Human Metabolism of Sirolimus Revisited
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Enzymes
2.2. Formation Kinetics of SRL Metabolites
2.3. Large-Scale Preparation of SRL Metabolites Using Pooled HLMs
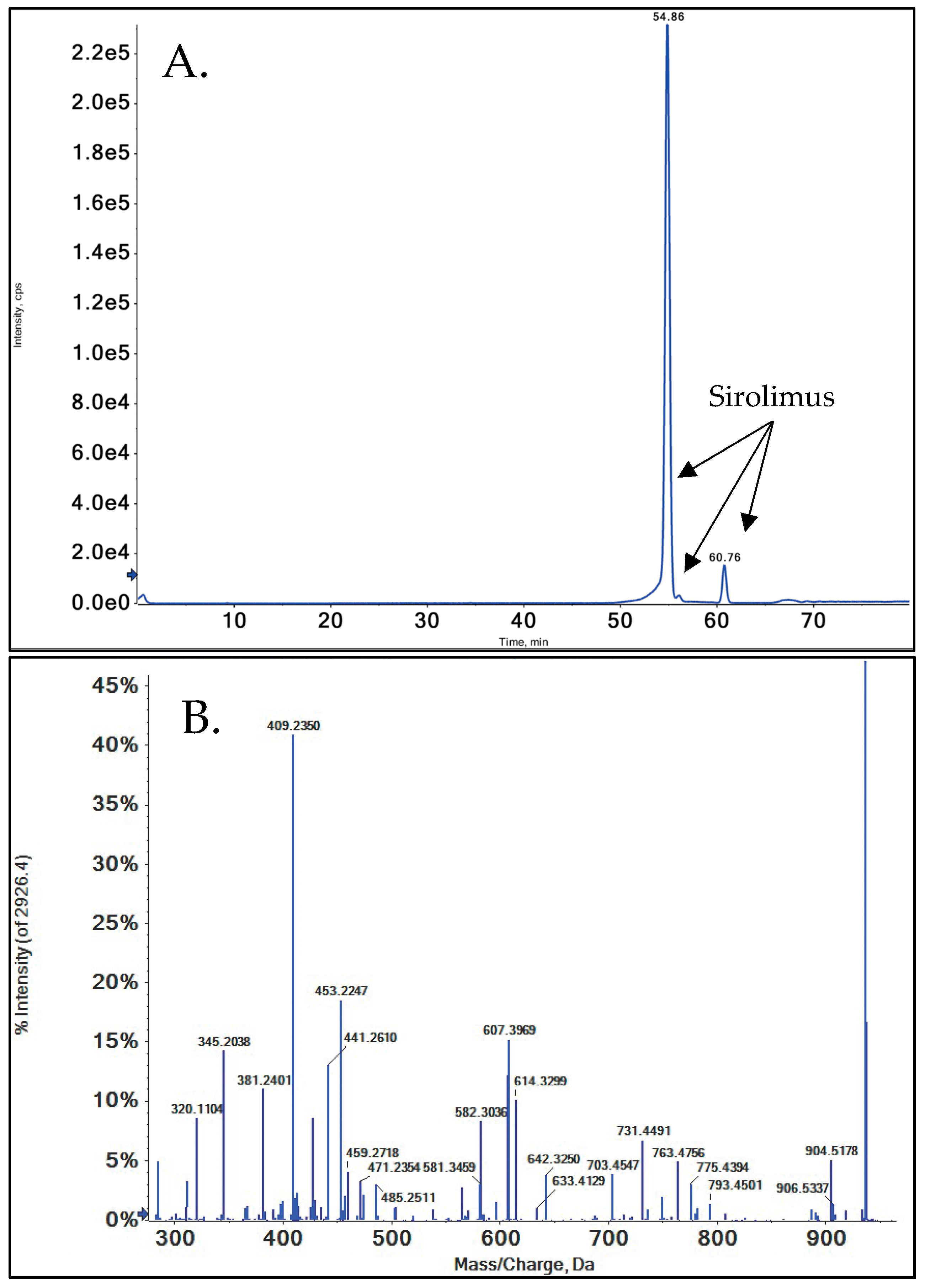
2.4. Separation, Isolation, and Mass Spectrometry Detection of SRL and Its Metabolites
2.5. Density Functional Theory Calculations
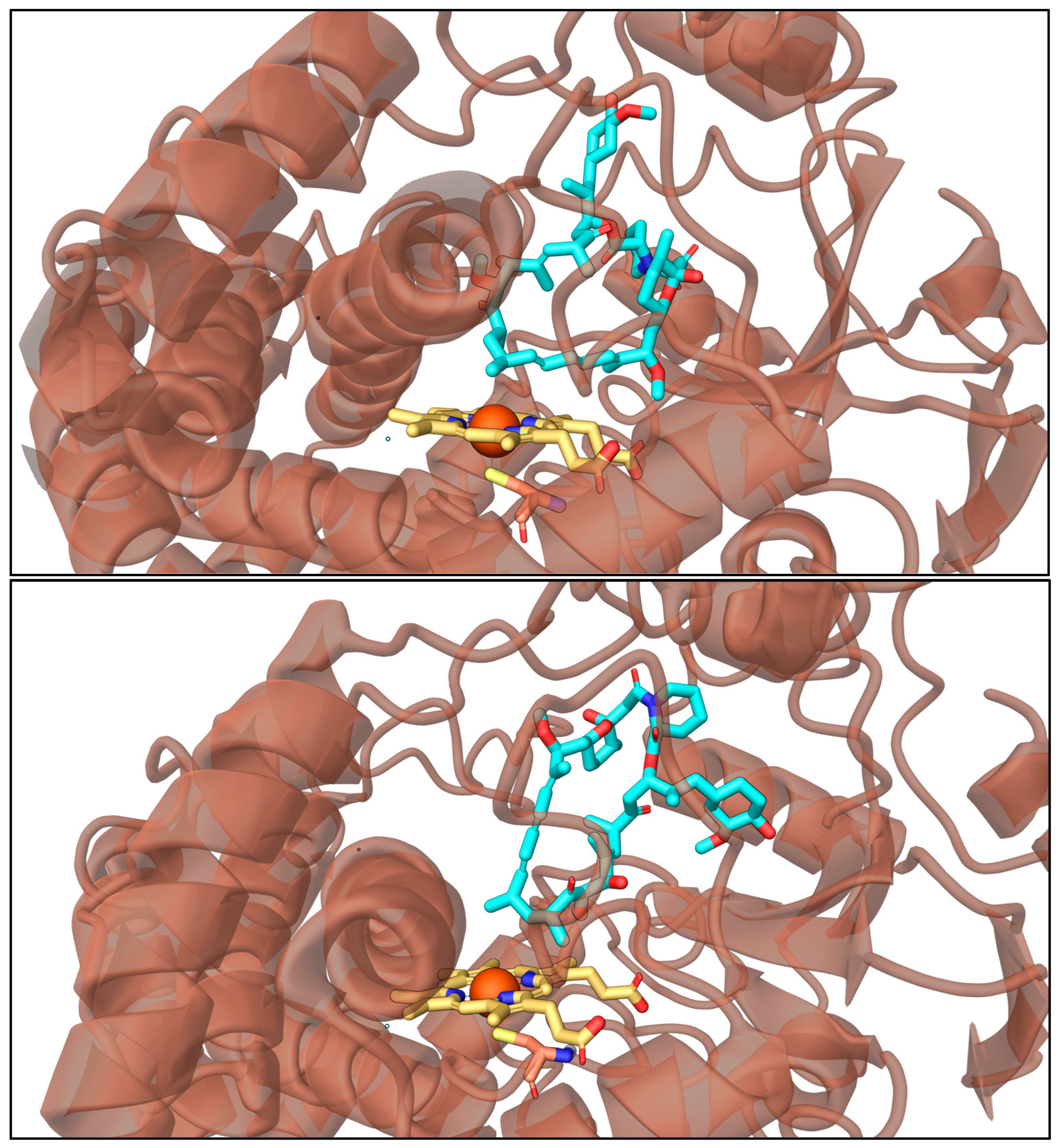
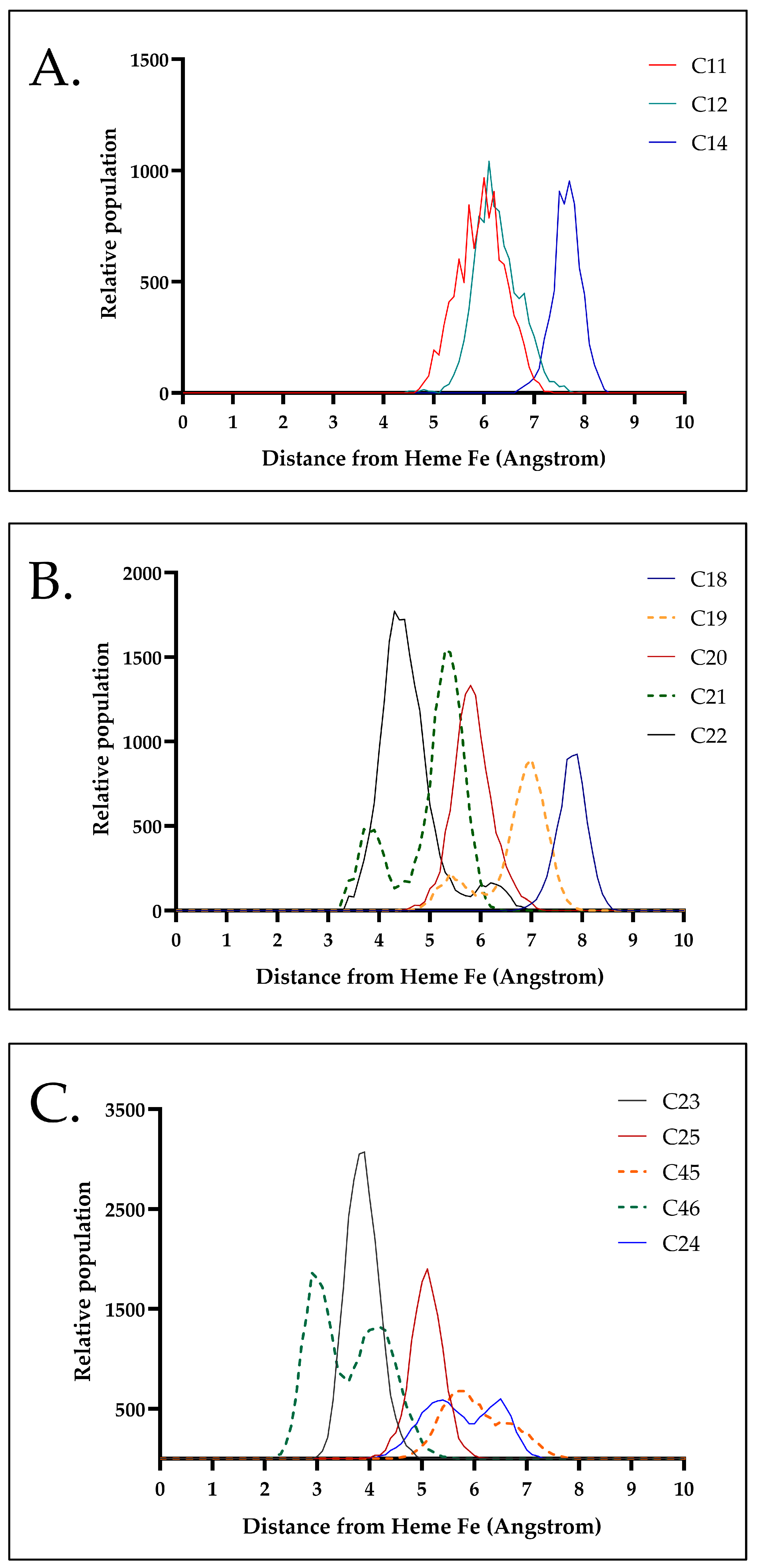
2.6. Molecular Dynamic Simulations
2.7. Statistics and Data Analysis
3. Results
3.1. HPLC-MS of SRL
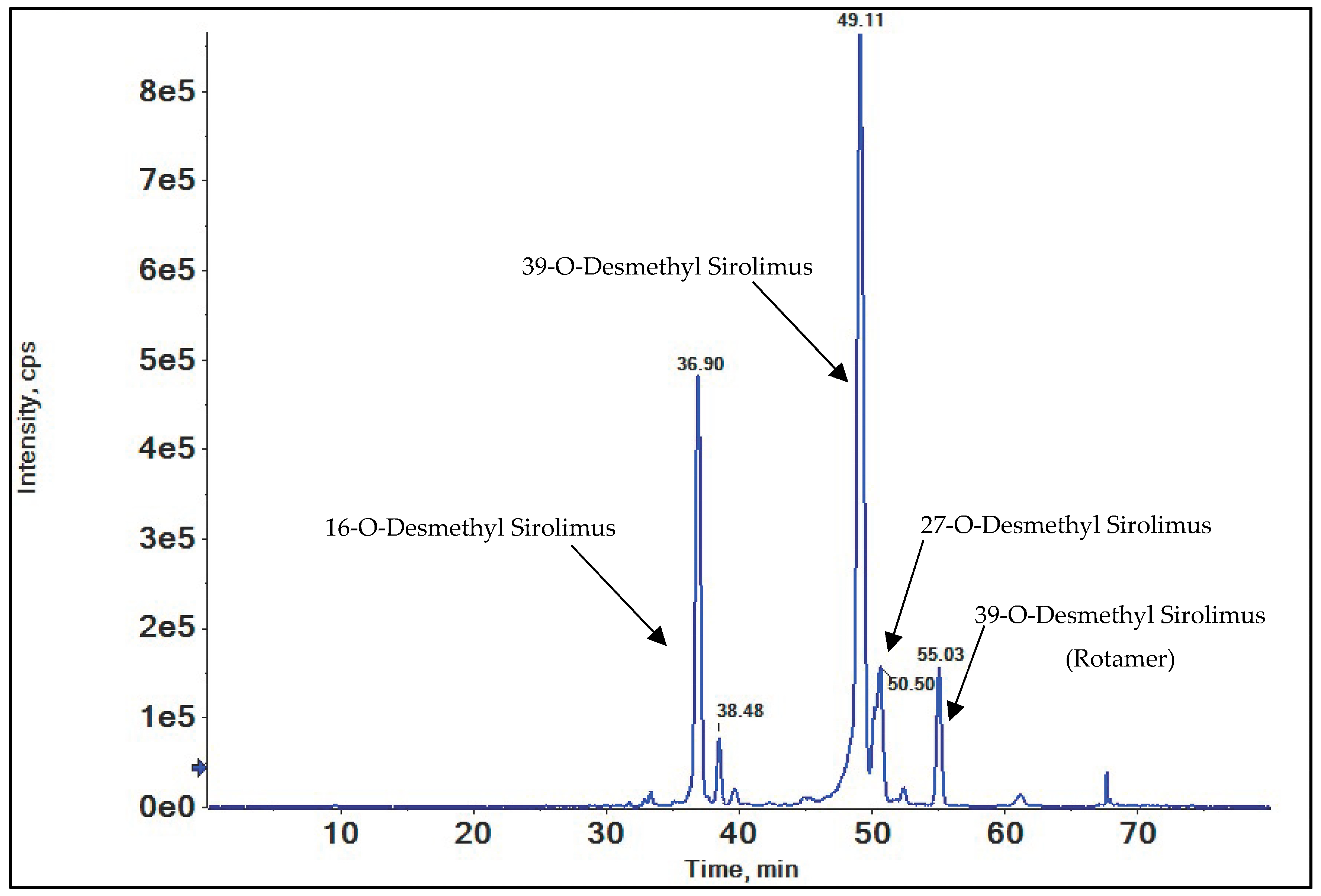
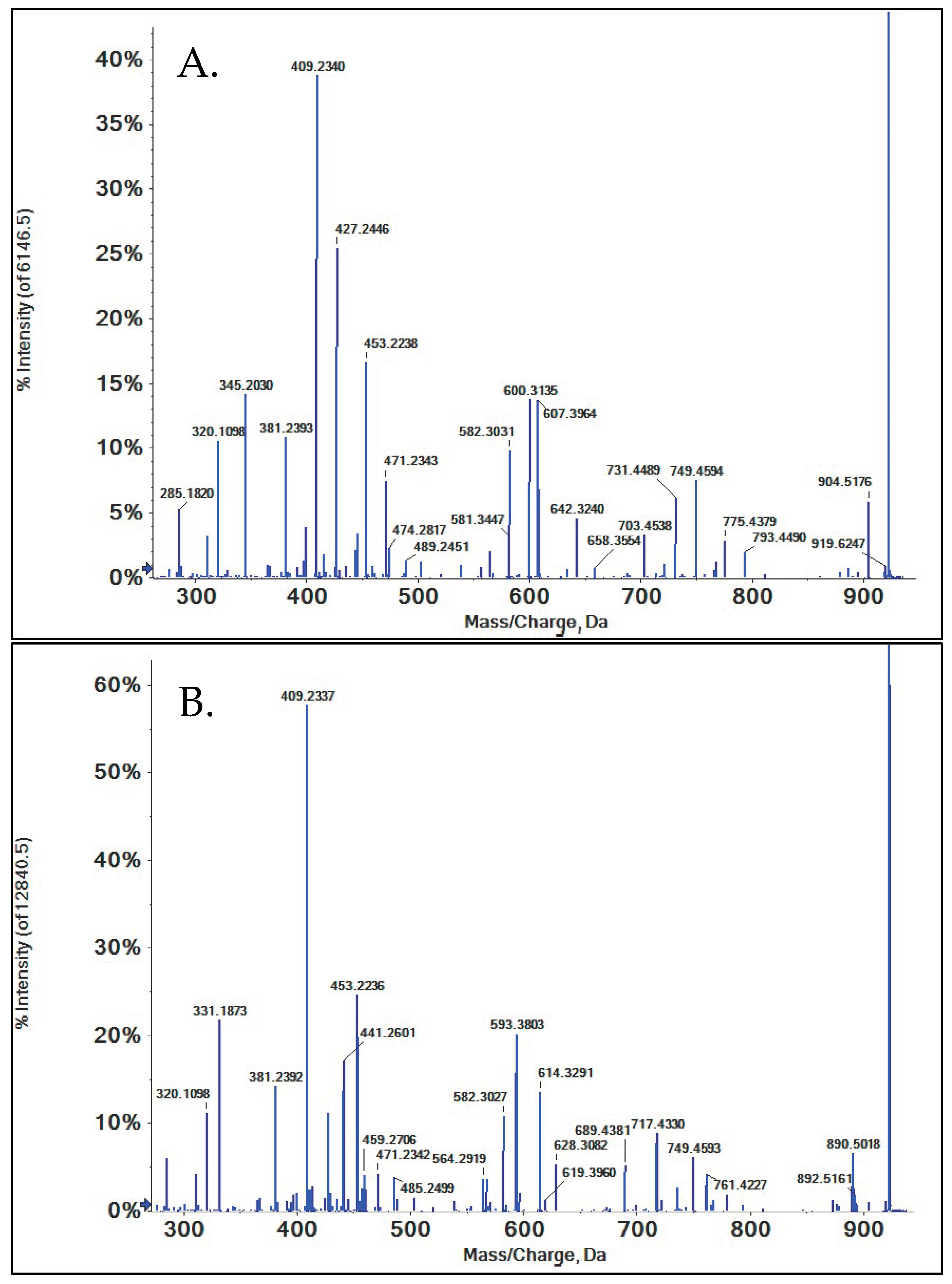
3.2. O-Demethylation
3.2.1. 16-O-Desmethyl SRL
3.2.2. 39-O-Desmethyl SRL
3.2.3. 27-O-Desmethyl SRL
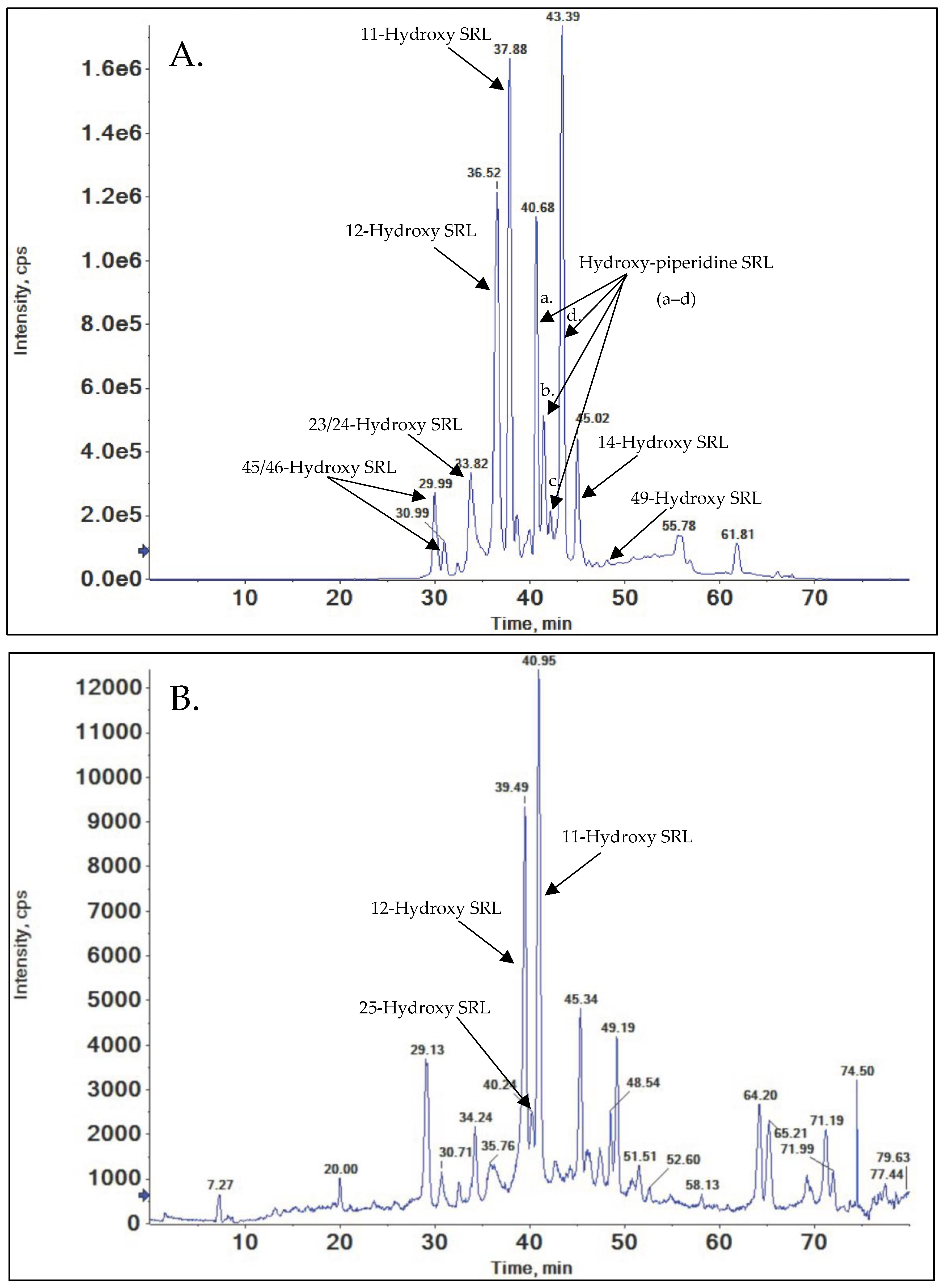
3.3. Hydroxylation
3.3.1. 45/46-Hydroxy SRL
3.3.2. 23/24-Hydroxy SRL
3.3.3. 12-Hydroxy-SRL
3.3.4. 25-Hydroxy-SRL
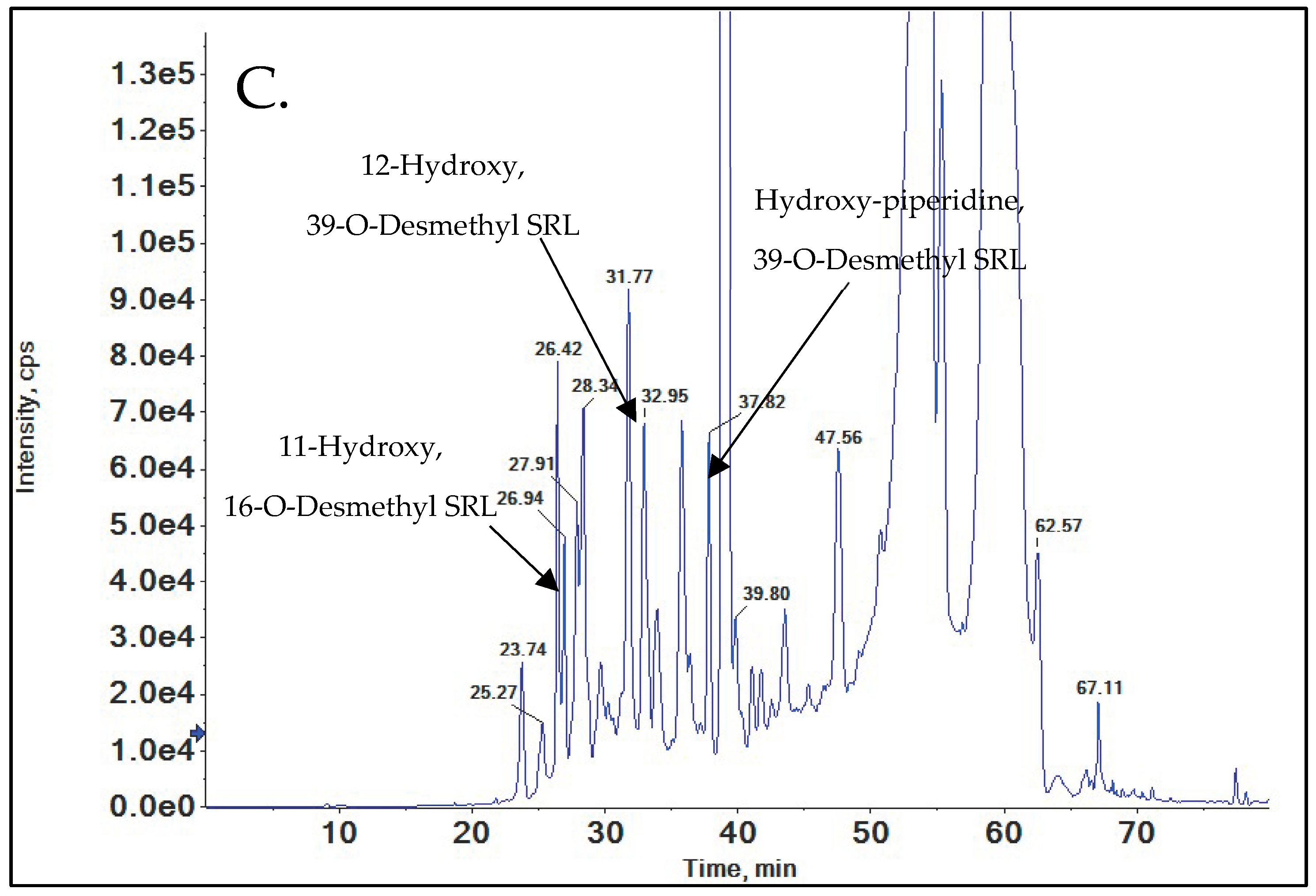
3.3.5. 11-Hydroxy-SRL
3.3.6. Hydroxy-Piperidine SRL
3.3.7. 14-Hydroxy SRL
3.3.8. 49-Hydroxy SRL
3.4. Second-Generation Metabolites
3.5. Di-Desmethylated Metabolites of SRL
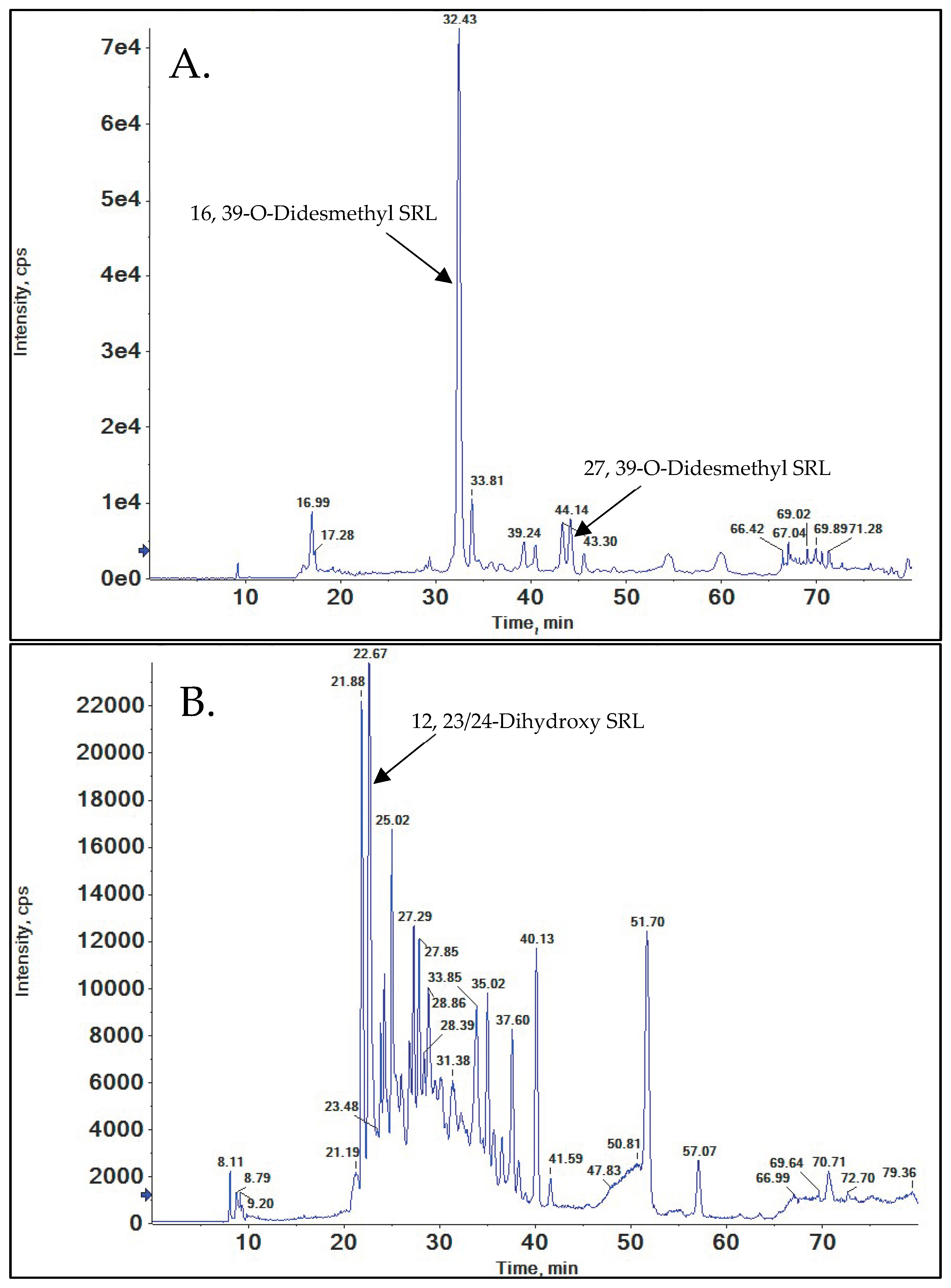
3.5.1. 16, 39-O-Didesmethyl SRL
3.5.2. 27, 39-O-Didesmethyl SRL
3.6. Di-Hydroxylated Metabolites of SRL
12, 23/24-Dihydroxy SRL
3.7. Hydroxy, O-Desmethyl Metabolites of SRL
3.7.1. Hydroxy-Piperidine, 39-O-Desmethyl SRL
3.7.2. 12-Hydroxy, 39-O-Desmethyl SRL
3.7.3. 11-Hydroxy, 16-O-Desmethyl SRL
3.8. Determination of Vmax, Km, and the Intrinsic Clearance (CLint) for the Formation of SRL Metabolites by Pooled HLM
3.9. DFT Calculations
3.10. Molecular Dynamics Simulations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sehgal, S.N.; Baker, H.; Vézina, C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot. 1975, 28, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, S.N. Sirolimus: Its discovery, biological properties, and mechanism of action. Transplant. Proc. 2003, 35, S7–S14. [Google Scholar] [CrossRef] [PubMed]
- Blazar, B.R.; Taylor, P.A.; Sehgal, S.N.; Vallera, D.A. Rapamycin, a potent inhibitor of T-cell function, prevents graft rejection in murine recipients of allogeneic T-cell-depleted donor marrow. Blood 1994, 83, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Morris, R.E. Rapamycins: Antifungal, antitumor, antiproliferative, and immunosuppressive macrolides. Transplant. Rev. 1992, 6, 39–87. [Google Scholar] [CrossRef]
- Wiseman, A.C. Immunosuppressive Medications. Clin. J. Am. Soc. Nephrol. 2016, 11, 332–343. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Nguyen, L.S.; Vautier, M.; Allenbach, Y.; Zahr, N.; Benveniste, O.; Funck-Brentano, C.; Salem, J.-E. Sirolimus and mTOR Inhibitors: A Review of Side Effects and Specific Management in Solid Organ Transplantation. Drug Saf. 2019, 42, 813–825. [Google Scholar] [CrossRef] [PubMed]
- Dumont, F.J.; Su, Q. Mechanism of action of the immunosuppressant rapamycin. Life Sci. 1995, 58, 373–395. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Kim, S.G.; Blenis, J. Rapamycin: One drug, many effects. Cell Metab. 2014, 19, 373–379. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Liu, G.Y.; Sabatini, D.M. Author Correction: mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 2020, 21, 246. [Google Scholar] [CrossRef]
- Augustine, J.J.; Bodziak, K.A.; Hricik, D.E. Use of Sirolimus in Solid Organ Transplantation. Drugs 2007, 67, 369–391. [Google Scholar] [CrossRef] [PubMed]
- McCormack, F.X.; Inoue, Y.; Moss, J.; Singer, L.G.; Strange, C.; Nakata, K.; Barker, A.F.; Chapman, J.T.; Brantly, M.L.; Stocks, J.M.; et al. Efficacy and Safety of Sirolimus in Lymphangioleiomyomatosis. N. Engl. J. Med. 2011, 364, 1595–1606. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abizaid, A. Sirolimus-eluting coronary stents: A review. Vasc. Health Risk Manag. 2007, 3, 191–201. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Abizaid, A.; Costa, J.R. New Drug-Eluting Stents. Circ. Cardiovasc. Interv. 2010, 3, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Adams, D.M.; Trenor, C.C.; Hammill, A.M.; Vinks, A.A.; Patel, M.N.; Chaudry, G.; Wentzel, M.S.; Mobberley-Schuman, P.S.; Campbell, L.M.; Brookbank, C.; et al. Efficacy and Safety of Sirolimus in the Treatment of Complicated Vascular Anomalies. Pediatrics 2016, 137, e20153257. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Klawitter, J.; Nashan, B.; Christians, U. Everolimus and sirolimus in transplantation-related but different. Expert Opin. Drug Saf. 2015, 14, 1055–1070. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Oaks, Z.; Winans, T.; Huang, N.; Banki, K.; Perl, A. Activation of the Mechanistic Target of Rapamycin in SLE: Explosion of Evidence in the Last Five Years. Curr. Rheumatol. Rep. 2016, 18, 73. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Castagnoli, R.; Delmonte, O.M.; Calzoni, E.; Notarangelo, L.D. Hematopoietic Stem Cell Transplantation in Primary Immunodeficiency Diseases: Current Status and Future Perspectives. Front. Pediatr. 2019, 7, 295. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Bride, K.L.; Vincent, T.; Smith-Whitley, K.; Lambert, M.P.; Bleesing, J.J.; Seif, A.E.; Manno, C.S.; Casper, J.; Grupp, S.A.; Teachey, D.T. Sirolimus is effective in relapsed/refractory autoimmune cytopenias: Results of a prospective multi-institutional trial. Blood 2016, 127, 17–28. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Filler, G.; Bendrick-Peart, J.; Strom, T.; Zhang, Y.L.; Johnson, G.; Christians, U. Characterization of sirolimus metabolites in pediatric solid organ transplant recipients. Pediatr. Transplant. 2009, 13, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Tao, T.; Li, H.; Zhu, X. mTOR signaling pathway and mTOR inhibitors in cancer: Progress and challenges. Cell Biosci. 2020, 10, 31. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Blagosklonny, M.V. Cancer prevention with rapamycin. Oncotarget 2023, 14, 342–350. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Cohen, E.E.; Wu, K.; Hartford, C.; Kocherginsky, M.; Eaton, K.N.; Zha, Y.; Nallari, A.; Maitland, M.L.; Fox-Kay, K.; Moshier, K.; et al. Phase I Studies of Sirolimus Alone or in Combination with Pharmacokinetic Modulators in Advanced Cancer Patients. Clin. Cancer Res. 2012, 18, 4785–4793. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yi, Z.; Liu, B.; Sun, X.; Rong, G.; Wang, W.; Li, H.; Guan, X.; Li, L.; Zhai, J.; Li, C.; et al. Safety and efficacy of sirolimus combined with endocrine therapy in patients with advanced hormone receptor-positive breast cancer and the exploration of biomarkers. Breast 2020, 52, 17–22. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Hu, M.; Ekshyyan, O.; Ferdinandez, L.H.; Rong, X.; Caldito, G.; Nathan, C.-A.O. Efficacy and comparative effectiveness of sirolimus as an anticancer drug. Laryngoscope 2011, 121, 978–982. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Zhou, Y.; Zhao, R.-H.; Tseng, K.-F.; Li, K.-P.; Lu, Z.-G.; Liu, Y.; Han, K.; Gan, Z.-H.; Lin, S.-C.; Hu, H.-Y.; et al. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA-34b expression. Acta Pharmacol. Sin. 2016, 37, 519–529. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Wagner, A.J.; Ravi, V.; Riedel, R.F.; Ganjoo, K.; Van Tine, B.A.; Chugh, R.; Cranmer, L.; Gordon, E.M.; Hornick, J.L.; Du, H.; et al. nab-Sirolimus for Patients With Malignant Perivascular Epithelioid Cell Tumors. J. Clin. Oncol. 2021, 39, 3660–3670. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Koenig, M.K.; Bell, C.S.; Hebert, A.A.; Roberson, J.; Samuels, J.A.; Slopis, J.M.; Tate, P.; Northrup, H.; Collaborators, F.T.T.T. Efficacy and Safety of Topical Rapamycin in Patients With Facial Angiofibromas Secondary to Tuberous Sclerosis Complex. JAMA Dermatol. 2018, 154, 773–780. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Mohammadpour, N.; Elyasi, S.; Vahdati, N.; Mohammadpour, A.H.; Shamsara, J. A review on therapeutic drug monitoring of immunosuppressant drugs. Iran. J. Basic Med. Sci. 2011, 14, 485–498. [Google Scholar] [PubMed] [PubMed Central]
- Shaw, L.M.; Holt, D.W.; Keown, P.; Venkataramanan, R.; Yatscoff, R.W. Current opinions on therapeutic drug monitoring of immunosuppressive drugs. Clin. Ther. 1999, 21, 1632–1652. [Google Scholar] [CrossRef] [PubMed]
- Ericson, J.E.; Zimmerman, K.O.; Gonzalez, D.; Melloni, C.M.; Guptill, J.T.; Hill, K.D.; Wu, H.; Cohen-Wolkowiez, M. A Systematic Literature Review Approach to Estimate the Therapeutic Index of Selected Immunosuppressant Drugs After Renal Transplantation. Ther. Drug Monit. 2017, 39, 13–20. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Kang, J.-S.; Lee, M.-H. Overview of therapeutic drug monitoring. Korean J. Intern. Med. 2009, 24, 1–10. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Simamora, P.; Alvarez, J.M.; Yalkowsky, S.H. Solubilization of rapamycin. Int. J. Pharm. 2001, 213, 25–29. [Google Scholar] [CrossRef]
- Sattler, M.; Guengerich, F.P.; Yun, C.H.; Christians, U.; Sewing, K.F. Cytochrome P-450 3A enzymes are responsible for biotransformation of FK506 and rapamycin in man and rat. Drug Metab. Dispos. 1992, 20, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Meur, Y.; Djebli, N.; Szelag, J.; Hoizey, G.; Toupance, O.; Rérolle, J.P.; Marquet, P.; Le Meur, Y. CYP3A5*3 influences sirolimus oral clearance in de novo and stable renal transplant recipients. Clin. Pharmacol. Ther. 2006, 80, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, W.; Serkova, N.; Hausen, B.; Morris, R.; Benet, L.; Christians, U. Comparison of the in vitro metabolism of the macrolide immunosuppressants sirolimus and RAD. Transplant. Proc. 2001, 33, 514–515. [Google Scholar] [CrossRef] [PubMed]
- Christians, U.; Sattler, M.; Schiebel, H.M.; Kruse, C.; Radeke, H.H.; Linck, A.; Sewing, K.F. Isolation of two immunosuppressive metabolites after in vitro metabolism of rapamycin. Drug Metab. Dispos. 1992, 20, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Streit, F.; Christians, U.; Schiebel, H.M.; Meyer, A.; Sewing, K.F. Structural identification of three metabolites and a degradation product of the macrolide immunosuppressant sirolimus (rapamycin) by electrospray-MS/MS after incubation with human liver microsomes. Drug Metab. Dispos. 1996, 24, 1272–1278. [Google Scholar] [CrossRef] [PubMed]
- Boernsen, K.O.; Egge-Jacobsen, W.; Inverardi, B.; Strom, T.; Streit, F.; Schiebel, H.; Benet, L.Z.; Christians, U. Assessment and validation of the MS/MS fragmentation patterns of the macrolide immunosuppressant everolimus. J. Mass Spectrom. 2007, 42, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Paine, M.F.; Leung, L.Y.; Lim, H.; Liao, K.; Oganesian, A.; Zhang, M.-Y.; Thummel, K.E.; Watkins, P.B. Identification of a novel route of extraction of sirolimus in human small intestine: Roles of metabolism and secretion. J. Pharmacol. Exp. Ther. 2002, 301, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Strom, T.; Shokati, T.; Klawitter, J.; Klawitter, J.; Hoffman, K.; Schiebel, H.; Christians, U. Structural identification of SAR-943 metabolites generated by human liver microsomes in vitro using mass spectrometry in combination with analysis of fragmentation patterns. J. Mass Spectrom. 2011, 46, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Shokati, T.; Drake, S.H.; Zhao, W.; Klawitter, J.; Klawitter, J.; Christians, U. Structural Identification of Zotarolimus (ABT-578) Metabolites Generated by Human Liver Microsomes Using Ion-Trap and High-Resolution Time-of-Flight Mass Spectrometry in Combination with the Analysis of Fragmentation Patterns. Metabolites 2023, 13, 1093. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gallant-Haidner, H.L.; Trepanier, D.J.; Freitag, D.G.; Yatscoff, R.W. Pharmacokinetics and metabolism of sirolimus. Ther. Drug Monit. 2000, 22, 31–35. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.; Jacobsen, W.; Christians, U.; Benet, L.Z.; Kollman, P.A. Metabolism of sirolimus and its derivative everolimus by cytochrome P450 3A4: Insights from docking, molecular dynamics, and quantum chemical calculations. J. Med. Chem. 2001, 44, 2027–2034. [Google Scholar] [CrossRef] [PubMed]
- Bochevarov, A.D.; Harder, E.; Hughes, T.F.; Greenwood, J.R.; Braden, D.A.; Philipp, D.M.; Rinaldo, D.; Halls, M.D.; Zhang, J.; Friesner, R.A. Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. Int. J. Quantum Chem. 2013, 113, 2110–2142. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 1993, 98, 5648–5652. [Google Scholar] [CrossRef]
- Lee, C.; Yang, W.; Parr, R.G. Development of the colle-salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B 1988, 37, 785–789. [Google Scholar] [CrossRef]
- Vosko, S.H.; Wilk, L.; Nusair, M. Accurate spin-dependent electron liquid correlation energies for local spin density calculations: A critical analysis. Can. J. Phys. 1980, 58, 1200–1211. [Google Scholar] [CrossRef]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab Initio Calculation of Vibrational Absorption and Circular Dichroism Spectra Using Density Functional Force Fields. J. Phys. Chem. 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Ditchfield, R.; Hehre, W.J.; Pople, J.A. Self-consistent molecular-orbital methods. IX. An extended gaussian-type basis for molecular-orbital studies of organic molecules. J. Chem. Phys. 1971, 54, 724–728. [Google Scholar] [CrossRef]
- Ekroos, M.; Sjögren, T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. USA 2006, 103, 13682–13687. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Shokati, T.; Hartmann, M.; Davari, B.; Klawitter, J.; Klawitter, J.; Christians, U. Temsirolimus metabolic pathways revisited. Xenobiotica 2020, 50, 640–653. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.; Shiraga, T.; Matsuda, H.; Nagase, K.; Tokuma, Y.; Hata, T.; Fujii, Y.; Sakuma, S.; Fujitsu, T.; Fujikawa, A. Further metabolism of FK506 (tacrolimus). Identification and biological activities of the metabolites oxidized at multiple sites of FK506. Drug Metab. Dispos. 1995, 23, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Iwasaki, K.; Shiraga, T.; Nagase, K.; Tozuka, Z.; Noda, K.; Sakuma, S.; Fujitsu, T.; Shimatani, K.; Sato, A.; Fujioka, M. Isolation, identification, and biological activities of oxidative metabolites of FK506, a potent immunosuppressive macrolide lactone. Drug Metab. Dispos. 1993, 21, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Maurer, G.; Loosli, H.R.; Schreier, E.; Keller, B. Disposition of cyclosporine in several animal species and man. I. Structural elucidation of its metabolites. Drug Metab. Dispos. 1984, 12, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Cai, P.; Tsao, R.; Ruppen, M.E. In vitro metabolic study of temsirolimus: Preparation, isolation, and identification of the metabolites. Drug Metab. Dispos. 2007, 35, 1554–1563. [Google Scholar] [CrossRef] [PubMed]
- Nickmilder, M.J.M.; Latinne, D.; De Houx, J.-P.; Verbeeck, R.K.; Lhoëst, G.J.J. Isolation and identification of a C39 demethylated metabolite of rapamycin from pig liver microsomes and evaluation of its immunosuppressive activity. Clin. Chem. 1998, 44, 532–538. [Google Scholar] [CrossRef] [PubMed]
- Nickmilder, M.J.M.; Latinne, D.; Verbeeck, R.K.; Janssens, W.; Svoboda, D.; Lhoest, G.J.J. Isolation and identification of new rapamycin dihydrodiol metabolites from dexamethasoneinduced rat liver microsomes. Xenobiotica 1997, 27, 869–883. [Google Scholar] [CrossRef]
- Naming and Indexing of Chemical Substances for Chemical AbstractsTM. American Chemical Society: Columbus, OH, USA, 2008. [Cited 2007 Edition]. Available online: https://web.cas.org/marketing/pdf/indexguideapp.pdf (accessed on 21 March 2024).
- Guidance for Industry and FDA Staff Class II Special Controls Guidance Document: Sirolimus Test Systems. U.S. Department of Health and Human Services Food and Drug Administration Center for Devices and Radiological Health, 2004; [Updated 30 September 2004]. Available online: https://www.fda.gov/media/71350/download (accessed on 21 March 2024).
- Mahalati, K.; Kahan, B.D. Clinical Pharmacokinetics of Sirolimus. Clin. Pharmacokinet. 2001, 40, 573–585. [Google Scholar] [CrossRef] [PubMed]
- Hallensleben, K.; Raida, M.; Habermehl, G. Identification of a new metabolite of macrolide immunosuppressant, like rapamycin and SDZ RAD, using high performance liquid chromatography and electrospray tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2000, 11, 516–525. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A. Immunoassays and Issues with Interference in Therapeutic Drug Monitoring. In Clinical Challenges in Therapeutic Drug Monitoring Special Populations, Physiological Conditions and Pharmacogenomics; Clarke, W., Dasgupta, A., Eds.; Elservier Inc.: Amsterdam, The Netherlands, 2016; pp. 17–44. [Google Scholar]
- Cattaneo, D.; Merlini, S.; Pellegrino, M.; Carrara, F.; Zenoni, S.; Murgia, S.; Baldelli, S.; Gaspari, F.; Remuzzi, G.; Perico, N. Therapeutic drug monitoring of sirolimus: Effect of concomitant immunosuppressive therapy and optimization of drug dosing. Am. J. Transplant. 2004, 4, 1345–1351. [Google Scholar] [CrossRef] [PubMed]
- Pieri, M.; Miraglia, N.; Gentile, A.; Polichetti, G.; Castiglia, L.; Federico, S.; Sabbatini, M.; Basile, V.; Tarantino, G.; Acampora, A.; et al. Quantification of sirolimus and everolimus by immunoassay techniques: Test specificity and cross-reactivity evaluation. Int. J. Immunopathol. Pharmacol. 2008, 21, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Luengo, J.I.; Yamashita, D.S.; Dunnington, D.; Beck, A.K.; Rozamus, L.W.; Yen, H.-K.; Bossard, M.J.; Levy, M.A.; Hand, A.; Newman-Tarr, T.; et al. Structure-activity studies of rapamycin analogs: Evidence that the C-7 methoxy group is part of the effector domain and positioned at the FKBP12-FRAP interface. Chem. Biol. 1995, 2, 471–481. [Google Scholar] [CrossRef] [PubMed]




| Hydroxylated metabolites | 46-Hydroxy sirolimus |
| 45-Hydroxy sirolimus | |
| 23/24-Hydroxy sirolimus | |
| 12-Hydroxy sirolimus | |
| 25-Hydroxy sirolimus | |
| 11-Hydroxy sirolimus | |
| Hydroxy-piperidine sirolimus (a, b, c, d) | |
| 14-Hydroxy sirolimus | |
| 49-Hydroxy sirolimus | |
| Dihydroxy metabolites | 12, 24-Dihydroxy sirolimus |
| Desmethyl metabolites | 16-O-Desmethyl sirolimus |
| 27-O-Desmethyl sirolimus | |
| 39-O-Desmethyl sirolimus | |
| Didesmethyl metabolites | 27, 39-O-Didesmethyl sirolimus |
| 16, 39-O-Didesmethyl sirolimus | |
| Hydroxy-desmethyl metabolites | Piperidine-hydroxy-39-O-desmethyl sirolimus |
| 12-Hydroxy-39-O-desmethyl sirolimus | |
| 11-Hydroxy-16-O-desmethyl sirolimus |
| Theoretical Mass | Measured Mass | Δppm | |
|---|---|---|---|
| Sirolimus | 936.5444 | 936.5444 | 0.0 |
| A | 731.4493 | 731.4491 | 0.3 |
| B | 763.4756 | 763.4756 | 0.0 |
| C | 642.3249 | 642.3250 | 0.2 |
| D | 345.2036 | 345.2038 | 0.6 |
| E | 703.4544 | 703.4547 | 0.4 |
| F | 399.2506 | 399.2508 | 0.5 |
| G | 614.3300 | 614.3298 | 0.3 |
| H | 485.2510 | 485.2511 | 0.2 |
| I | 459.2717 | 459.2718 | 0.2 |
| J | 409.2349 | 409.2350 | 0.2 |
| K | 607.3969 | 607.3969 | 0.0 |
| L | 397.2349 | 397.2363 | 3.5 |
| M | 582.3037 | 582.3035 | 0.3 |
| N | 453.2248 | 453.2247 | 0.2 |
| O | 441.2611 | 441.2610 | 0.5 |
| P | 381.2400 | 381.2401 | 0.3 |
| Q | 320.1105 | 320.1104 | 0.3 |
| R | 413.2662 | 413.2662 | 0.0 |
| Metabolite | Vmax (pmol/mg protein/min) | Km (µM) | CLint (µL/mg protein/min) |
|---|---|---|---|
| [95% CI] | [95% CI] | ||
| 23/24-Hydroxy SRL | 2.221 | 20.82 | 0.11 |
| [1.8–2.8] | [11.35–38.27] | ||
| 12-Hydroxy SRL | 4.264 | 7.211 | 0.6 |
| [3.674–4.955] | [3.4–13.01] | ||
| 11-Hydroxy SRL | 6.203 | 9.577 | 0.65 |
| [5.673–6.799] | [6.6–13.43] | ||
| Hydroxy-Piperidine SRL | 2.781 | 13.15 | 0.21 |
| [2.350–3.333] | [7.1–23.14] | ||
| 16-O-desmethyl SRL | 1.828 | 24.37 | 0.08 |
| [1.452–2.393] | [12.20–50.43] | ||
| 39-O-desmthyl SRL | 9.647 | 14.95 | 0.65 |
| [8.692–10.76] | [10.39–21.51] | ||
| 27-O-desmethyl SRL | 0.552 | 9.647 | 0.06 |
| [0.47–0.65] | [8.7–10.76] |
| MO-CH3 → MO + CH3 | MO + H → MOH | MO-CH3 + H → MO-H + CH3 | |
|---|---|---|---|
| Demethylation Site | ΔGdemethylation (kcal/mol) | ΔGhydrogenation (kcal/mol) | ΔGOverall (kcal/mol) |
| C16 | 84.43 | −108.48 | −24.05 |
| C27 | 78.12 | −90.82 | −12.71 |
| C39 | 85.62 | −107.51 | −21.89 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davari, B.; Shokati, T.; Ward, A.M.; Nguyen, V.; Klawitter, J.; Klawitter, J.; Christians, U. Human Metabolism of Sirolimus Revisited. Metabolites 2025, 15, 489. https://doi.org/10.3390/metabo15070489
Davari B, Shokati T, Ward AM, Nguyen V, Klawitter J, Klawitter J, Christians U. Human Metabolism of Sirolimus Revisited. Metabolites. 2025; 15(7):489. https://doi.org/10.3390/metabo15070489
Chicago/Turabian StyleDavari, Baharak, Touraj Shokati, Alexandra M. Ward, Vu Nguyen, Jost Klawitter, Jelena Klawitter, and Uwe Christians. 2025. "Human Metabolism of Sirolimus Revisited" Metabolites 15, no. 7: 489. https://doi.org/10.3390/metabo15070489
APA StyleDavari, B., Shokati, T., Ward, A. M., Nguyen, V., Klawitter, J., Klawitter, J., & Christians, U. (2025). Human Metabolism of Sirolimus Revisited. Metabolites, 15(7), 489. https://doi.org/10.3390/metabo15070489







