ESI-TOFMS | The frozen tissue was homogenized in methanol. Liquid-liquid extraction with water and chloroform. Filtered through a 5-kDa cutoff filter. The filtrate was lyophilized and dissolved in water. | Anionic metabolites: 50 mM ammonium acetate solution (pH 8.5) | [55] |
| Cationic metabolites: 1 M formic acid | |||||
| Rat plasma | Anionic and cationic metabolites | CE-ESI-TOFMS | The plasma was plunged into methanol. Liquid-liquid extraction with water and chloroform. Filtered through a 5-kDa cutoff filter. The filtrate was lyophilized and dissolved in water. | Anionic metabolites: 50 mM ammonium acetate solution (pH 8.5) | [56] |
| Cationic metabolites: 1 M formic acid | |||||
| Human HT29 colon cancer cell | Non-targeted | CE-ESI-TOFMS | SPE | 1 M formic acid | [57] |
| Protein precipitation | |||||
| Ultracentrifugation | |||||
| Human urine | Non-targeted | CE-ESI-MS | Urine sample was diluted and centrifuged. | 10% acetic acid (pH 2.2) | [47] |
3.2. Gas Chromatography
3.3. High Performance Liquid Chromatography
| Sample | Analyte | Separation method | Sample preparation | Separation condition | Reference |
|---|---|---|---|---|---|
| Human plasma | Non-targeted | LC-MS/MS | Human plasma was mixed with water, extraction buffer, and extraction solvent. The sample was shaken and centrifuged; then, supernatant was transferred onto a plate, dried down under nitrogen flow, and reconstituted in acetonitrile/water (20/80) with 0.1% formic acid. | 0.1% formic acid in water and 0.1% formic acid in acetonitrile | [[80] |
| Acquity UPLC BEH Shield RP18 column (1.7 μm, 50 mm × 2.1 mm) | |||||
| Human urine | Non-targeted | HILIC-MS | Urine was mixed with acetonitrile before being centrifuged. | 13 mM ammonium acetate buffer (pH 9.1) and acetonitrile | [81] |
| Aphera NH2 polymer (5 μm, 150 mm × 2 mm) | |||||
| Human serum | Non targeted | LC-TOFMS | Serum was mixed with acetonitrile and centrifuged. The supernatant was lyophilized and reconstituted. | 0.1% formic acid in water and acetonitrile (98:2), and acetonitrile. | [82] |
| BEH C18 column (1.7 μm, 50 mm × 2.1 mm) | |||||
| Trypanosoma brucei brucei | Non-targeted | HILIC-Orbitrap LC-MS | Metabolites were extracted from the cell pellet by addition of chloroform/methanol/ water (1:3:1). | 0.1% formic acid in water and 0.08% formic acid in acetonitrile | [83] |
| (strain 427) | ZIC-HILIC (5 μm, 150 mm × 4.6 mm) | ||||
| Human aortic endothelial cells | Amine | LC-MS/MS | Cell sample was lysed with a sonic dismembranator. | 10 mM formic acid in water and 10 mM formic acid in methanol | [84] |
| Capillary column with in-house made (3 μm, 200 mm × 0.075 mm) | |||||
| Human urine | Carnitine and acylcarnitine | LC-MS/MS | Urine sample was extracted using methanol. | Eluent A, acetonitrile/water (80/20); eluent B, acetonitrile/water (20/80); eluent C, acetonitrile/ water/acetic acid/triethylamine (20/80/0.5/0.5); and eluent D, acetonitrile/water/acetic acid/ triethylamine (90/10/0.5/0.5) | [85] |
| Hypersil MOS-1 C8 (3 μm, 100 mm × 4.6 mm) | |||||
| Extracellular metabolites from acute lymphoblastic leukemia cell | Purine and pyrimidine metabolite | HILIC-TOFMS | T Culture medium sample was processed by adding of acetonitrile, vortexing, and centrifuging. | 0.1% formic acid in water and 0.1% formic acid in acetonitrile | [86] |
| Acquity amide column (1.7 μm, 150 mm × 2.1 mm). | |||||
| Human serum | Non-targeted | LC-TOFMS | Deproteinization | 0.1% formic acid in water and 0.1% formic acid in acetonitrile | [87] |
| Liquid-liquid extraction | Mediterranea reversed-phase C18 analytical column (3 μm, 100 mm × 0.46 mm) | ||||
| Solid-phase extraction | |||||
| Human platelet | Non-targeted | HILIC-MS | Cell sample was extracted using several solvents. | 0.1% formic acid in water and 0.1% formic acid in acetonitrile | [88] |
| Acquity amide column (1.7 μm, 150 mm × 2.1 mm) | |||||
| Saccharomyces cerevisiae | Phospholipid | HILIC-MS/MS | Liquid-liquid extraction with methanol and chloroform. Aliquot of the upper phase was collected. | Acetonitrile/water, 70:30 (v/v) with 10 mM ammonium acetate adjusted to pH 4.5 | [89] |
| Xbridge HILIC (5 μm, 150 mm × 4.6 mm) |
4. Applications
4.1. Amino Acids
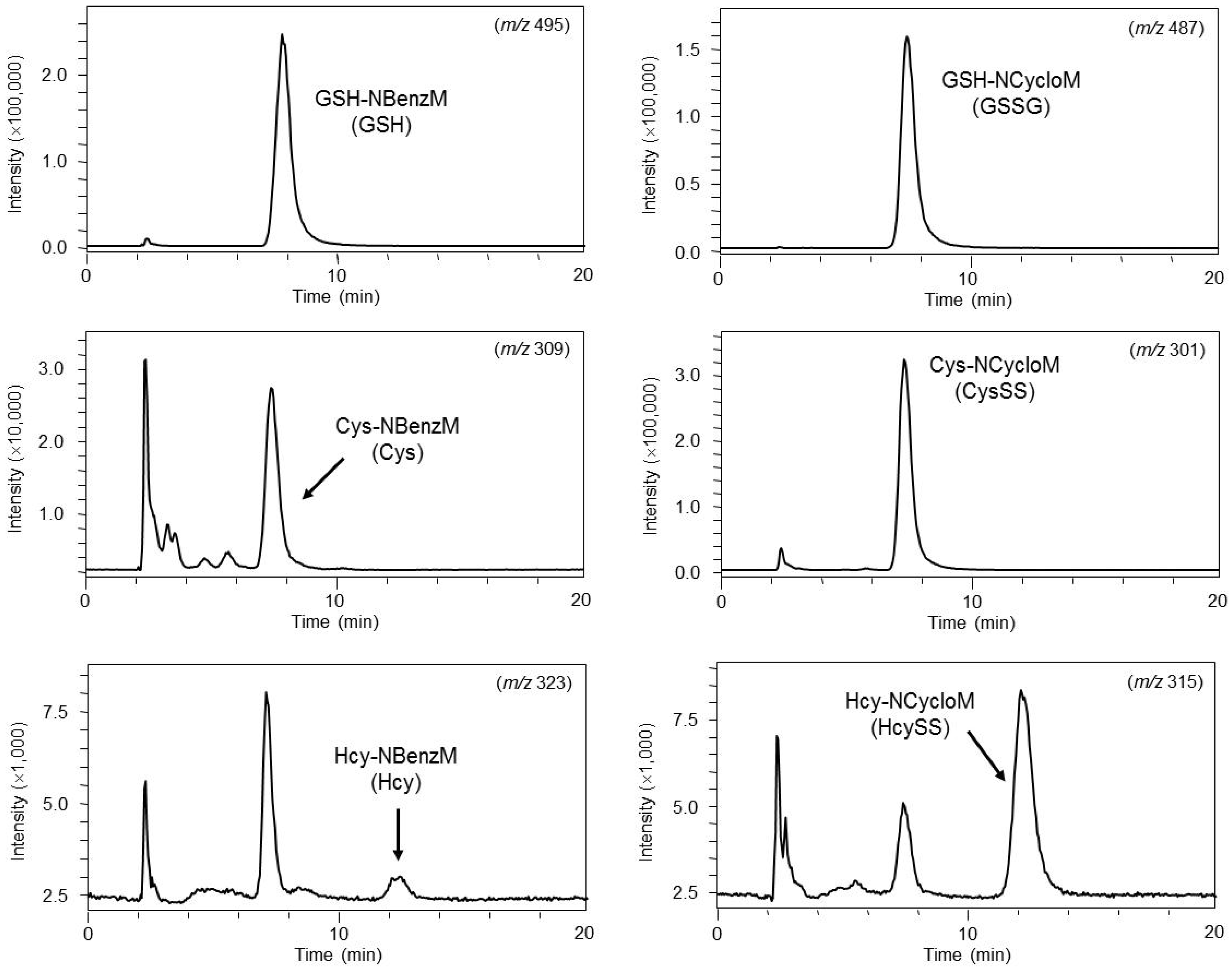
4.2. Glutathione and Related Compounds
4.3. Neurochemicals
4.4. Tricarboxylic Acid Cycle
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Hollywood, K.; Brison, D.R.; Goodacre, R. Metabolomics: current technologies and future trends. Proteomics 2006, 6, 4716–4723. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, M.C.; Wang, H.; Chan, C.Y.; Jiang, L.; Ngai, S.M.; Yu, J.; He, M.L.; Shaw, P.C.; Yew, D.T.; et al. Proteomic analysis of EZH2 downstream target proteins in hepatocellular carcinoma. Proteomics 2007, 7, 3097–3104. [Google Scholar] [CrossRef]
- Struck, W.; Waszczuk-Jankowska, M.; Kaliszan, R.; Markuszewski, M.J. The state-of-the-art determination of urinary nucleosides using chromatographic techniques "hyphenated" with advanced bioinformatic methods. Anal. Bioanal. Chem. 2011, 401, 2039–2050. [Google Scholar] [CrossRef]
- Ishiwata, J.; Kasamatsu, A.; Sakuma, K.; Iyoda, M.; Yamatoji, M.; Usukura, K.; Ishige, S.; Shimizu, T.; Yamano, Y.; Ogawara, K.; et al. State of heat shock factor 1 expression as a putative diagnostic marker for oral squamous cell carcinoma. Int. J. Oncol. 2012, 40, 47–52. [Google Scholar]
- Nordström, A.; O'Maille, G.; Qin, C.; Siuzdak, G. Nonlinear data alignment for UPLC-MS and HPLC-MS based metabolomics: quantitative analysis of endogenous and exogenous metabolites in human serum. Anal. Chem. 2006, 78, 3289–3295. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, Y.; Gao, J.; Chen, T.; Zhao, A.; Yan, Y.; Jia, W. Metabolite profiling of hemodialysate using gas chromatography time-of-flight mass spectrometry. J. Pharm. Biomed. Anal. 2011, 55, 1142–1147. [Google Scholar] [CrossRef]
- Li, S.; Liu, H.; Jin, Y.; Lin, S.; Cai, Z.; Jiang, Y. Metabolomics study of alcohol-induced liver injury and hepatocellular carcinoma xenografts in mice. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 2369–2375. [Google Scholar] [CrossRef]
- Rhee, E.P.; Gerszten, R.E. Metabolomics and cardiovascular biomarker discovery. Clin. Chem. 2012, 58, 139–147. [Google Scholar] [CrossRef]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom. Rev. 2011, 30, 884–906. [Google Scholar]
- Viant, M.R.; Rosenblum, E.S.; Tieerdema, R.S. NMR-based metabolomics: a powerful approach for characterizing the effects of environmental stressors on organism health. Environ. Sci. Technol. 2003, 37, 4982–4989. [Google Scholar] [CrossRef]
- Chylla, R.A.; Hu, K.; Ellinger, J.J.; Markley, J.L. Deconvolution of two-dimensional NMR spectra by fast maximum likelihood reconstruction: application to quantitative metabolomics. Anal. Chem. 2011, 83, 4871–4880. [Google Scholar]
- Zhang, A.; Sun, H.; Wang, P.; Han, Y.; Wang, X. Modern analytical techniques in metabolomics analysis. Analyst 2012, 137, 293–300. [Google Scholar] [CrossRef]
- Araníbar, N.; Ott, K.H.; Roongta, V.; Mueller, L. Metabolomic analysis using optimized NMR and statistical methods. Anal. Biochem. 2006, 355, 62–70. [Google Scholar]
- Dalsgaard, P.W.; Petersen, B.O.; Duus, J.Ø.; Zidorn, C.; Frisvad, J.C.; Christophersen, C.; Larsen, T.O. Atlantinone A, a meroterpenoid produced by Penicillium ribeum and several cheese associated Penicillium species. Metabolites 2012, 2, 214–220. [Google Scholar] [CrossRef]
- Theodoridis, G.A.; Gika, H.G.; Want, E.J.; Wilson, I.D. Liquid chromatography-mass spectrometry based global metabolite profiling: a review. Anal. Chim. Acta 2012, 711, 7–16. [Google Scholar] [CrossRef]
- Barbas, C.; Moraes, E.P.; Villaseñor, A. Capillary electrophoresis as a metabolomics tool for non-targeted fingerprinting of biological samples. J. Pharm. Biomed. Anal. 2011, 55, 823–831. [Google Scholar] [CrossRef]
- Jonsson, P.; Gullberg, J.; Nordström, A.; Kusano, M.; Kowalczyk, M.; Sjöström, M.; Moritz, T. A strategy for identifying differences in large series of metabolomic samples analyzed by GC/MS. Anal. Chem. 2004, 76, 1738–1745. [Google Scholar] [CrossRef]
- Guy, P.A.; Tavazzi, I.; Bruce, S.J.; Ramadan, Z.; Kochhar, S. Global metabolic profiling analysis on human urine by UPLC-TOFMS: issues and method validation in nutritional metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2008, 871, 253–260. [Google Scholar] [CrossRef]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef]
- Haag, M.; Schmidt, A.; Sachsenheimer, T.; Brügger, B. Quantification of signaling lipids by nano-electrospray ionization tandem mass spectrometry (Nano-ESI MS/MS). Metabolites 2012, 2, 57–76. [Google Scholar] [CrossRef]
- Sreekumar, A.; Poisson, L.M.; Rajendiran, T.M.; Khan, A.P.; Cao, Q.; Yu, J.; Laxman, B.; Mehra, R.; Lonigro, R.J.; Li, Y.; Nyati, M.K.; Ahsan, A.; Kalyana-Sundaram, S.; Han, B.; Cao, X.; Byun, J.; Omenn, G.S.; Ghosh, D.; Pennathur, S.; Alexander, D.C.; Berger, A.; Shuster, J.R.; Wei, J.T.; Varambally, S.; Beecher, C.; Chinnaiyan, A.M. Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 2009, 457, 910–914. [Google Scholar]
- Wu, H.; Xue, R.; Lu, C.; Deng, C.; Liu, T.; Zeng, H.; Wang, Q.; Shen, X. Metabolomic study for diagnostic model of oesophageal cancer using gas chromatography/mass spectrometry. J. Chromatogr. B. 2009, 877, 3111–3117. [Google Scholar] [CrossRef]
- Blekherman, G.; Laubenbacher, R.; Cortes, D.F.; Mendes, P.; Torti, F.M.; Akman, S.; Torti, S.V.; Shulaev, V. Bioinformatics tools for cancer metabolomics. Metabolomics 2011, 7, 329–343. [Google Scholar] [CrossRef]
- Huo, T.; Cai, S.; Lu, X.; Sha, Y.; Yu, M.; Li, F. Metabonomic study of biochemical changes in the serum of type 2 diabetes mellitus patients after the treatment of metformin hydrochloride. J. Pharm. Biomed. Anal. 2009, 49, 976–982. [Google Scholar] [CrossRef]
- Wang, T.J.; Larson, M.G.; Vasan, R.S.; Cheng, S.; Rhee, E.P.; McCabe, E.; Lewis, G.D.; Fox, C.S.; Jacques, P.F.; Fernandez, C.; et al. Metabolite profiles and the risk of developing diabetes. Nat. Med. 2011, 17, 448–453. [Google Scholar]
- Chu, F.L.; Pirastru, L.; Popovic, R.; Sleno, L. Carotenogenesis up-regulation in Scenedesmus sp. using a targeted metabolomics approach by liquid chromatography-high-resolution mass spectrometry. J. Agric. Food Chem. 2011, 59, 3004–3013. [Google Scholar] [CrossRef]
- Zhang, H.M.; Li, S.L.; Zhang, H.; Wang, Y.; Zhao, Z.L.; Chen, S.L.; Xu, H.X. Holistic quality evaluation of commercial white and red ginseng using a UPLC-QTOF-MS/MS-based metabolomics approach. J. Pharm. Biomed. Anal. 2012, 62, 258–273. [Google Scholar] [CrossRef]
- Watkins, S.M.; German, J.B. Metabolomics and biochemical profiling in drug discovery and development. Curr. Opin. Mol. Ther. 2002, 4, 224–228. [Google Scholar]
- Chen, C.; Gonzalez, F.J.; Idle, J.R. LC-MS-based metabolomics in drug metabolism. Drug Metab. Rev. 2007, 39, 581–597. [Google Scholar] [CrossRef]
- Chen, X.; Liu, L.; Palacios, G.; Gao, J.; Zhang, N.; Li, G.; Lu, J.; Song, T.; Zhang, Y.; Lv, H. Plasma metabolomics reveals biomarkers of the atherosclerosis. J. Sep. Sci. 2010, 33, 2776–2783. [Google Scholar] [CrossRef]
- Mamas, M.; Dunn, W.B.; Neyses, L.; Goodacre, R. The role of metabolites and metabolomics in clinically applicable biomarkers of disease. Arch. Toxicol. 2011, 85, 5–17. [Google Scholar] [CrossRef]
- Kotłowska, A.; Sworczak, K.; Stepnowski, P. Urine metabolomics analysis for adrenal incidentaloma activity detection and biomarker discovery. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 359–363. [Google Scholar] [CrossRef]
- Patti, G.J. Separation strategies for untargeted metabolomics. J. Sep. Sci. 2011, 34, 3460–3469. [Google Scholar] [CrossRef]
- Monton, M.R.; Soga, T. Metabolome analysis by capillary electrophoresis-mass spectrometry. J. Chromatogr.A. 1168, 237–246. [Google Scholar]
- Breadmore, M.C.; Quirino, J.P. 100,000-fold concentration of anions in capillary zone electrophoresis using electroosmotic flow controlled counterflow isotachophoretic stacking under field amplified conditions. Anal. Chem. 2008, 80, 6373–6381. [Google Scholar] [CrossRef]
- Wang, T.; Ma, J.; Wu, S.; Sun, L.; Yuan, H.; Zhang, L.; Liang, Z.; Zhang, Y. On-line combination of monolithic immobilized pH gradient-based capillary isoelectric focusing and capillary zone electrophoresis via a partially etched porous interface for protein analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 804–810. [Google Scholar] [CrossRef]
- Kitagawa, F.; Otsuka, K. Recent progress in capillary electrophoretic analysis of amino acid enantiomers. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 3078–3095. [Google Scholar] [CrossRef]
- Chiesl, T.N.; Chu, W.K.; Stockton, A.M.; Amashukeli, X.; Grunthaner, F.; Mathies, R.A. Enhanced amine and amino acid analysis using Pacific Blue and the Mars Organic Analyzer microchip capillary electrophoresis system. Anal. Chem. 2009, 81, 2537–2544. [Google Scholar]
- Riekkola, M.L. Recent advances in nonaqueous capillary electrophoresis. Electrophoresis 2002, 23, 3865–3883. [Google Scholar] [CrossRef]
- Olivares, J.A.; Nguyen, N.T.; Yonker, C.R.; Smith, R.D. On-line mass spectrometric detection for capillary zone electrophoresis. Anal. Chem. 1987, 59, 1230–1232. [Google Scholar] [CrossRef]
- Schmitt-Kopplin, P.; Frommberger, M. Capillary electrophoresis-mass spectrometry: 15 years of developments and applications. Electrophoresis 2003, 24, 3837–3867. [Google Scholar] [CrossRef]
- Sisu, E.; Flangea, C.; Serb, A.; Rizzi, A.; Zamfir, A.D. High-performance separation techniques hyphenated to mass spectrometry for ganglioside analysis. Electrophoresis 2011, 32, 1591–1609. [Google Scholar]
- Bjørnsdottir, I.; Tjørnelund, J.; Hansen, S.H. Nonaqueous capillary electrophoresis-its applicability in the analysis of food, pharmaceuticals and biological fluids. Electrophoresis 1998, 19, 2179–2186. [Google Scholar] [CrossRef]
- Bianco, G.; Schmitt-Kopplin, P.; De Benedetto, G.; Kettrup, A.; Cataldi, T.R. Determination of glycoalkaloids and relative aglycones by nonaqueous capillary electrophoresis coupled with electrospray ionization-ion trap mass spectrometry. Electrophoresis 2002, 23, 2904–2912. [Google Scholar] [CrossRef]
- Ramautar, R.; Mayboroda, O.A.; Somsen, G.W.; de Jong, G.J. CE-MS for metabolomics: Developments and applications in the period 2008-2010. Electrophoresis 2011, 32, 52–65. [Google Scholar] [CrossRef]
- Ramautar, R.; Toraño, J.S.; Somsen, G.W.; de Jong, G.J. Evaluation of CE methods for global metabolic profiling of urine. Electrophoresis 2010, 31, 2319–2327. [Google Scholar] [CrossRef]
- Ramautar, R.; Busnel, J.M.; Deelder, A.M.; Mayboroda, O.A. Enhancing the coverage of the urinary metabolome by sheathless capillary electrophoresis-mass spectrometry. Anal. Chem. 2012, 84, 885–892. [Google Scholar] [CrossRef]
- Peng, J.; Gygi, S.P. Proteomics: the move to mixtures. J. Mass Spectrom. 2001, 36, 1083–1091. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 1999, 29, 1181–1189. [Google Scholar] [CrossRef]
- Ruperez, F.J.; Garcia-Martinez, D.; Baena, B.; Maeso, N.; Vallejo, M.; Angulo, S.; Garcia, A.; Ibañez, E.; Señorans, F.J.; Cifuentes, A.; et al. Dunaliella salina extract effect on diabetic rats: metabolic fingerprinting and target metabolite analysis. J. Pharm. Biomed. Anal. 2009, 49, 786–792. [Google Scholar] [CrossRef]
- Chalcraft, K.R.; Lee, R.; Mills, C.; Britz-McKibbin, P. Virtual quantification of metabolites by capillary electrophoresis-electrospray ionization-mass spectrometry: predicting ionization efficiency without chemical standards. Anal. Chem. 2009, 81, 2506–2515. [Google Scholar] [CrossRef]
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M.; et al. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925. [Google Scholar]
- Lapainis, T.; Rubakhin, S.S.; Sweedler, J.V. Capillary electrophoresis with electrospray ionization mass spectrometric detection for single-cell metabolomics. Anal. Chem. 2009, 81, 5858–5864. [Google Scholar] [CrossRef]
- Hasunuma, T.; Harada, K.; Miyazawa, S.; Kondo, A.; Fukusaki, E.; Miyake, C. Metabolic turnover analysis by a combination of in vivo 13C-labelling from 13CO2 and metabolic profiling with CE-MS/MS reveals rate-limiting steps of the C3 photosynthetic pathway in Nicotiana tabacum leaves. J. Exp. Bot. 2010, 61, 1041–1051. [Google Scholar] [CrossRef]
- Kato, Y.; Kubo, Y.; Iwata, D.; Kato, S.; Sudo, T.; Sugiura, T.; Kagaya, T.; Wakayama, T.; Hirayama, A.; Sugimoto, M.; et al. Gene knockout and metabolome analysis of carnitine/organic cation transporter OCTN1. Pharm. Res. 2010, 27, 832–840. [Google Scholar] [CrossRef]
- Toyohara, T.; Suzuki, T.; Akiyama, Y.; Yoshihara, D.; Takeuchi, Y.; Mishima, E.; Kikuchi, K.; Suzuki, C.; Tanemoto, M.; Ito, S.; et al. Metabolomic profiling of the autosomal dominant polycystic kidney disease rat model. Clin. Exp. Nephrol. 2011, 15, 676–687. [Google Scholar] [CrossRef]
- Simó, C.; Ibáñez, C.; Gómez-Martínez, A.; Ferragut, J.A.; Cifuentes, A. Is metabolomics reachable? Different purification strategies of human colon cancer cells provide different CE-MS metabolite profiles. Electrophoresis 2011, 32, 1765–1777. [Google Scholar]
- Krone, N.; Hughes, B.A.; Lavery, G.G.; Stewart, P.M.; Arlt, W.; Shackleton, C.H. Gas chromatography/mass spectrometry (GC/MS) remains a pre-eminent discovery tool in clinical steroid investigations even in the era of fast liquid chromatography tandem mass spectrometry (LC/MS/MS). J. Steroid Biochem. Mol. Biol. 2010, 121, 496–504. [Google Scholar] [CrossRef]
- Halket, J.M.; Waterman, D.; Przyborowska, A.M.; Patel, R.K.; Fraser, P.D.; Bramley, P.M. Chemical derivatization and mass spectral libraries in metabolic profiling by GC/MS and LC/MS/MS. J. Exp. Bot. 2005, 56, 219–243. [Google Scholar]
- Farré, M.; Petrovic, M.; Barceló, D. Recently developed GC/MS and LC/MS methods for determining NSAIDs in water samples. Anal. Bioanal. Chem. 2007, 387, 1203–1214. [Google Scholar] [CrossRef]
- Buiarelli, F.; Giannetti, L.; Jasionowska, R.; Cruciani, C.; Neri, B. Determination of nandrolone metabolites in human urine: comparison between liquid chromatography/tandem mass spectrometry and gas chromatography/mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 1881–1894. [Google Scholar]
- Iwasaki, Y.; Nakano, Y.; Mochizuki, K.; Nomoto, M.; Takahashi, Y.; Ito, R.; Saito, K.; Nakazawa, H. A new strategy for ionization enhancement by derivatization for mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2011, 879, 1159–1165. [Google Scholar] [CrossRef]
- Gao, S.; Zhang, Z.P.; Karnes, H.T. Sensitivity enhancement in liquid chromatography/ atmospheric pressure ionization mass spectrometry using derivatization and mobile phase additives. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2005, 825, 98–110. [Google Scholar] [CrossRef]
- Gautam, U.G.; Gautam, M.P.; Sawada, T.; Takafuji, M.; Ihara, H. Thermodynamic investigations on shape selective separation behaviors of poly(4-vinylpyridine)-grafted silica for polycyclic aromatic hydrocarbons in both normal-phase and reversed-phase high-performance liquid chromatography. J. Chromatogr. A. 1216, 3571–3577. [Google Scholar]
- Canelas, A.B.; ten Pierick, A.; Ras, C.; Seifar, R.M.; van Dam, J.C.; van Gulik, W.M.; Heijnen, J.J. Quantitative evaluation of intracellular metabolite extraction techniques for yeast metabolomics. Anal. Chem. 2009, 81, 7379–7389. [Google Scholar]
- Coulier, L.; Bas, R.; Jespersen, S.; Verheij, E.; van der Werf, M.J.; Hankemeier, T. Simultaneous quantitative analysis of metabolites using ion-pair liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 2006, 78, 6573–6582. [Google Scholar]
- Kiefer, P.; Delmotte, N.; Vorholt, J.A. Nanoscale ion-pair reversed-phase HPLC-MS for sensitive metabolome analysis. Anal. Chem. 2011, 83, 850–855. [Google Scholar]
- Apfelthaler, E.; Bicker, W.; Lämmerhofer, M.; Sulyok, M.; Krska, R.; Lindner, W.; Schuhmacher, R. Retention pattern profiling of fungal metabolites on mixed-mode reversed-phase/weak anion exchange stationary phases in comparison to reversed-phase and weak anion exchange separation materials by liquid chromatography-electrospray ionisation-tandem mass spectrometry. J. Chromatogr. A. 2008, 1191, 171–181. [Google Scholar] [CrossRef]
- Alpert, A.J. Hydrophilic-interaction chromatography for the separation of peptides, nucleic acids and other polar compounds. J. Chromatogr. 1990, 499, 177–196. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Hoshi, M.; Ito, R.; Saito, K.; Nakazawa, H. Analysis of glutathione and glutathione disulfide in human saliva using hydrophilic interaction chromatography with mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2006, 839, 74–79. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Ishii, Y.; Ito, R.; Saito, K.; Nakazawa, H. New approaches for analysis of metabolism compounds in hydrophilic interaction chromatography. J. Liq. Chromatogr. Relat. Technol. 2007, 30, 2117–2126. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Goto, M.; Mochizuki, K.; Terayama, E.; Ito, R.; Saito, K.; Sugino, N.; Makino, T.; Nakazawa, H. Development and validation of a hydrophilic interaction chromatography-tandem mass spectrometry for quantification of nicotine and its metabolites in human maternal and cord sera. Biomed. Chromatogr. 2011, 25, 503–510. [Google Scholar] [CrossRef]
- Spagou, K.; Tsoukali, H.; Raikos, N.; Gika, H.; Wilson, I.D.; Theodoridis, G. Hydrophilic interaction chromatography coupled to MS for metabonomic/metabolomic studies. J. Sep. Sci. 2010, 33, 716–727. [Google Scholar] [CrossRef]
- Zhou, T.; Lucy, C.A. Hydrophilic interaction chromatography of nucleotides and their pathway intermediates on titania. J. Chromatogr. A. 2008, 1187, 87–93. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Nakano, Y.; Mochizuki, K.; Ogawa, T.; Oda, M.; Ito, R.; Saito, K.; Nakazawa, H. Quantification of reduced and oxidized thiols in mouse serum by column-switching hydrophilic interaction chromatography coupled with mass spectrometry. J. Pharm. Biomed. Anal. 2011, 56, 103–113. [Google Scholar] [CrossRef]
- Schiesel, S.; Lämmerhofer, M.; Lindner, W. Multitarget quantitative metabolic profiling of hydrophilic metabolites in fermentation broths of β-lactam antibiotics production by HILIC-ESI-MS/MS. Anal. Bioanal. Chem. 2010, 396, 1655–1679. [Google Scholar] [CrossRef]
- Preinerstorfer, B.; Schiesel, S.; Lämmerhofer, M.; Lindner, W. Metabolic profiling of intracellular metabolites in fermentation broths from beta-lactam antibiotics production by liquid chromatography-tandem mass spectrometry methods. J. Chromatogr. A. 2010, 1217, 312–328. [Google Scholar] [CrossRef]
- Fan, T.W.; Lorkiewicz, P.K.; Sellers, K.; Moseley, H.N.; Higashi, R.M.; Lane, A.N. Stable isotope-resolved metabolomics and applications for drug development. Pharmacol. Ther. 2012, 133, 366–391. [Google Scholar] [CrossRef]
- Cubbon, S.; Antonio, C.; Wilson, J.; Thomas-Oates, J. Metabolomic applications of HILIC-LC-MS. Mass Spectrom. Rev. 2010, 29, 671–684. [Google Scholar] [CrossRef]
- Liu, G.; Snapp, H.M.; Ji, Q.C.; Arnold, M.E. Strategy of accelerated method development for high-throughput bioanalytical assays using ultra high-performance liquid chromatography coupled with mass spectrometry. Anal. Chem. 2009, 81, 9225–9232. [Google Scholar]
- Kim, K.; Aronov, P.; Zakharkin, S.O.; Anderson, D.; Perroud, B.; Thompson, I.M.; Weiss, R.H. Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol. Cell Proteomics 2009, 8, 558–570. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, X.; Cao, R.; Lu, X.; Zhao, S.; Fekete, A.; Huang, Q.; Schmitt-Kopplin, P.; Wang, Y.; Xu, Z.; Wan, X.; Wu, X.; Zhao, N.; Xu, C.; Xu, G. Serum 27-nor-5β-cholestane-3,7,12,24,25 pentol glucuronide discovered by metabolomics as potential diagnostic biomarker for epithelium ovarian cancer. J. Proteome Res. 2011, 10, 2625–2632. [Google Scholar] [CrossRef]
- Creek, D.J.; Jankevics, A.; Breitling, R.; Watson, D.G.; Barrett, M.P.; Burgess, K.E. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: improved metabolite identification by retention time prediction. Anal. Chem. 2011, 83, 8703–8710. [Google Scholar]
- Yuan, W.; Zhang, J.; Li, S.; Edwards, J.L. Amine metabolomics of hyperglycemic endothelial cells using capillary LC-MS with isobaric tagging. J. Proteome Res. 2011, 10, 5242–5250. [Google Scholar] [CrossRef]
- Ganti, S.; Taylor, S.L.; Kim, K.; Hoppel, C.L.; Guo, L.; Yang, J.; Evans, C.; Weiss, R.H. Urinary acylcarnitines are altered in human kidney cancer. Int. J. Cancer 2012, 130, 2791–2800. [Google Scholar] [CrossRef]
- Paglia, G.; Hrafnsdóttir, S.; Magnúsdóttir, M.; Fleming, R.M.; Thorlacius, S.; Palsson, B.Ø.; Thiele, I. Monitoring metabolites consumption and secretion in cultured cells using ultra-performance liquid chromatography quadrupole-time of flight mass spectrometry (UPLC-Q-ToF-MS). Anal. Bioanal. Chem. 2012, 402, 1183–1198. [Google Scholar] [CrossRef]
- Ferreiro-Vera, C.; Priego-Capote, F.; Luque de Castro, M.D. Comparison of sample preparation approaches for phospholipids profiling in human serum by liquid chromatography-tandem mass spectrometry. J. Chromatogr. A. 2012, 1240, 21–28. [Google Scholar] [CrossRef]
- Paglia, G.; Magnúsdóttir, M.; Thorlacius, S.; Sigurjónsson, O.E.; Guðmundsson, S.; Palsson, B.Ø.; Thiele, I. Intracellular metabolite profiling of platelets: Evaluation of extraction processes and chromatographic strategies. J. Chromatogr. B, Analyt. Technol. Biomed. Life Sci. 2012, 898, 111–120. [Google Scholar] [CrossRef]
- Sun, T.; Wetzel, S.J.; Johnson, M.E.; Surlow, B.A.; Patton-Vogt, J. Development and validation of a hydrophilic interaction liquid chromatography-tandem mass spectrometry method for the quantification of lipid-related extracellular metabolites in Saccharomyces cerevisiae. J. Chromatogr. B 2012, 897, 1–9. [Google Scholar] [CrossRef]
- Li, P.; Yin, Y.L.; Li, D.; Kim, S.W.; Wu, G. Amino acids and immune function. Br. J. Nutr. 2007, 98, 237–252. [Google Scholar] [CrossRef]
- Galli, F. Amino acid and protein modification by oxygen and nitrogen species. Amino Acids 2012, 42, 1–4. [Google Scholar] [CrossRef]
- Janečková, H.; Hron, K.; Wojtowicz, P.; Hlídková, E.; Barešová, A.; Friedecký, D.; Zídková, L.; Hornik, P.; Behúlová, D.; Procházková, D.; Vinohradská, H.; Pešková, K.; Bruheim, P.; Smolka, V.; Sťastná, S.; Adam, T. Targeted metabolomic analysis of plasma samples for the diagnosis of inherited metabolic disorders. J. Chromatogr. A. 2012, 1226, 11–17. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Mochizuki, K.; Nakano, Y.; Maruya, N.; Goto, M.; Maruyama, Y.; Ito, R.; Saito, K.; Nakazawa, H. Comparison of fluorescence reagents for simultaneous determination of hydroxylated phenylalanine and nitrated tyrosine by high-performance liquid chromatography with fluorescence detection. Biomed. Chromatogr. 2012, 26, 41–50. [Google Scholar] [CrossRef]
- Bollard, M.E.; Stanley, E.G.; Lindon, J.C.; Nicholson, J.K.; Holmes, E. NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed. 2005, 18, 143–162. [Google Scholar] [CrossRef]
- Waldhier, M.C.; Almstetter, M.F.; Nürnberger, N.; Gruber, M.A.; Dettmer, K.; Oefner, P.J. Improved enantiomer resolution and quantification of free D-amino acids in serum and urine by comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J. Chromatogr. A 2011, 1218, 4537–4544. [Google Scholar] [CrossRef]
- Williams, B.J.; Cameron, C.J.; Workman, R.; Broeckling, C.D.; Sumner, L.W.; Smith, J.T. Amino acid profiling in plant cell cultures: an inter-laboratory comparison of CE-MS and GC-MS. Electrophoresis 2007, 28, 1371–1379. [Google Scholar] [CrossRef]
- Liu, J.L.; Wang, H.L.; Zhang, L.F.; Xu, Y.F.; Deng, W.; Zhu, H.; Qin, C. Metabonomics study of brain-specific human S100B transgenic mice by using high-performance liquid chromatography coupled with quadrupole time of flight mass spectrometry. Biol. Pharm. Bull. 2011, 34, 871–876. [Google Scholar] [CrossRef]
- Dernovics, M.; Far, J.; Lobinski, R. Identification of anionic selenium species in Se-rich yeast by electrospray QTOF MS/MS and hybrid linear ion trap/orbitrap MSn. Metallomics 2009, 1, 317–329. [Google Scholar] [CrossRef]
- D'Agostino, L.A.; Lam, K.P.; Lee, R.; Britz-McKibbin, P. Comprehensive plasma thiol redox status determination for metabolomics. J. Proteome Res. 2011, 10, 592–603. [Google Scholar] [CrossRef]
- Husain, A.; Sato, D.; Jeelani, G.; Mi-ichi, F.; Ali, V.; Suematsu, M.; Soga, T.; Nozaki, T. Metabolome analysis revealed increase in S-methylcysteine and phosphatidylisopropanolamine synthesis upon L-cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. J. Biol. Chem. 2010, 285, 39160–39170. [Google Scholar]
- Zhang, X.; Rauch, A.; Xiao, H.; Rainer, G.; Logothetis, N.K. Mass spectrometry-based neurochemical analysis: perspectives for primate research. Expert Rev. Proteomics 2008, 5, 641–652. [Google Scholar] [CrossRef]
- Nirogi, R.; Mudigonda, K.; Kandikere, V.; Ponnamaneni, R. Quantification of acetylcholine, an essential neurotransmitter, in brain microdialysis samples by liquid chromatography mass spectrometry. Biomed. Chromatogr. 2010, 24, 39–48. [Google Scholar] [CrossRef]
- Diao, P.; Yuan, H.; Huo, F.; Chen, L.; Xiao, D.; Paau, M.C.; Choi, M.M. A simple and sensitive CE method for the simultaneous determination of catecholamines in urine with in-column optical fiber light-emitting diode-induced fluorescence detection. Talanta 2011, 85, 1279–1284. [Google Scholar] [CrossRef]
- Zhang, Y.; Tingley, F.D., III; Tseng, E.; Tella, M.; Yang, X.; Groeber, E.; Liu, J.; Li, W.; Schmidt, C.J.; Steenwyk, R. Development and validation of a sample stabilization strategy and a UPLC-MS/MS method for the simultaneous quantitation of acetylcholine (ACh), histamine (HA), and its metabolites in rat cerebrospinal fluid (CSF). J. Chromatogr. B 2011, 879, 2023–2033. [Google Scholar] [CrossRef]
- Li, J.; von Pföstl, V.; Zaldivar, D.; Zhang, X.; Logothetis, N.; Rauch, A. Measuring multiple neurochemicals and related metabolites in blood and brain of the rhesus monkey by using dual microdialysis sampling and capillary hydrophilic interaction chromatography-mass spectrometry. Anal. Bioanal. Chem. 2012, 402, 2545–2554. [Google Scholar] [CrossRef]
- Gao, P.; Shi, C.; Tian, J.; Shi, X.; Yuan, K.; Lu, X.; Xu, G. Investigation on response of the metabolites in tricarboxylic acid cycle of Escherichia coli and Pseudomonas aeruginosa to antibiotic perturbation by capillary electrophoresis. J. Pharm. Biomed. Anal. 2007, 44, 180–187. [Google Scholar] [CrossRef]
- Bando, K.; Kunimatsu, T.; Sakai, J.; Kimura, J.; Funabashi, H.; Seki, T.; Bamba, T.; Fukusaki, E. GC-MS-based metabolomics reveals mechanism of action for hydrazine induced hepatotoxicity in rats. J. Appl. Toxicol. 2011, 31, 524–535. [Google Scholar] [CrossRef]
- Buescher, J.M.; Moco, S.; Sauer, U.; Zamboni, N. Ultrahigh performance liquid chromatography-tandem mass spectrometry method for fast and robust quantification of anionic and aromatic metabolites. Anal. Chem. 2010, 82, 4403–4412. [Google Scholar] [CrossRef]
- Lorenz, M.A.; Burant, C.F.; Kennedy, R.T. Reducing time and increasing sensitivity in sample preparation for adherent mammalian cell metabolomics. Anal. Chem. 2011, 83, 3406–3414. [Google Scholar]
- Büscher, J.M.; Czernik, D.; Ewald, J.C.; Sauer, U.; Zamboni, N. Cross-platform comparison of methods for quantitative metabolomics of primary metabolism. Anal. Chem. 2009, 81, 2135–2143. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Iwasaki, Y.; Sawada, T.; Hatayama, K.; Ohyagi, A.; Tsukuda, Y.; Namekawa, K.; Ito, R.; Saito, K.; Nakazawa, H. Separation Technique for the Determination of Highly Polar Metabolites in Biological Samples. Metabolites 2012, 2, 496-515. https://doi.org/10.3390/metabo2030496
Iwasaki Y, Sawada T, Hatayama K, Ohyagi A, Tsukuda Y, Namekawa K, Ito R, Saito K, Nakazawa H. Separation Technique for the Determination of Highly Polar Metabolites in Biological Samples. Metabolites. 2012; 2(3):496-515. https://doi.org/10.3390/metabo2030496
Chicago/Turabian StyleIwasaki, Yusuke, Takahiro Sawada, Kentaro Hatayama, Akihito Ohyagi, Yuri Tsukuda, Kyohei Namekawa, Rie Ito, Koichi Saito, and Hiroyuki Nakazawa. 2012. "Separation Technique for the Determination of Highly Polar Metabolites in Biological Samples" Metabolites 2, no. 3: 496-515. https://doi.org/10.3390/metabo2030496
APA StyleIwasaki, Y., Sawada, T., Hatayama, K., Ohyagi, A., Tsukuda, Y., Namekawa, K., Ito, R., Saito, K., & Nakazawa, H. (2012). Separation Technique for the Determination of Highly Polar Metabolites in Biological Samples. Metabolites, 2(3), 496-515. https://doi.org/10.3390/metabo2030496




