The Causal Role of the Gut Microbiota–Plasma Metabolome Axis in Myeloproliferative Neoplasm Pathogenesis: A Mendelian Randomization and Mediation Analysis
Abstract
1. Introduction
2. Materials and Methods
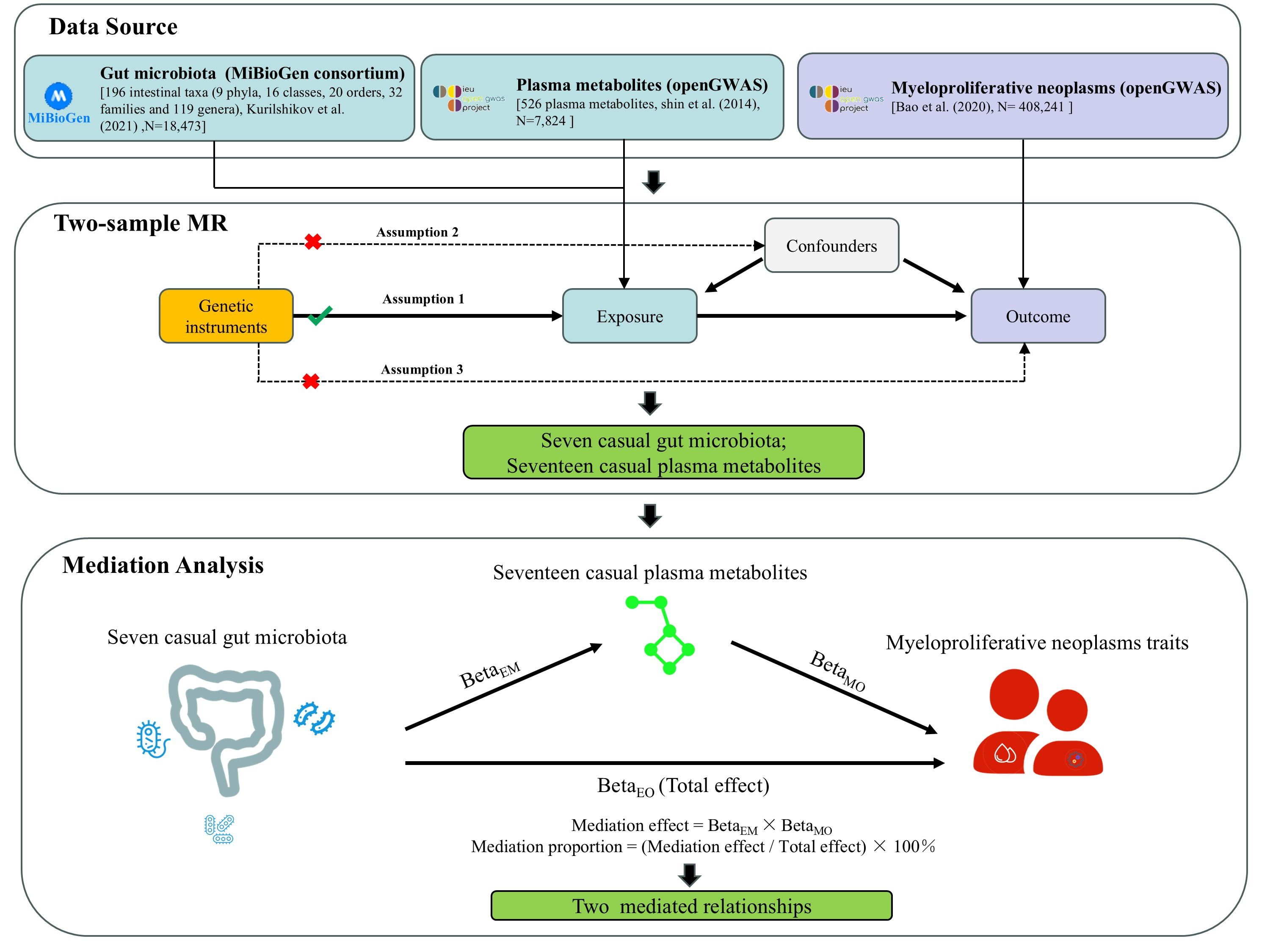
2.1. Study Design
2.2. Data Sources
2.3. SNP Selection
2.4. Statistical Analysis Strategy
2.5. Ethical Approval and Consent to Participate
3. Results
3.1. Genetic Instrumentation
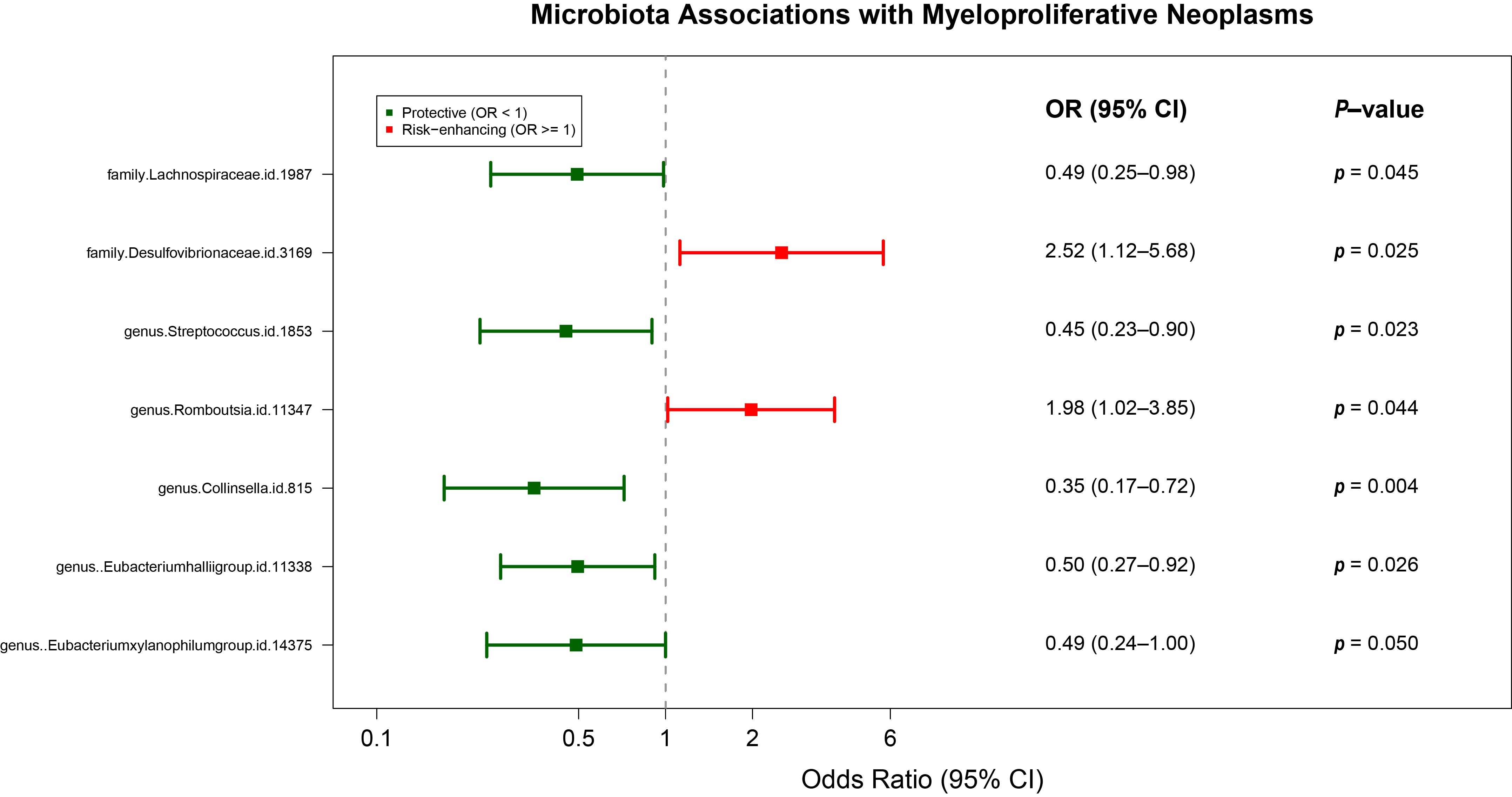
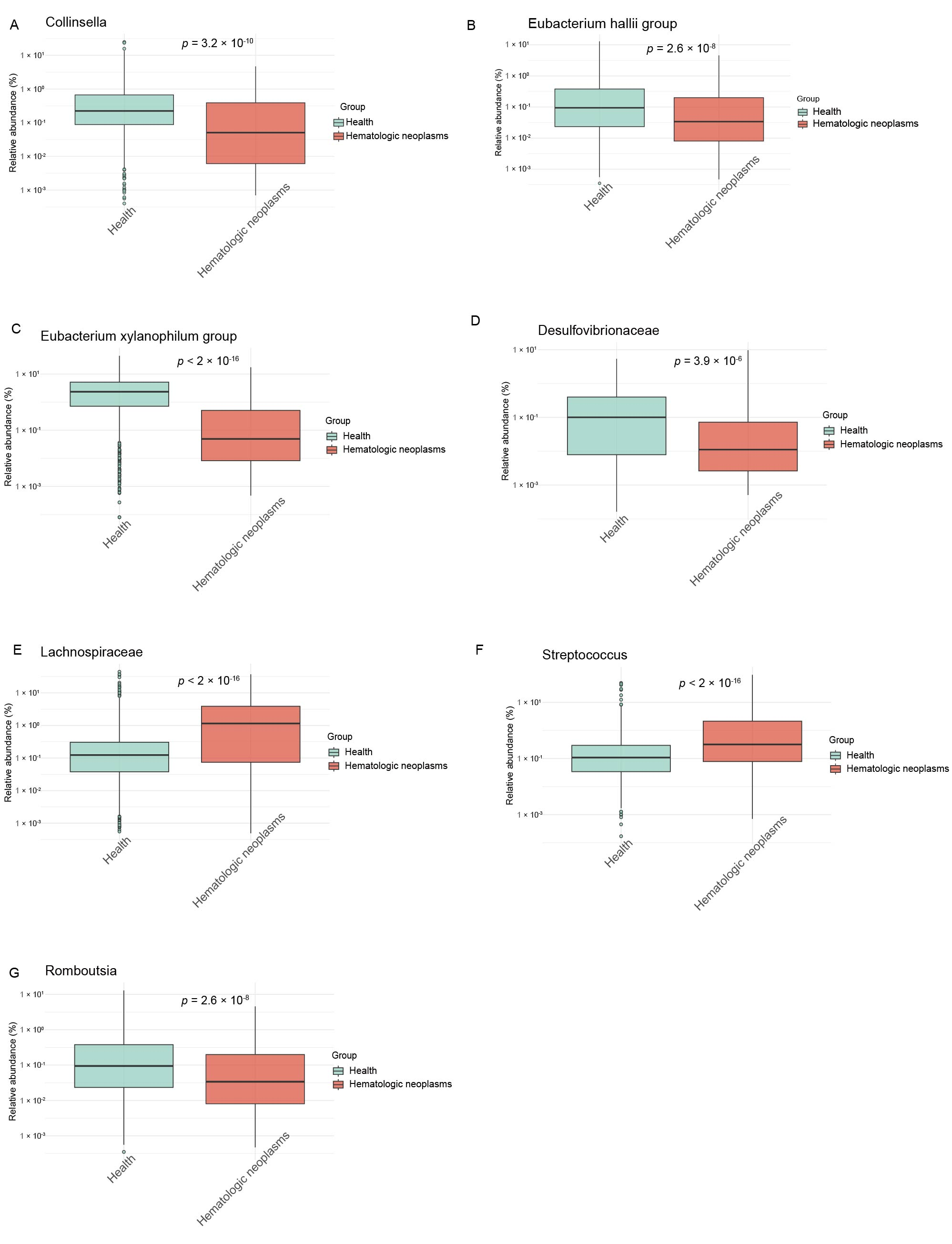
3.2. Causal GM Taxa Associated with MPN
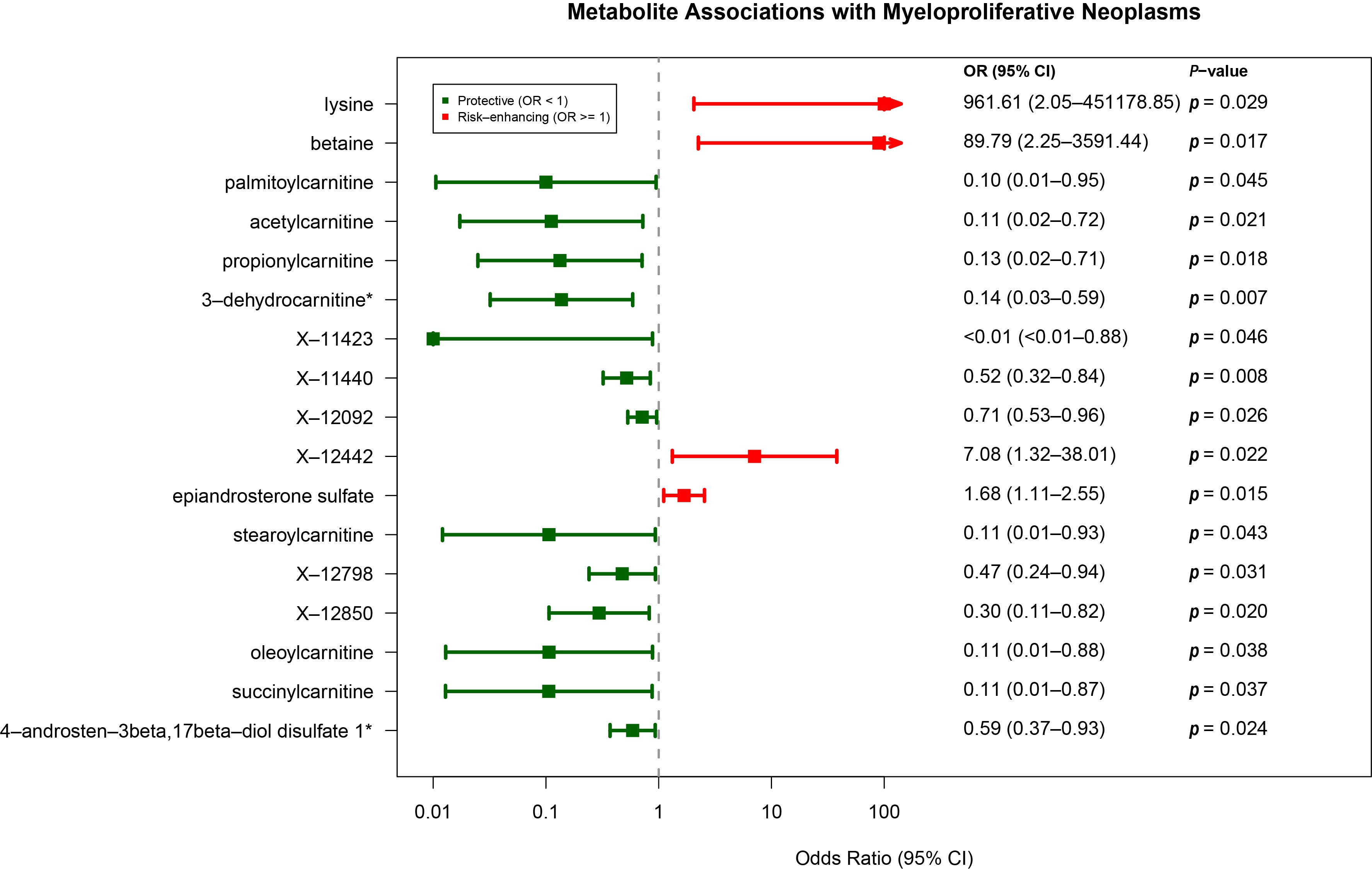
3.3. Causal Plasma Metabolites Associated with MPN
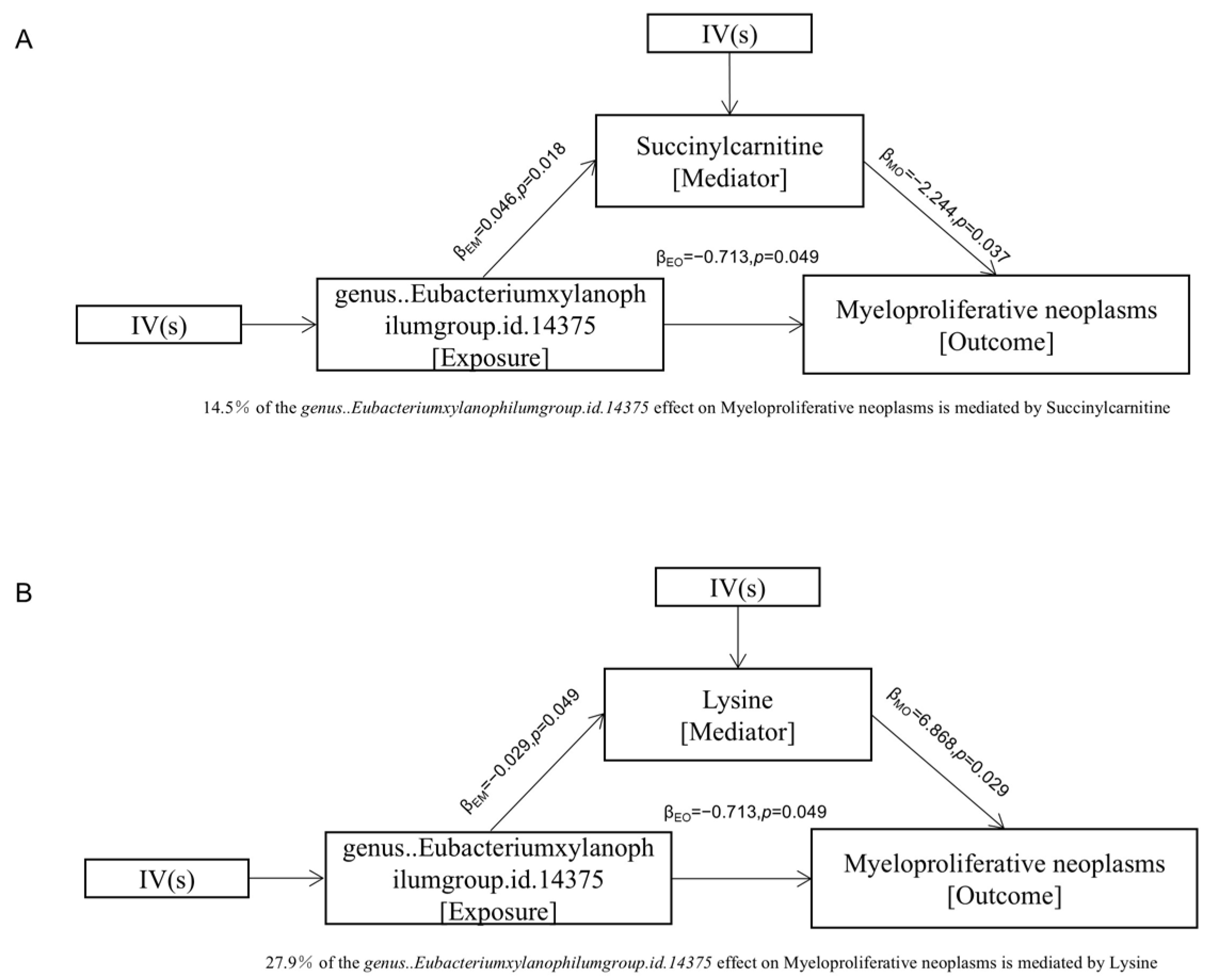
3.4. Mediation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sud, A.; Chattopadhyay, S.; Thomsen, H.; Sundquist, K.; Sundquist, J.; Houlston, R.S.; Hemminki, K. Familial risks of acute myeloid leukemia, myelodysplastic syndromes, and myeloproliferative neoplasms. Blood 2018, 132, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Bao, E.L.; Nandakumar, S.K.; Liao, X.; Bick, A.G.; Karjalainen, J.; Tabaka, M.; Gan OI; Havulinna, A.S.; Kiiskinen, T.T.J.; Lareau, C.A.; et al. Inherited myeloproliferative neoplasm risk affects haematopoietic stem cells. Nature 2020, 586, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Tapper, W.; Jones, A.V.; Kralovics, R.; Harutyunyan, A.S.; Zoi, K.; Leung, W.; Godfrey, A.L.; Guglielmelli, P.; Callaway, A.; Ward, D.; et al. Genetic variation at MECOM, TERT, JAK2 and HBS1L-MYB predisposes to myeloproliferative neoplasms. Nat. Commun. 2015, 6, 6691. [Google Scholar] [CrossRef] [PubMed]
- Hinds, D.A.; Barnholt, K.E.; Mesa, R.A.; Kiefer, A.K.; Do, C.B.; Eriksson, N.; Mountain, J.L.; Francke, U.; Tung, J.Y.; Nguyen, H.M.; et al. Germ line variants predispose to both JAK2 V617F clonal hematopoiesis and myeloproliferative neoplasms. Blood 2016, 128, 1121–1128. [Google Scholar] [CrossRef] [PubMed]
- Mason, K.L.; Huffnagle, G.B.; Noverr, M.C.; Kao, J.Y. Overview of gut immunology. Adv. Exp. Med. Biol. 2008, 635, 1–14. [Google Scholar] [PubMed]
- Margolis, K.G.; Gershon, M.D.; Bogunovic, M. Cellular Organization of Neuroimmune Interactions in the Gastrointestinal Tract. Trends Immunol. 2016, 37, 487–501. [Google Scholar] [CrossRef] [PubMed]
- McDermott, A.J.; Huffnagle, G.B. The microbiome and regulation of mucosal immunity. Immunology 2014, 142, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Vereecke, L.; Beyaert, R.; van Loo, G. Enterocyte death and intestinal barrier maintenance in homeostasis and disease. Trends Mol. Med. 2011, 17, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.; El Alaoui, K.; Haunschild, C.; Avelar-Barragan, J.; Mendez Luque, L.F.; Whiteson, K.; Fleischman, A.G. Fecal Microbial Community Composition in Myeloproliferative Neoplasm Patients Is Associated with an Inflammatory State. Microbiol. Spectr. 2022, 10, e0003222. [Google Scholar] [CrossRef] [PubMed]
- Eickhardt-Dalbøge, C.S.; Ingham, A.C.; Andersen, L.O.; Nielsen, H.V.; Fuursted, K.; Stensvold, C.R.; Larsen, M.K.; Kjær, L.; Christensen, S.F.; Knudsen, T.A.; et al. The gut microbiota in patients with polycythemia vera is distinct from that of healthy controls and varies by treatment. Blood Adv. 2023, 7, 3326–3337. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Fu, Y.; Sun, T.Y.; Jiang, Z.; Miao, Z.; Shuai, M.; Gou, W.; Ling, C.W.; Yang, J.; Wang, J.; et al. The interplay between host genetics and the gut microbiome reveals common and distinct microbiome features for complex human diseases. Microbiome 2020, 8, 145. [Google Scholar] [CrossRef] [PubMed]
- McKenney, A.S.; Lau, A.N.; Somasundara, A.V.H.; Spitzer, B.; Intlekofer, A.M.; Ahn, J.; Shank, K.; Rapaport, F.T.; Patel, M.A.; Papalexi, E.; et al. JAK2/IDH-mutant-driven myeloproliferative neoplasm is sensitive to combined targeted inhibition. J. Clin. Investig. 2018, 128, 789–804. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- de Almeida, F.C.; Berzoti-Coelho, M.G.; Toro, D.M.; Cacemiro, M.D.C.; Bassan, V.L.; Barretto, G.D.; Garibaldi, P.M.M.; Palma, L.C.; de Figueiredo-Pontes, L.L.; Sorgi, C.A.; et al. Bioactive Lipids as Chronic Myeloid Leukemia’s Potential Biomarkers for Disease Progression and Response to Tyrosine Kinase Inhibitors. Front. Immunol. 2022, 13, 840173. [Google Scholar] [CrossRef] [PubMed]
- Tefferi, A.; Pardanani, A. Myeloproliferative Neoplasms: A Contemporary Review. JAMA Oncol. 2015, 1, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Abdo, K.M.; Cunningham, M.L.; Snell, M.L.; Herbert, R.A.; Travlos, G.S.; Eldridge, S.R.; Bucher, J.R. 14-Week toxicity and cell proliferation of methyleugenol administered by gavage to F344 rats and B6C3F1 mice. Food. Chem. Toxicol. 2001, 39, 303–316. [Google Scholar] [CrossRef] [PubMed]
- Emdin, C.A.; Khera, A.V.; Kathiresan, S. Mendelian Randomization. JAMA 2017, 318, 1925–1926. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.A.; Harbord, R.M.; Sterne, J.A.; Timpson, N.; Davey Smith, G. Mendelian randomization: Using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008, 27, 1133–1163. [Google Scholar] [CrossRef] [PubMed]
- Sanna, S.; van Zuydam, N.R.; Mahajan, A.; Kurilshikov, A.; Vich Vila, A.; Võsa, U.; Mujagic, Z.; Masclee, A.A.M.; Jonkers, D.M.A.E.; Oosting, M.; et al. Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 2019, 51, 600–605. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021, 19, 55–71. [Google Scholar] [CrossRef] [PubMed]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le, C.I.; Raygoza, J.A.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.Y.; Fauman, E.B.; Petersen, A.K.; Krumsiek, J.; Santos, R.; Huang, J.; Arnold, M.; Erte I,, I.; Forgetta, V.; Yang, T.P.; et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014, 46, 543–550. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, E.; Glymour, M.M.; Holmes, M.V.; Kang, H.; Morrison, J.; Munafò, M.R.; Palmer, T.; Schooling, C.M.; Wallace, C.; Zhao, Q.; et al. Mendelian randomization. Nat. Rev. Methods Primers 2022, 2, 7. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Dai, H.; Hou, T.; Hou, Y.; Wang, T.; Lin, H.; Zhao, Z.; Li, M.; Zheng, R.; Wang, S.; et al. Dissecting Causal Relationships Between Gut Microbiota, Blood Metabolites, and Stroke: A Mendelian Randomization Study. J. Stroke 2023, 25, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Sudmant, P.H.; Rausch, T.; Gardner, E.J.; Handsaker, R.E.; Abyzov, A.; Huddleston, J.; Zhang, Y.; Ye, K.; Jun, G.; Fritz, M.H.; et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015, 526, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Thompson, S.G. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhao, S.; Huang, H.X.; Xie, P.; Cai, X.H.; Qu, Y.D.; Zhang, W.; Luo, J.Q.; Zhang, L.; Li, X. Systematic Mendelian randomization study of the effect of gut microbiome and plasma metabolome on severe COVID-19. Front. Immunol. 2023, 14, 1211612. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.R.; Sanderson, E.; Hammerton, G.; Richmond, R.C.; Davey, G.; Heron, J.; Taylor, A.E.; Davies, N.M.; Howe, L.D. Mendelian randomisation for mediation analysis: Current methods and challenges for implementation. Eur. J. Epidemiol. 2021, 36, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Peters, J.E.; Prins, B.; Persyn, E.; Traylor, M.; Surendran, P.; Karthikeyan, S.; Yonova-Doing, E.; Di Angelantonio, E.; Roberts, D.J.; et al. Systematic Mendelian randomization using the human plasma proteome to discover potential therapeutic targets for stroke. Nat. Commun. 2022, 13, 6143. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Pan, L.; Qu, S.; Qin, T.; Xiao, Z.; Xu, Z. Intra-abdominal Streptococcus agalactiae infection associated with myelofibrosis treated with ruxolitinib: A case report of an atypical clinical presentation. Curr. Med. Res. Opin. 2022, 38, 371–374. [Google Scholar] [CrossRef] [PubMed]
- de Vos, W.M.; Tilg, H.; Van Hul, M.; Cani, P.D. Gut microbiome and health: Mechanistic insights. Gut 2022, 71, 1020–1032. [Google Scholar] [CrossRef] [PubMed]
- McCulloch, J.A.; Davar, D.; Rodrigues, R.R.; Badger, J.H.; Fang, J.R.; Cole, A.M.; Balaji, A.K.; Vetizou, M.; Prescott, S.M.; Fernandes, M.R.; et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med. 2022, 28, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wu, Y.; Dai, P.; Wang, D.; Liu, L.; Chai, B. Gut microbial signatures of patients with primary hepatocellular carcinoma and their healthy first-degree relatives. J. Appl. Microbiol. 2023, 134, lxad221. [Google Scholar] [CrossRef] [PubMed]
- Barger, J.F.; Gallo, C.A.; Tandon, P.; Liu, H.; Sullivan, A.; Grimes, H.L.; Plas, D.R. S6K1 determines the metabolic requirements for BCR-ABL survival. Oncogene 2013, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Iriyama, N.; Miura, K.; Uchino, Y.; Takahashi, H.; Nakagawa, M.; Iizuka, K.; Hamada, T.; Koike, T.; Kurihara, K.; Nakayama, T.; et al. Relationship between Carnitine Deficiency and Tyrosine Kinase Inhibitor Use in Patients with Chronic Myeloid Leukemia. Chemotherapy 2022, 67, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Tronstad, K.J.; Bruserud, Ø.; Berge, K.; Berge, R.K. Antiproliferative effects of a non-beta-oxidizable fatty acid, tetradecylthioacetic acid, in native human acute myelogenous leukemia blast cultures. Leukemia 2002, 16, 2292–2301. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.L.; Simmons, P.A.; Vehige, J.; Willcox, M.D.; Garrett, Q. Role of carnitine in disease. Nutr. Metab. 2010, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu , X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Dugas, M.A.; Proulx, F.; de Jaeger, A.; Lacroix, J.; Lambert, M. Markers of tissue hypoperfusion in pediatric septic shock. Intensive Care Med. 2000, 26, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Zhang, I.W.; Curto, A.; López-Vicario, C.; Casulleras, M.; Duran-Güell, M.; Flores-Costa, R.; Colsch, B.; Aguilar, F.; Aransay, A.M.; Lozano, J.J.; et al. Mitochondrial dysfunction governs immunometabolism in leukocytes of patients with acute-on-chronic liver failure. J. Hepatol. 2022, 76, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Lyman, G.H.; Greenlee, H.; Bohlke, K.; Bao , T.; DeMichele, A.M.; Deng, G.E.; Fouladbakhsh, J.M.; Gil, B.; Hershman, D.L.; Mansfield, S.; et al. Integrative Therapies During and After Breast Cancer Treatment: ASCO Endorsement of the SIO Clinical Practice Guideline. J. Clin. Oncol. 2018, 36, 2647–2655. [Google Scholar] [CrossRef] [PubMed]
- Witkowski, M.; Weeks, T.L.; Hazen, S.L. Gut Microbiota and Cardiovascular Disease. Circ. Res. 2020, 127, 553–570. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Wu, X.; Wu, Q.; Chatoff, A.; Megill, E.; Gao, J.; Huang, T.; Duan, T.; Yang, K.; Jin, C.; et al. Lysine catabolism reprograms tumour immunity through histone crotonylation. Nature 2023, 617, 818–826. [Google Scholar] [CrossRef] [PubMed]
- Lieu, E.L.; Nguyen, T.; Rhyne, S.; Kim, J. Amino acids in cancer. Exp. Mol. Med. 2020, 52, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Cebrián, N.; Rojas-Benedicto, A.; Albors-Vaquer, A.; Bellosillo, B.; Besses, C.; Martínez-López, J.; Pineda-Lucena, A.; Puchades-Carrasco, L. Polycythemia Vera and Essential Thrombocythemia Patients Exhibit Unique Serum Metabolic Profiles Compared to Healthy Individuals and Secondary Thrombocytosis Patients. Cancers 2021, 13, 482. [Google Scholar] [CrossRef] [PubMed]
- Violante, S.; Ijlst, L.; Ruiter, J.; Koster, J.; van Lenthe, H.; Duran, M.; de Almeida, I.T.; Wanders, R.J.; Houten, S.M.; Ventura, F.V. Substrate specificity of human carnitine acetyltransferase: Implications for fatty acid and branched-chain amino acid metabolism. Biochim. Biophys. Acta 2013, 1832, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Makarova, E.; Makrecka-Kuka, M.; Vilks, K.; Volska, K.; Sevostjanovs, E.; Grinberga, S.; Zarkova-Malkova, O.; Dambrova, M.; Liepinsh, E. Decreases in Circulating Concentrations of Long-Chain Acylcarnitines and Free Fatty Acids During the Glucose Tolerance Test Represent Tissue-Specific Insulin Sensitivity. Front. Endocrinol. 2019, 10, 870. [Google Scholar] [CrossRef] [PubMed]
- Meslier, V.; Laiola, M.; Roager, H.M.; De Filippis, F.; Roume, H.; Quinquis, B.; Giacco, R.; Mennella, I.; Ferracane, R.; Pons, N.; et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut 2020, 69, 1258–1268. [Google Scholar] [CrossRef] [PubMed]
- Mossad, O.; Batut, B.; Yilmaz, B.; Dokalis, N.; Mezö, C.; Nent, E.; Nabavi, L.S.; Mayer, M.; Maron, F.J.M.; Buescher, J.M.; et al. Gut microbiota drives age-related oxidative stress and mitochondrial damage in microglia via the metabolite N(6)-carboxymethyllysine. Nat. Neurosci. 2022, 25, 295–305. [Google Scholar] [CrossRef] [PubMed]
| Mediated | Exposure | Method | Beta | SE | P | Q-Statistics | Ph | Egger Intercept | Ppleio |
|---|---|---|---|---|---|---|---|---|---|
| Succinyl carnitine | Eubacterium Xylanophilum group | Inverse variance weighted | 0.046 | 0.019 | 0.0184 | 2.845 | 0.416 | NA | NA |
| MR Egger | 0.126 | 0.059 | 0.169 | 0.832 | 0.659 | −0.006 | 0.291 | ||
| Lysine | Inverse variance weighted | −0.029 | 0.014 | 0.049 | 0.231 | 0.972 | NA | NA | |
| MR Egger | −0.031 | 0.042 | 0.543 | 0.227 | 0.892 | 0.001 | 0.957 |
| Exposure | Univariate | Multivariate | ||||||
|---|---|---|---|---|---|---|---|---|
| Beta | SE | P | OR (95% CI) | Beta | SE | P | OR (95% CI) | |
| Lysine | 6.868 | 3.138 | 0.028 | 961.61 (2.05 to 451,178.85) | 5.714 | 2.780 | 0.039 | 303.18 (1.30 to 70,536.17) |
| Succinylcarnitine | −2.244 | 1.076 | 0.037 | 0.11 (0.01 to 0.87) | −2.342 | 0.823 | 0.004 | 0.10 (0.02 to 0.48) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kan, H.; Zhang, K.; Mao, A.; Geng, L. The Causal Role of the Gut Microbiota–Plasma Metabolome Axis in Myeloproliferative Neoplasm Pathogenesis: A Mendelian Randomization and Mediation Analysis. Metabolites 2025, 15, 501. https://doi.org/10.3390/metabo15080501
Kan H, Zhang K, Mao A, Geng L. The Causal Role of the Gut Microbiota–Plasma Metabolome Axis in Myeloproliferative Neoplasm Pathogenesis: A Mendelian Randomization and Mediation Analysis. Metabolites. 2025; 15(8):501. https://doi.org/10.3390/metabo15080501
Chicago/Turabian StyleKan, Hao, Ka Zhang, Aiqin Mao, and Li Geng. 2025. "The Causal Role of the Gut Microbiota–Plasma Metabolome Axis in Myeloproliferative Neoplasm Pathogenesis: A Mendelian Randomization and Mediation Analysis" Metabolites 15, no. 8: 501. https://doi.org/10.3390/metabo15080501
APA StyleKan, H., Zhang, K., Mao, A., & Geng, L. (2025). The Causal Role of the Gut Microbiota–Plasma Metabolome Axis in Myeloproliferative Neoplasm Pathogenesis: A Mendelian Randomization and Mediation Analysis. Metabolites, 15(8), 501. https://doi.org/10.3390/metabo15080501





