Exploring Postharvest Metabolic Shifts and NOX2 Inhibitory Potential in Strawberry Fruits and Leaves via Untargeted LC-MS/MS and Chemometric Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Strawberry Fruit and Leaf Samples and Storage Conditions
2.2. Metabolite Extraction from Strawberry Fruits and Leaves
2.3. LC-MS/MS Analysis
2.4. Statistical Analysis
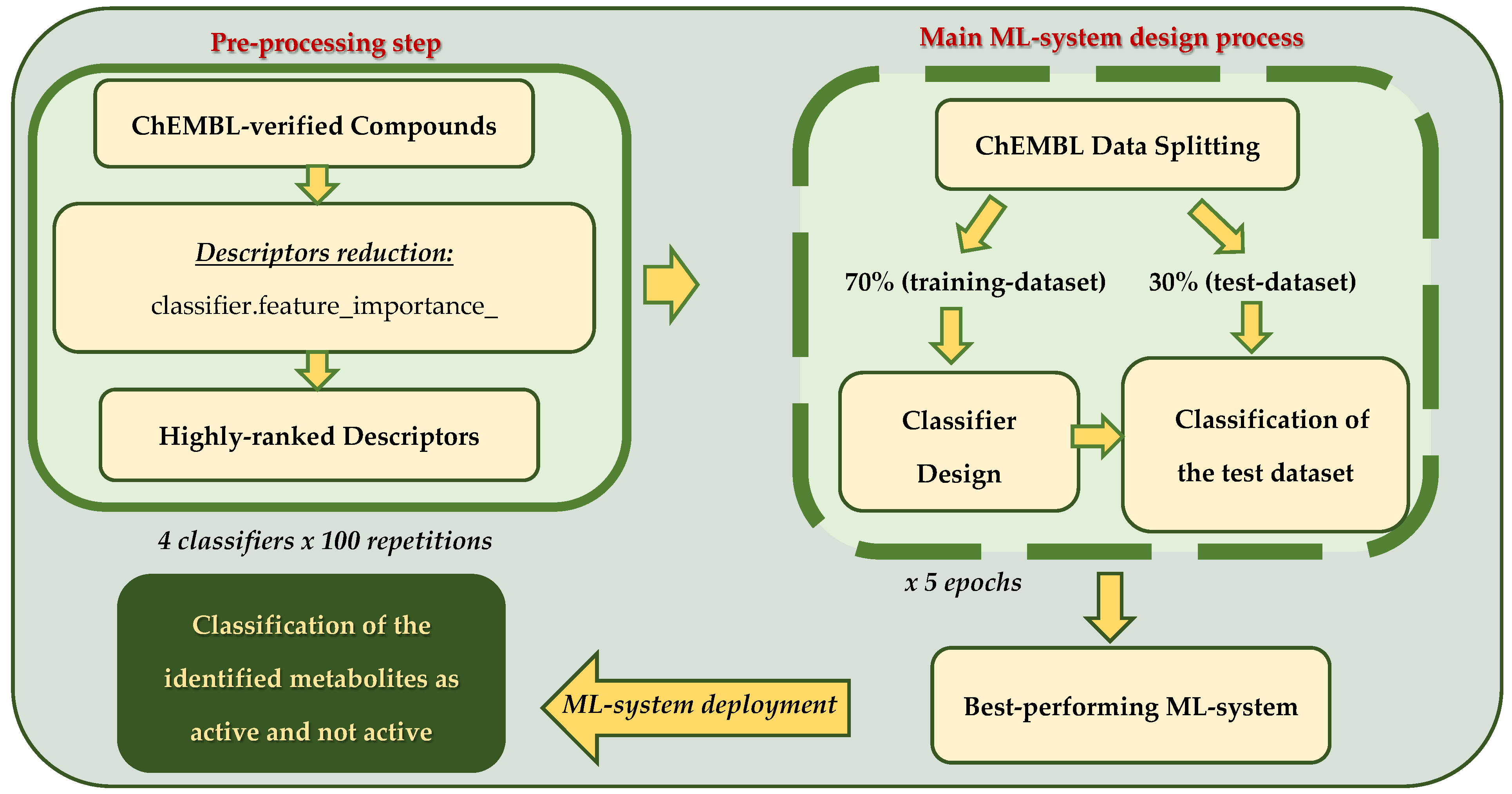
2.5. Machine Learning-Based Evaluation of Strawberry Metabolites for NOX2 Inhibitory Potential
2.5.1. Curation of NOX2 Inhibitor Dataset and Molecular Descriptors Calculation
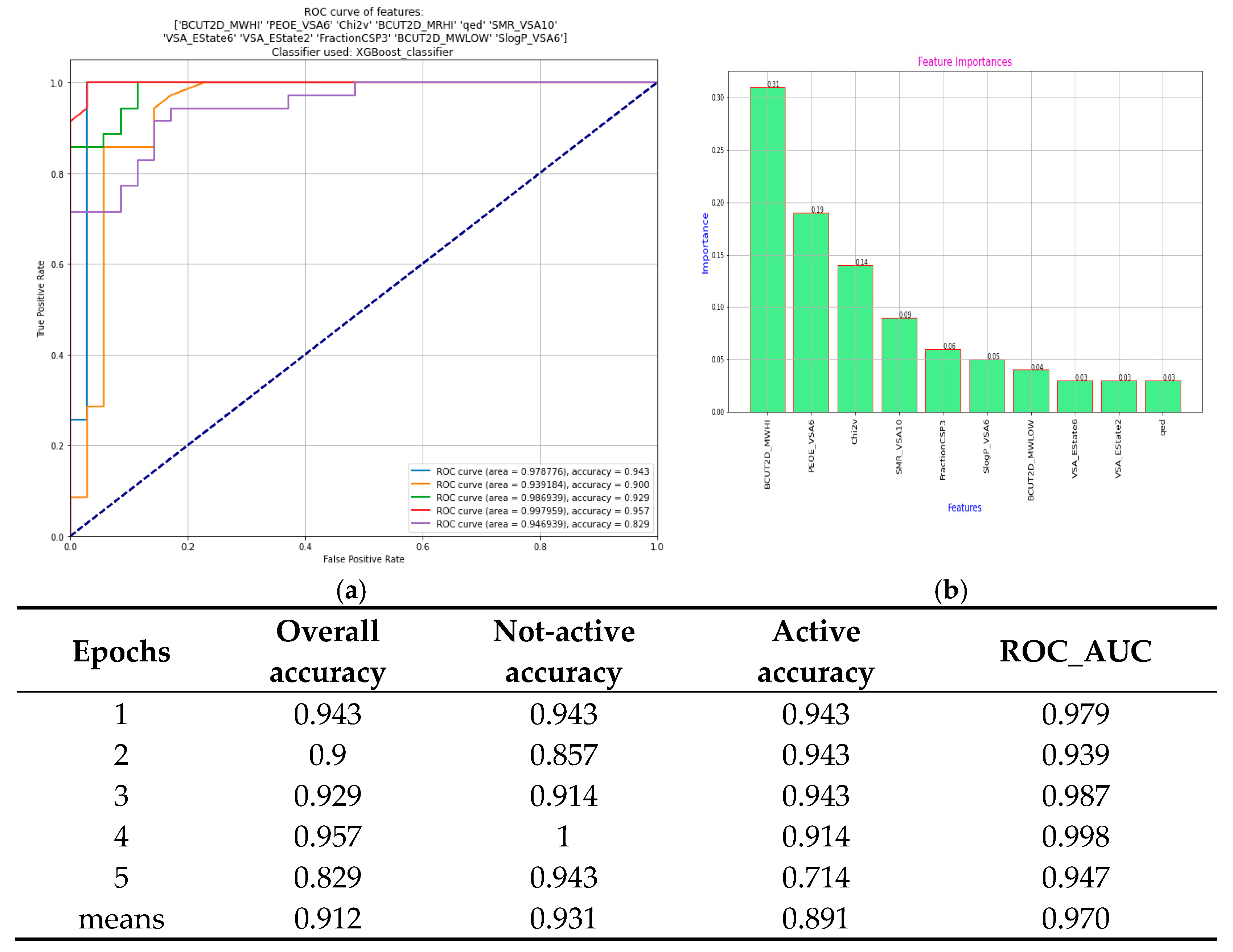
2.5.2. Machine Learning Analysis
2.5.3. Machine Learning System Deployment to Predict Bioactivity of Identified Strawberry Metabolites by LC-MS/MS Analysis
3. Results
3.1. Metabolite Profile of Strawberry Fruits and Leaves
3.2. Statistical Analysis of Metabolite Changes During Storage in Strawberry Fruits
3.2.1. ANOVA Post Hoc Analysis of Metabolite Fluctuations in Strawberry Fruits
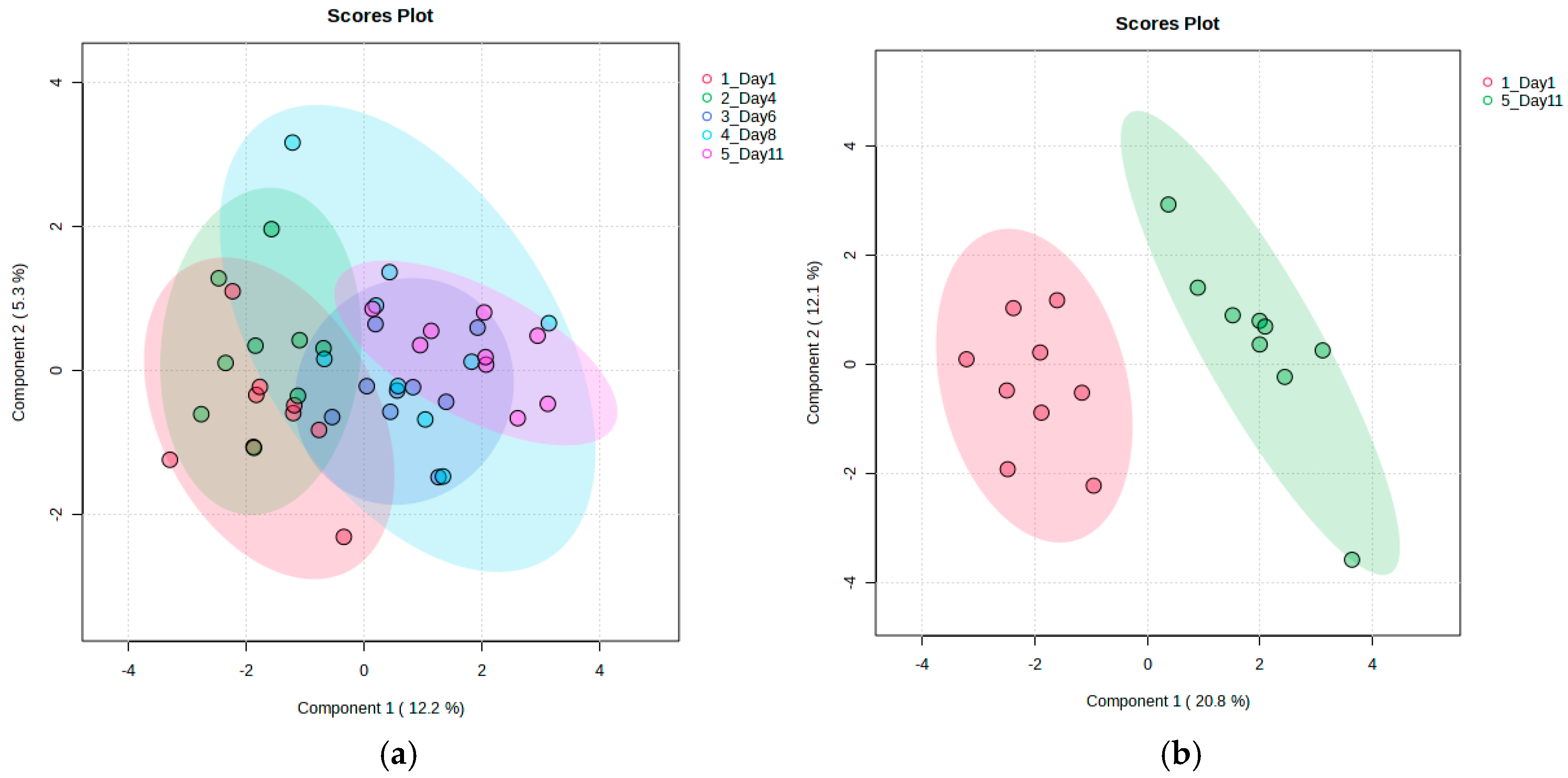
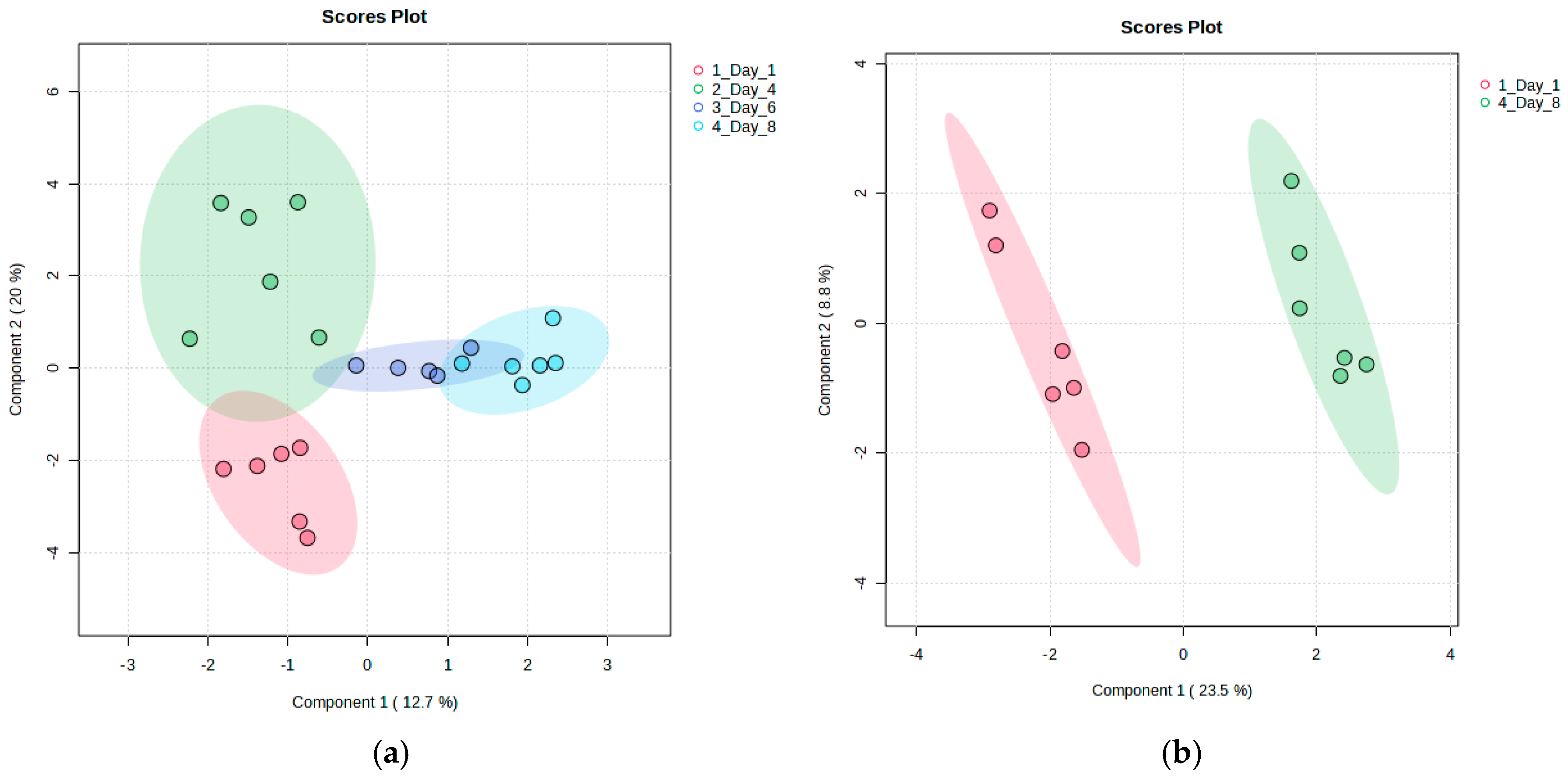
3.2.2. Discriminant Analysis of Metabolite Variability in Strawberry Fruits During Storage
3.3. Statistical Analysis of Metabolite Changes During Storage in Strawberry Leaves
3.3.1. ANOVA Post Hoc Analysis of Metabolic Alterations in Strawberry Leaves
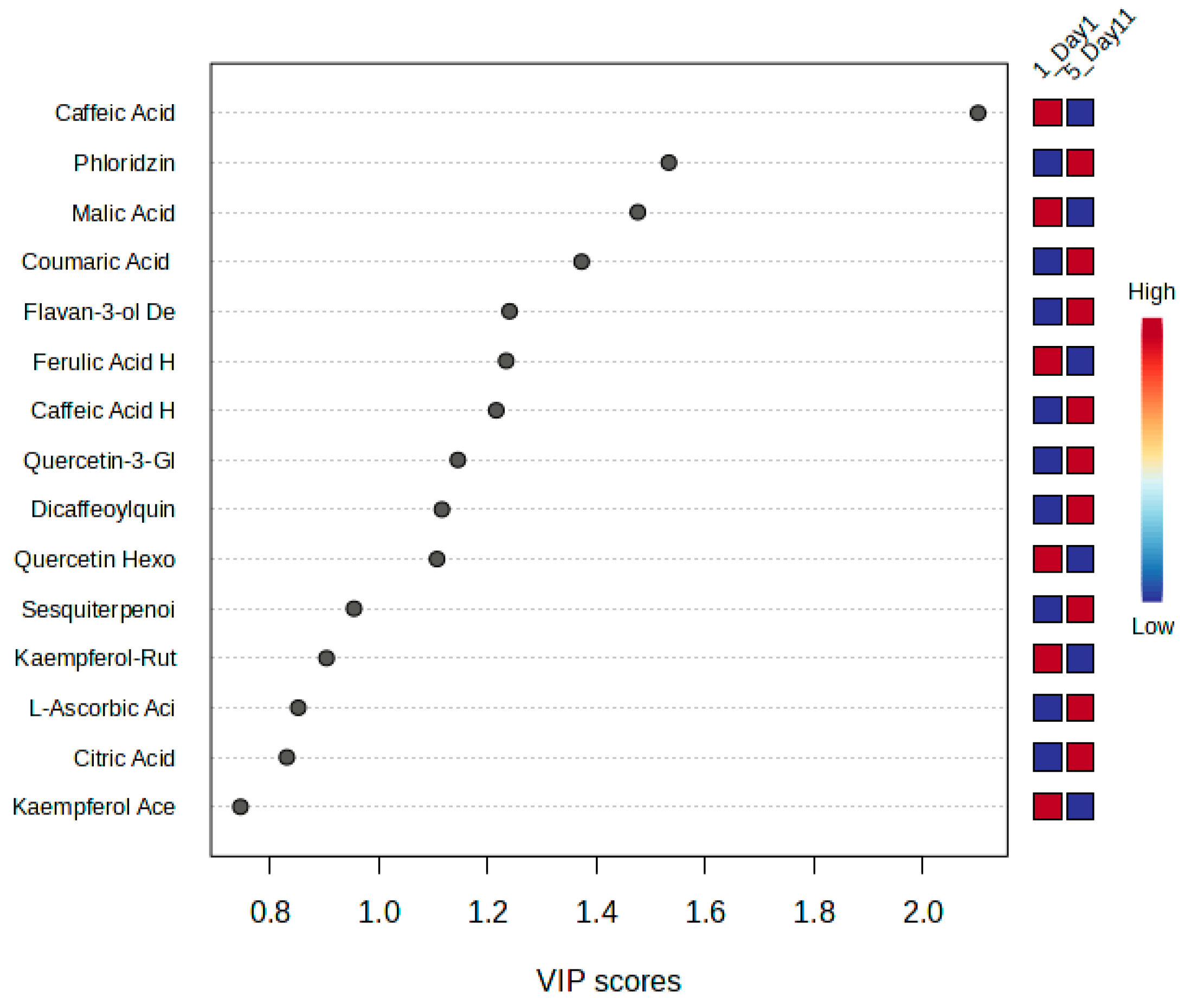
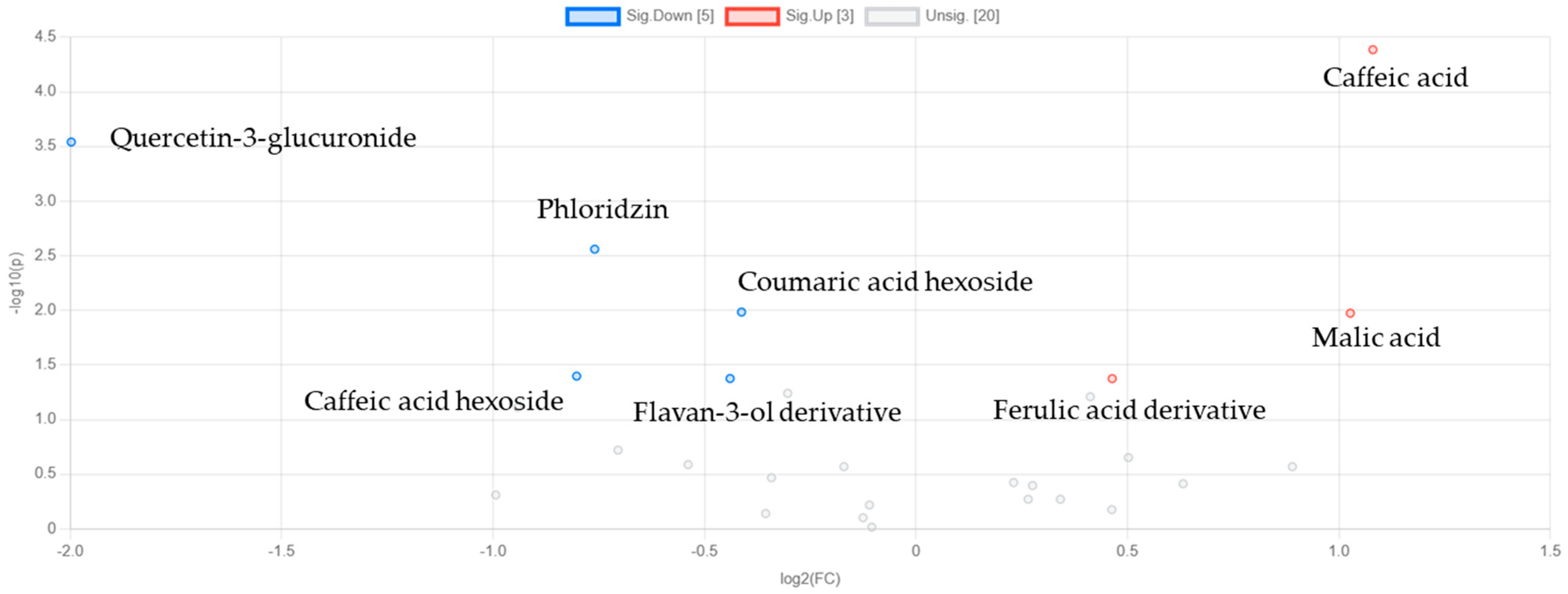
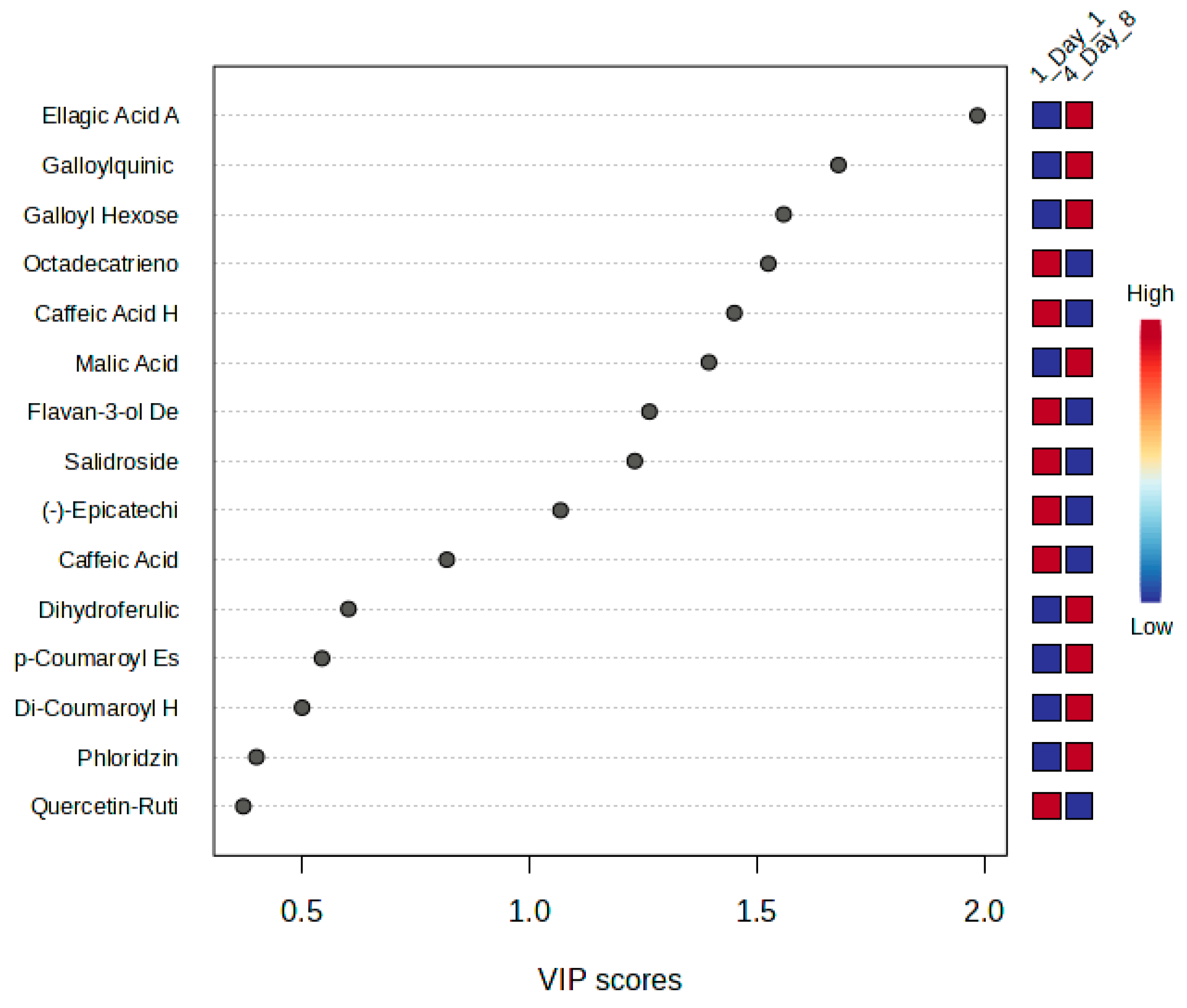
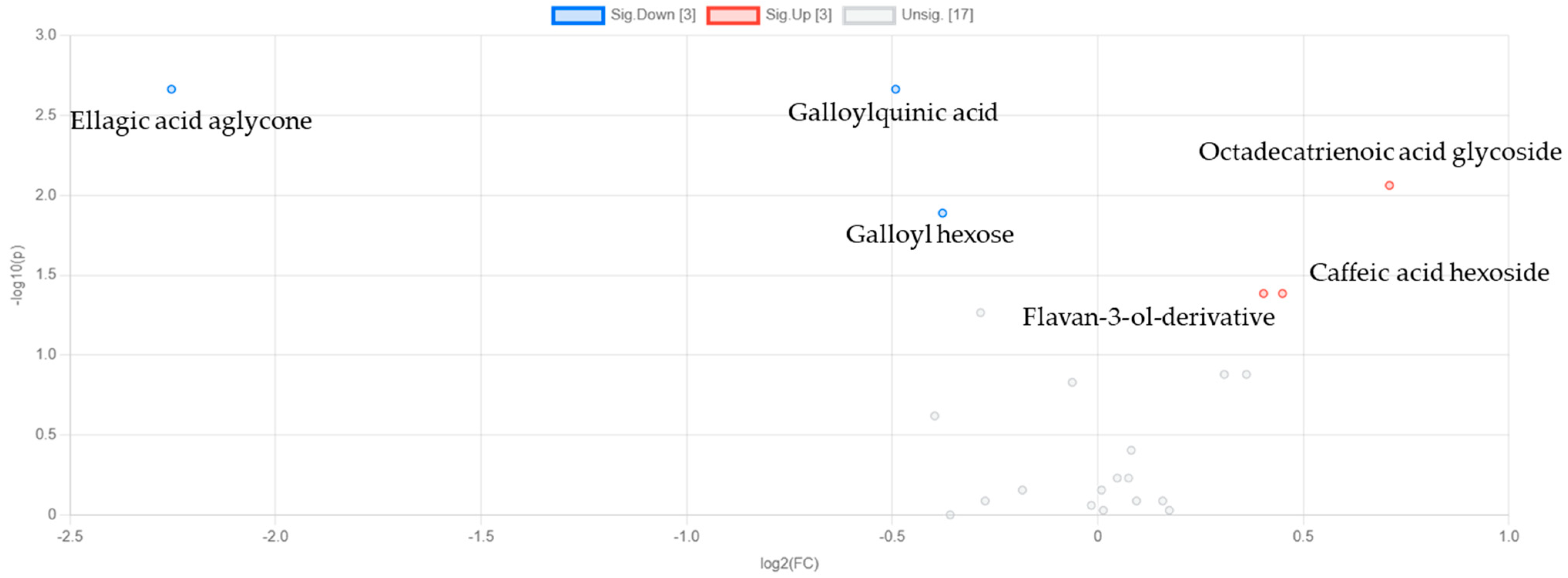
3.3.2. Discriminant Analysis of Metabolite Variability in Strawberry Leaves During Storage
3.3.3. Machine Learning-Based Evaluation of NOX2 Inhibitory Potential
4. Discussion
4.1. Metabolic Changes in Strawberry Fruits During Storage
4.2. Metabolic Changes in Strawberry Leaves During Storage
4.3. NOX2 Inhibitory Potential of Strawberry Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Giampieri, F.; Alvarez-Suarez, J.M.; Battino, M. Strawberry and Human Health: Effects beyond Antioxidant Activity. J. Agric. Food Chem. 2014, 62, 3867–3876. [Google Scholar] [CrossRef]
- Liu, Z.; Liang, T.; Kang, C. Molecular Bases of Strawberry Fruit Quality Traits: Advances, Challenges, and Opportunities. Plant Physiol. 2023, 193, 900–914. [Google Scholar] [CrossRef]
- Newerli-Guz, J.; Śmiechowska, M.; Drzewiecka, A.; Tylingo, R. Bioactive Ingredients with Health-Promoting Properties of Strawberry Fruit (Fragaria x Ananassa Duchesne). Molecules 2023, 28, 2711. [Google Scholar] [CrossRef] [PubMed]
- Priyadarshi, R.; Jayakumar, A.; De Souza, C.K.; Rhim, J.; Kim, J.T. Advances in Strawberry Postharvest Preservation and Packaging: A Comprehensive Review. Comp. Rev. Food Sci. Food Safe 2024, 23, e13417. [Google Scholar] [CrossRef] [PubMed]
- Ktenioudaki, A.; O’Donnell, C.P.; Do Nascimento Nunes, M.C. Modelling the Biochemical and Sensory Changes of Strawberries during Storage under Diverse Relative Humidity Conditions. Postharvest Biol. Technol. 2019, 154, 148–158. [Google Scholar] [CrossRef]
- Baicu, A.; Popa, M.E. Trends in Prolonging the Post-Harvest Life of Strawberries—A Review. Ann. Univ. Dunarea Jos Galati Fascicle VI Food Technol. 2018, 42, 9–16. [Google Scholar]
- Zhang, W.; Jiang, Y.; Zhang, Z. The Role of Different Natural Organic Acids in Postharvest Fruit Quality Management and Its Mechanism. Food Front. 2023, 4, 1127–1143. [Google Scholar] [CrossRef]
- Urün, I.; Attar, S.H.; Sönmez, D.A.; Gündeşli, M.A.; Ercişli, S.; Kafkas, N.E.; Bandić, L.M.; Duralija, B. Comparison of Polyphenol, Sugar, Organic Acid, Volatile Compounds, and Antioxidant Capacity of Commercially Grown Strawberry Cultivars in Turkey. Plants 2021, 10, 1654. [Google Scholar] [CrossRef]
- Badmi, R.; Gogoi, A.; Doyle Prestwich, B. Secondary Metabolites and Their Role in Strawberry Defense. Plants 2023, 12, 3240. [Google Scholar] [CrossRef]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An Overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef]
- Yan, Y.; Pico, J.; Sun, B.; Pratap-Singh, A.; Gerbrandt, E.; Diego Castellarin, S. Phenolic Profiles and Their Responses to Pre- and Post-Harvest Factors in Small Fruits: A Review. Crit. Rev. Food Sci. Nutr. 2023, 63, 3574–3601. [Google Scholar] [CrossRef] [PubMed]
- Kårlund, A.; Hanhineva, K.; Lehtonen, M.; McDougall, G.J.; Stewart, D.; Karjalainen, R.O. Non-targeted Metabolite Profiling Highlights the Potential of Strawberry Leaves as a Resource for Specific Bioactive Compounds. J. Sci. Food Agric. 2017, 97, 2182–2190. [Google Scholar] [CrossRef] [PubMed]
- Thirumurugan, D.; Cholarajan, A.; Raja, S.S.S.; Vijayakumar, R. An Introductory Chapter: Secondary Metabolites. In Secondary Metabolites—Sources and Applications; Vijayakumar, R., Raja, S.S.S., Eds.; InTech: Houston, TX, USA, 2018; ISBN 978-1-78923-642-2. [Google Scholar]
- Andrés, C.M.C.; Pérez De La Lastra, J.M.; Juan, C.A.; Plou, F.J.; Pérez-Lebeña, E. Polyphenols as Antioxidant/Pro-Oxidant Compounds and Donors of Reducing Species: Relationship with Human Antioxidant Metabolism. Processes 2023, 11, 2771. [Google Scholar] [CrossRef]
- Morawietz, H.; Brendel, H.; Diaba-Nuhoho, P.; Catar, R.; Perakakis, N.; Wolfrum, C.; Bornstein, S.R. Cross-Talk of NADPH Oxidases and Inflammation in Obesity. Antioxidants 2023, 12, 1589. [Google Scholar] [CrossRef]
- Battino, M.; Giampieri, F.; Cianciosi, D.; Ansary, J.; Chen, X.; Zhang, D.; Gil, E.; Forbes-Hernández, T. The Roles of Strawberry and Honey Phytochemicals on Human Health: A Possible Clue on the Molecular Mechanisms Involved in the Prevention of Oxidative Stress and Inflammation. Phytomedicine 2021, 86, 153170. [Google Scholar] [CrossRef] [PubMed]
- Ladika, G.; Strati, I.F.; Tsiaka, T.; Cavouras, D.; Sinanoglou, V.J. On the Assessment of Strawberries’ Shelf-Life and Quality, Based on Image Analysis, Physicochemical Methods, and Chemometrics. Foods 2024, 13, 234. [Google Scholar] [CrossRef]
- Marcus, Y. Extraction by Subcritical and Supercritical Water, Methanol, Ethanol and Their Mixtures. Separations 2018, 5, 4. [Google Scholar] [CrossRef]
- Sultana, B.; Anwar, F.; Ashraf, M. Effect of Extraction Solvent/Technique on the Antioxidant Activity of Selected Medicinal Plant Extracts. Molecules 2009, 14, 2167–2180. [Google Scholar] [CrossRef]
- Tsiaka, T.; Kritsi, E.; Bratakos, S.M.; Sotiroudis, G.; Petridi, P.; Savva, I.; Christodoulou, P.; Strati, I.F.; Zoumpoulakis, P.; Cavouras, D.; et al. Quality Assessment of Ground Coffee Samples from Greek Market Using Various Instrumental Analytical Methods, In Silico Studies and Chemometrics. Antioxidants 2023, 12, 1184. [Google Scholar] [CrossRef]
- Kritsi, E.; Tsiaka, T.; Sotiroudis, G.; Mouka, E.; Aouant, K.; Ladika, G.; Zoumpoulakis, P.; Cavouras, D.; Sinanoglou, V.J. Potential Health Benefits of Banana Phenolic Content during Ripening by Implementing Analytical and In Silico Techniques. Life 2023, 13, 332. [Google Scholar] [CrossRef]
- Aaby, K.; Mazur, S.; Nes, A.; Skrede, G. Phenolic Compounds in Strawberry (Fragaria x Ananassa Duch.) Fruits: Composition in 27 Cultivars and Changes during Ripening. Food Chem. 2012, 132, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Seeram, N.P.; Lee, R.; Scheuller, H.S.; Heber, D. Identification of Phenolic Compounds in Strawberries by Liquid Chromatography Electrospray Ionization Mass Spectroscopy. Food Chem. 2006, 97, 1–11. [Google Scholar] [CrossRef]
- Kårlund, A.; Hanhineva, K.; Lehtonen, M.; Karjalainen, R.O.; Sandell, M. Nontargeted Metabolite Profiles and Sensory Properties of Strawberry Cultivars Grown Both Organically and Conventionally. J. Agric. Food Chem. 2015, 63, 1010–1019. [Google Scholar] [CrossRef] [PubMed]
- Sinanoglou, V.J.; Kavga, A.; Strati, I.F.; Sotiroudis, G.; Lantzouraki, D.; Zoumpoulakis, P. Effects of Infrared Radiation on Eggplant (Solanum melongena L.) Greenhouse Cultivation and Fruits’ Phenolic Profile. Foods 2019, 8, 630. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Fernández, M.A.; Hornedo-Ortega, R.; Cerezo, A.B.; Troncoso, A.M.; García-Parrilla, M.C. Determination of Nonanthocyanin Phenolic Compounds Using High-Resolution Mass Spectrometry (UHPLC-Orbitrap-MS/MS) and Impact of Storage Conditions in a Beverage Made from Strawberry by Fermentation. J. Agric. Food Chem. 2016, 64, 1367–1376. [Google Scholar] [CrossRef]
- Roy, S.; Wu, B.; Liu, W.; Archbold, D.D. Comparative Analyses of Polyphenolic Composition of Fragaria spp. Color Mutants. Plant Physiol. Biochem. 2018, 125, 255–261. [Google Scholar] [CrossRef]
- D’Urso, G.; Pizza, C.; Piacente, S.; Montoro, P. Combination of LC–MS Based Metabolomics and Antioxidant Activity for Evaluation of Bioactive Compounds in Fragaria Vesca Leaves from Italy. J. Pharm. Biomed. Anal. 2018, 150, 233–240. [Google Scholar] [CrossRef]
- Álvarez-Fernández, M.A.; Hornedo-Ortega, R.; Cerezo, A.B.; Troncoso, A.M.; García-Parrilla, M.C. Effects of the Strawberry (Fragaria Ananassa) Purée Elaboration Process on Non-Anthocyanin Phenolic Composition and Antioxidant Activity. Food Chem. 2014, 164, 104–112. [Google Scholar] [CrossRef]
- Lantzouraki, D.Z.; Sinanoglou, V.J.; Zoumpoulakis, P.G.; Glamočlija, J.; Ćirić, A.; Soković, M.; Heropoulos, G.; Proestos, C. Antiradical–Antimicrobial Activity and Phenolic Profile of Pomegranate (Punica granatum L.) Juices from Different Cultivars: A Comparative Study. RSC Adv. 2015, 5, 2602–2614. [Google Scholar] [CrossRef]
- Simirgiotis, M.J.; Schmeda-Hirschmann, G. Determination of Phenolic Composition and Antioxidant Activity in Fruits, Rhizomes and Leaves of the White Strawberry (Fragaria chiloensis spp. Chiloensis Form Chiloensis) Using HPLC-DAD–ESI-MS and Free Radical Quenching Techniques. J. Food Compos. Anal. 2010, 23, 545–553. [Google Scholar] [CrossRef]
- Suh, D.H.; Jung, E.S.; Lee, G.M.; Lee, C.H. Distinguishing Six Edible Berries Based on Metabolic Pathway and Bioactivity Correlations by Non-Targeted Metabolite Profiling. Front. Plant Sci. 2018, 9, 1462. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Hu, L.; Li, Y.; Chen, X.; Zhang, Z.; Liu, B.; Li, P.; Gong, X.; Ma, F. MdUGT88F1-Mediated Phloridzin Biosynthesis Regulates Apple Development and Valsa Canker Resistance. Plant Physiol. 2019, 180, 2290–2305. [Google Scholar] [CrossRef]
- Li, W.; Chen, H.; Xu, B.; Wang, Y.; Zhang, C.; Cao, Y.; Xing, X. Research Progress on Classification, Sources and Functions of Dietary Polyphenols for Prevention and Treatment of Chronic Diseases. J. Future Foods 2023, 3, 289–305. [Google Scholar] [CrossRef]
- Simirgiotis, M.J.; Theoduloz, C.; Caligari, P.D.S.; Schmeda-Hirschmann, G. Comparison of Phenolic Composition and Antioxidant Properties of Two Native Chilean and One Domestic Strawberry Genotypes. Food Chem. 2009, 113, 377–385. [Google Scholar] [CrossRef]
- Martău, G.A.; Bernadette-Em ˝oke, T.; Odocheanu, R.; Soporan, D.A.; Bochis, M.; Simon, E.; Vodnar, D.C. Vaccinium Species (Ericaceae): Phytochemistry and Biological Properties of Medicinal Plants. Molecules 2023, 28, 1533. [Google Scholar] [CrossRef]
- Bouyahya, A.; Omari, N.E.; El Hachlafi, N.; Jemly, M.E.; Hakkour, M.; Balahbib, A.; El Menyiy, N.; Bakrim, S.; Naceiri Mrabti, H.; Khouchlaa, A.; et al. Chemical Compounds of Berry-Derived Polyphenols and Their Effects on Gut Microbiota, Inflammation, and Cancer. Molecules 2022, 27, 3286. [Google Scholar] [CrossRef] [PubMed]
- Mauri, A.; Consonni, V.; Todeschini, R. Molecular Descriptors. In Handbook of Computational Chemistry; Leszczynski, J., Kaczmarek-Kedziera, A., Puzyn, T.G., Papadopoulos, M., Reis, H.K., Shukla, M., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 2065–2093. ISBN 978-3-319-27281-8. [Google Scholar]
- Koyuncu, M.A.; Dilmacunal, T. Determination of Vitamin C and Organic Acid Changes in Strawberry by HPLC during Cold Storage. Not. Bot. Horti Agrobot. Cluj-Napoca 2010, 38, 95–98. [Google Scholar]
- Ikegaya, A.; Oishi, R. Differences in Analytical Methods Affect the Analytical Values of Soluble Solids, Free Sugar, and Organic Acid Content of Strawberries. LWT-Food Sci. Technol. 2024, 201, 116258. [Google Scholar] [CrossRef]
- Olennikov, D.N.; Khandy, M.T.; Chirikova, N.K. Oriental Strawberry Metabolites: LC–MS Profiling, Antioxidant Potential, and Postharvest Changes of Fragaria Orientalis Fruits. Horticulturae 2022, 8, 975. [Google Scholar] [CrossRef]
- Sun, X.-H.; Xiong, J.-J.; Zhu, A.-D.; Zhang, L.; Ma, Q.-L.; Xu, J.; Cheng, Y.-J.; Deng, X.-X. Sugars and Organic Acids Changes in Pericarp and Endocarp Tissues of Pumelo Fruit during Postharvest Storage. Sci. Hortic. 2012, 142, 112–117. [Google Scholar] [CrossRef]
- Lin, S.-D.; Sung, J.-M.; Chen, C.-L. Effect of Drying and Storage Conditions on Caffeic Acid Derivatives and Total Phenolics of Echinacea Purpurea Grown in Taiwan. Food Chem. 2011, 125, 226–231. [Google Scholar] [CrossRef]
- Yan, J.; Luo, Z.; Ban, Z.; Lu, H.; Li, D.; Yang, D.; Aghdam, M.S.; Li, L. The Effect of the Layer-by-Layer (LBL) Edible Coating on Strawberry Quality and Metabolites during Storage. Postharvest Biol. Technol. 2019, 147, 29–38. [Google Scholar] [CrossRef]
- Kim, D.-S.; Park, K.-J.; Choi, J.H.; Lim, J.-H.; Kim, H.-J. Metabolomic Analysis of Strawberries at Different Maturities According to Postharvest Storage Period. Sci. Hortic. 2023, 321, 112283. [Google Scholar] [CrossRef]
- Marčetić, M.; Samardžić, S.; Ilić, T.; Božić, D.D.; Vidović, B. Phenolic Composition, Antioxidant, Anti-Enzymatic, Antimicrobial and Prebiotic Properties of Prunus spinosa L. Fruits. Foods 2022, 11, 3289. [Google Scholar] [CrossRef]
- Pott, D.M.; De Abreu E Lima, F.; Soria, C.; Willmitzer, L.; Fernie, A.R.; Nikoloski, Z.; Osorio, S.; Vallarino, J.G. Metabolic Reconfiguration of Strawberry Physiology in Response to Postharvest Practices. Food Chem. 2020, 321, 126747. [Google Scholar] [CrossRef] [PubMed]
- Managa, M.G.; Sultanbawa, Y.; Sivakumar, D. Effects of Different Drying Methods on Untargeted Phenolic Metabolites, and Antioxidant Activity in Chinese Cabbage (Brassica rapa L. Subsp. Chinensis) and Nightshade (Solanum retroflexum Dun.). Molecules 2020, 25, 1326. [Google Scholar] [CrossRef]
- Qaderi, R.; Mezzetti, B.; Capocasa, F.; Mazzoni, L. Influence of Different Storage Conditions on Strawberry Fruit Nutritional Quality. Acta Hortic. 2023, 409–416. [Google Scholar] [CrossRef]
- Ni, T.; Zhang, S.; Rao, J.; Zhao, J.; Huang, H.; Liu, Y.; Ding, Y.; Liu, Y.; Ma, Y.; Zhang, S.; et al. Phlorizin, an Important Glucoside: Research Progress on Its Biological Activity and Mechanism. Molecules 2024, 29, 741. [Google Scholar] [CrossRef]
- Qaderi, R.; Mezzetti, B.; Capocasa, F.; Mazzoni, L. Stability of Strawberry Fruit (Fragaria x Ananassa Duch.) Nutritional Quality at Different Storage Conditions. Appl. Sci. 2022, 13, 313. [Google Scholar] [CrossRef]
- Sun, X.; Han, G.; Meng, Z.; Lin, L.; Sui, N. Roles of Malic Enzymes in Plant Development and Stress Responses. Plant Signal. Behav. 2019, 14, e1644596. [Google Scholar] [CrossRef]
- Libik-Konieczny, M.; Surówka, E.; Nosek, M.; Goraj, S.; Miszalski, Z. Pathogen-Induced Changes in Malate Content and NADP-Dependent Malic Enzyme Activity in C3 or CAM Performing Mesembryanthemum crystallinum L. Plants. Acta Physiol. Plant. 2012, 34, 1471–1477. [Google Scholar] [CrossRef]
- Shabbir, A.; Shah, A.A.; Usman, S.; Ahmend, S.; Kaleem, M.; Shafique, S.; Gatasheh, M.K. Efficacy of Malic and Tartaric Acid in Mitigation of Cadmium Stress in Spinacia oleracea L. via Modulations in Physiological and Biochemical Attributes. Sci Rep. 2025, 15, 3366. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, H.; Huang, Z.; Zhang, C.; Lyu, L.; Li, W.; Wu, W. Metabolite Profiling and Classification of Highbush Blueberry Leaves under Different Shade Treatments. Metabolites 2022, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Shen, C.-Y.; Jiang, J.-G. The Sources of Salidroside and Its Targeting for Multiple Chronic Diseases. J. Funct. Foods 2020, 64, 103648. [Google Scholar] [CrossRef]
- Hager, T.J.; Howard, L.R.; Prior, R.L. Processing and Storage Effects on the Ellagitannin Composition of Processed Blackberry Products. J. Agric. Food Chem. 2010, 58, 11749–11754. [Google Scholar] [CrossRef]
- Serra, S.; Anthony, B.; Boscolo Sesillo, F.; Masia, A.; Musacchi, S. Determination of Post-Harvest Biochemical Composition, Enzymatic Activities, and Oxidative Browning in 14 Apple Cultivars. Foods 2021, 10, 186. [Google Scholar] [CrossRef]
- Simões, A.D.N.; Moreira, S.I.; Mosquim, P.R.; Soares, N.D.F.F.; Puschmann, R. The Effects of Storage Temperature on the Quality and Phenolic Metabolism of Whole and Minimally Processed Kale Leaves. Acta Sci. Agron. 2014, 37, 101. [Google Scholar] [CrossRef]
- Dussling, S.; Will, F.; Schweiggert, R.; Steingass, C.B. Non-Enzymatic Degradation of Flavan-3-Ols by Ascorbic Acid- and Sugar-Derived Aldehydes during Storage of Apple Juices and Concentrates Produced with the Innovative Spiral Filter Press. Food Res. Int. 2024, 193, 114827. [Google Scholar] [CrossRef]
- Shubina, V.S.; Kozina, V.I.; Shatalin, Y.V. A Comparative Study of the Inhibitory Effect of Some Flavonoids and a Conjugate of Taxifolin with Glyoxylic Acid on the Oxidative Burst of Neutrophils. Int. J. Mol. Sci. 2023, 24, 15068. [Google Scholar] [CrossRef]
- Basu, A.; Nguyen, A.; Betts, N.M.; Lyons, T.J. Strawberry As a Functional Food: An Evidence-Based Review. Crit. Rev. Food Sci. Nutr. 2014, 54, 790–806. [Google Scholar] [CrossRef]
- Charlton, N.C.; Mastyugin, M.; Török, B.; Török, M. Structural Features of Small Molecule Antioxidants and Strategic Modifications to Improve Potential Bioactivity. Molecules 2023, 28, 1057. [Google Scholar] [CrossRef] [PubMed]
- Kiokias, S.; Oreopoulou, V. A Review of the Health Protective Effects of Phenolic Acids against a Range of Severe Pathologic Conditions (Including Coronavirus-Based Infections). Molecules 2021, 26, 5405. [Google Scholar] [CrossRef] [PubMed]
- Losada-Barreiro, S.; Sezgin-Bayindir, Z.; Paiva-Martins, F.; Bravo-Díaz, C. Biochemistry of Antioxidants: Mechanisms and Pharmaceutical Applications. Biomedicines 2022, 10, 3051. [Google Scholar] [CrossRef] [PubMed]
- Waring, M.J. Lipophilicity in Drug Discovery. Expert Opin. Drug Discov. 2010, 5, 235–248. [Google Scholar] [CrossRef]
- Cipriano, A.; Viviano, M.; Feoli, A.; Milite, C.; Sarno, G.; Castellano, S.; Sbardella, G. NADPH Oxidases: From Molecular Mechanisms to Current Inhibitors. J. Med. Chem. 2023, 66, 11632–11655. [Google Scholar] [CrossRef]
- Bode, K.; Hauri-Hohl, M.; Jaquet, V.; Weyd, H. Unlocking the Power of NOX2: A Comprehensive Review on Its Role in Immune Regulation. Redox Biol. 2023, 64, 102795. [Google Scholar] [CrossRef]
- Chaudhary, P.; Janmeda, P.; Docea, A.O.; Yeskaliyeva, B.; Abdull Razis, A.F.; Modu, B.; Calina, D.; Sharifi-Rad, J. Oxidative Stress, Free Radicals and Antioxidants: Potential Crosstalk in the Pathophysiology of Human Diseases. Front. Chem. 2023, 11, 1158198. [Google Scholar] [CrossRef]
- Gęgotek, A.; Skrzydlewska, E. Antioxidative and Anti-Inflammatory Activity of Ascorbic Acid. Antioxidants 2022, 11, 1993. [Google Scholar] [CrossRef]
- Obrenovich, M.; Li, Y.; Tayahi, M.; Reddy, V.P. Polyphenols and Small Phenolic Acids as Cellular Metabolic Regulators. Curr. Issues Mol. Biol. 2022, 44, 4152–4166. [Google Scholar] [CrossRef]
- Tebay, L.E.; Robertson, H.; Durant, S.T.; Vitale, S.R.; Penning, T.M.; Dinkova-Kostova, A.T.; Hayes, J.D. Mechanisms of Activation of the Transcription Factor Nrf2 by Redox Stressors, Nutrient Cues, and Energy Status and the Pathways through Which It Attenuates Degenerative Disease. Free Radic. Biol. Med. 2015, 88, 108–146. [Google Scholar] [CrossRef]
- Muthukumaran, S.; Tranchant, C.; Shi, J.; Ye, X.; Xue, S.J. Ellagic Acid in Strawberry (Fragaria spp.): Biological, Technological, Stability, and Human Health Aspects. Food Qual. Saf. 2017, 1, 227–252. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, J.; Pan, G.; Huang, D.; Zhu, S. Ellagic Acid Enhances Antioxidant System Activity and Maintains the Quality of Strawberry Fruit during Storage. Phyton 2024, 93, 15–28. [Google Scholar] [CrossRef]
| No | Tentative Identification | Group | MW | MS (m/z), ID | MS/MS | Retention Time (min) | Reference | Strawberry Fruits /Leaves ** |
|---|---|---|---|---|---|---|---|---|
| 1 | Malic acid | Organic acids | 134.0578 | 133.12 | 115.31; 89.30; 71.22 | 0.654 | [24], https://hmdb.ca/spectra/ms_ms/2254556, accessed on 8 February 2025 | +/+ |
| 2 | Dehydroascorbic acid | Vitamin and vitamin derivatives | 174.0165 | 173.01 | 155.22; 111.31; 87.20; 85.23 | 0.836 | [24], https://www.hmdb.ca/spectra/ms_ms/283274, accessed on 8 February 2025 | +/− |
| 3 | L-ascorbic acid | Vitamin and vitamin derivatives | 176.0685 | 175.05 | 131.01; 115.20; 113.10; 87.42 | 1.584 | [24], https://hmdb.ca/spectra/ms_ms/1473014, accessed on 8 February 2025 | +/− |
| 4 | Caffeic acid | Phenolic acids | 180.0423 | 179.04 | 135.11; 107.21; 91.10 | 11.856 | [25], https://hmdb.ca/spectra/ms_ms/2234953, accessed on 8 February 2025 | +/+ |
| 5 | Gallic acid monohydrate | Phenolic acids | 188.0321 | 187.16 | 169.24; 125.40; 97.45 | 4.781 | [25] | +/− |
| 6 | Citric acid | Organic acids | 192.0271 | 191.12 | 173.42; 129.40; 111.22 | 0.737 | [24,26] | +/+ |
| 7 | Tryptophan | Amino acids | 204.0899 | 203.16 | 186.61; 142.32; 116.33 | 1.335 | [24] | +/− |
| 8 | (-) Epicatechin | Proanthocyanidins | 290.2681 | 289.20 | 245.80; 205.50; 203.21; 179.60; 109.12 | 1.671 | [27,28] | +/+ |
| 9 | Salidroside | Phenolic glycosides | 300.0852 | 299.27 | 179.54; 137.00; 89.43 | 1.062 | [12] | −/+ |
| 10 | Ellagic acid aglycone | Phenolic acids | 302.0067 | 301.31 | 300.51; 284.52, 257.51; 229.53; 200.34 | 4.101 | [24] | +/+ |
| 11 | Coumaric acid hexose | Phenolic acids | 326.1011 | 325.24 | 163.42; 145.31 | 1.855 | [24] | +/− |
| 12 | Galloyl hexose | Phenolic acids | 332.0752 | 331.10 | 169.31; 123.50 | 0.837 | [12] | −/+ |
| 13 | Coumaroylquinic acid | Phenolic acids | 338.1013 | 337.27 | 191.52; 173.50 | 4.100 | [12] | −/+ |
| 14 | Caffeic acid hexoside | Phenolic acids | 342.0951 | 341.32 | 179.31; 161.30; 135.12 | 0.611 | [29] | +/+ |
| 15 | Galloylquinic acid | Phenolic acids | 344.0753 | 343.23 | 191.00; 169.61; 93.42 | 0.800 | [12] | −/+ |
| 16 | p-Coumaroyl-ester | Phenolic acids | 356 * | 355.24 | 295.91; 193.41; 175.10; 134.51 | 3.486 | [23] | −/+ |
| 17 | Dihydroferulic acid 4-O-glucuronide | Hydroxycinnamic acids | 372.1056 | 371.31 | 209.82; 193.51 | 3.564 | [26] | −/+ |
| 18 | Apigenin-7-O-glucoside | Flavonols | 432.3775 | 431.28 | 270.82; 269.54; 225.22 | 2.927 | [26,29] | +/− |
| 19 | Phloridzin | Flavonoid glycosides | 436.1369 | 435.24 | 273.51; 167.33; 125.23 | 5.061 | [26] | +/+ |
| 20 | Ellagic acid deoxyhexoside | Hydroxycinnamic acid derivatives | 448.0641 | 447.29 | 302.20; 301.50; 300.51; 257.52; 229.51 | 3.892 | [27,30] | +/+ |
| 21 | Ferulic acid hexose derivative | Hydroxycinnamic acids | 450 * | 449.29 | 355.61; 287.52; 269.52; 193.83 | 3.147 | [22,29] | +/− |
| 22 | Kaempferol glucuronide | Flavonols | 462.3604 | 461.25 | 285.50; 257.51; 229.60; 175.60; 163.51; 113.21 | 4.609 | [24,31] | +/− |
| 23 | Quercetin hexoside | Flavonoid glycosides | 464.3763 | 463.53 | 300.50; 271.61; 255.72; 179.52 | 4.232 | [27,29,31] | +/− |
| 24 | Sesquiterpenoid | Terpenoids and related compounds | 464.2628 | 463.55 | 417.81; 255.51; 161.31 | 5.308 | [24] | +/− |
| 25 | Kaempferol hexose | Flavonols | 466.1118 | 465.28 | 447.30; 285.62; 241.63; 151.40 | 3.333 | [12,24] | −/+ |
| 26 | Di-coumaroyl hexose | Hydroxycinnamic acid derivatives | 472.1383 | 471.24 | 163.52; 145.31 | 5.236 | [12] | −/+ |
| 27 | Quercetin-3-glucuronide | Flavonoid glycosides | 478.3598 | 477.27 | 301.63; 255.71; 179.51; 151.34; 121.30 | 4.253 | [12,29] | +/+ |
| 29 | Sapogenin | Terpenoids and related compounds | 488.3515 | 487.54 | 469.81; 407.80; 135.51 | 6.451 | [24] | +/− |
| 30 | Kaempferol acetyl glucoside | Flavonols | 490.4136 | 489.28 | 447.93; 285.62; 255.42 | 4.957 | [27] | +/− |
| 31 | Octadecatrienoic acid glycoside | Fatty acid derivatives | 560.3221 | 559.47 | 513.90; 277.71; 253.71; 161.50 | 7.670 | [12] | −/+ |
| 32 | Dicaffeoylquinic acid | Phenolic acids | 562.2996 | 561.61 | 515.30; 191.82; 161.42 | 6.961 | [24] | +/− |
| 33 | Flavan-3-ol derivative | Proanthocyanidins | 578.1647 | 577.27 | 425.94; 407.82; 289.60; 269.51; 147.60 | 3.374 | [24,27] | +/+ |
| 34 | Kaempferol coumaroyl hexoside | Flavonols | 594.5196 | 593.34 | 447.52; 285.84; 255.41 | 6.126 | [27] | +/+ |
| 35 | Kaempferol-rutinoside | Flavonols | 594.1585 | 593.12 | 547.53; 327.51; 308.80; 285.60 | 6.111 | [32] | +/− |
| 36 | Kaempferol pentose glucuronide | Flavonols | 594.1244 | 593.24 | 307.63; 285.62; 113.31 | 6.008 | [12] | −/+ |
| 37 | Q-rutinoside | Flavonoid glycosides | 610.1533 | 609.26 | 301.50; 179.41; 151.42 | 3.856 | [23] | −/+ |
| Metabolites | p-Value | Significant Differences (Tukey’s HSD) * |
|---|---|---|
| Caffeic Acid | 2.59 × 10−7 | Day 11 vs. Day 1, Day 6 vs. Day 4, Day 11 vs. Day 4, Day 11 vs. Day 6, Day 11 vs. Day 8 |
| Malic Acid | 6.12 × 10−5 | Day 8 vs. Day 1, Day 11 vs. Day 1, Day 8 vs. Day 4, Day 11 vs. Day 4, Day 8 vs. Day 6 |
| Citric Acid | 7.50 × 10−5 | Day 6 vs. Day 1, Day 8 vs. Day 1, Day 6 vs. Day 4, Day 8 vs. Day 4, Day 11 vs. Day 8 |
| Ferulic Acid Hexose Derivative | 0.000481 | Day 6 vs. Day 4, Day 8 vs. Day 4, Day 11 vs. Day 4 |
| Coumaric Acid Hexose | 0.003591 | Day 11 vs. Day 1, Day 11 vs. Day 4 |
| Dicaffeoylquinic Acid | 0.008077 | Day 6 vs. Day 1, Day 8 vs. Day 6 |
| Kaempferol Glucuronide | 0.008119 | Day 4 vs. Day 1, Day 11 vs. Day 4 |
| Metabolites | p-Value | Significant Differences (Tukey’s HSD) * |
|---|---|---|
| Galloyl Hexose | 5.8522 × 10−8 | Day 4 vs. Day 1, Day 6 vs. Day 4, Day 8 vs. Day 4 |
| Ellagic Acid Aglycone | 8.7371 × 10−8 | Day 4 vs. Day 1, Day 6 vs. Day 1, Day 8 vs. Day 1, Day 6 vs. Day 4, Day 8 vs. Day 4 |
| Caffeic Acid Hexoside | 1.0295 × 10−5 | Day 6 vs. Day 1, Day 6 vs. Day 4, Day 8 vs. Day 6 |
| Salidroside | 2.6579 × 10−5 | Day 4 vs. Day1, Day 6 vs. Day 1, Day 8 vs. Day 4, Day 8 vs. Day 6 |
| Phloridzin | 2.8896 × 10−5 | Day 4 vs. Day 1, Day 6 vs. Day 4, Day 8 vs. Day 4 |
| Malic Acid | 0.00079905 | Day 4 vs. Day 1, Day 6 vs. Day 4 |
| Galloylquinic Acid | 0.0035448 | Day 4 vs. Day 1, Day 6 vs. Day 1 |
| Citric Acid | 0.0076111 | Day 4 vs. Day 1, Day 8 vs. Day 4 |
| Flavan-3-ol Derivative | 0.018281 | Day 8 vs. Day 4 |
| Features’ Ranking by Importance | Feature Names (Descriptors) | Frequency of Appearance |
|---|---|---|
| 1 | BCUT2D_MWHI | 381 |
| 2 | PEOE_VSA6 | 375 |
| 3 | Chi2v | 357 |
| 4 | BCUT2D_MRHI | 298 |
| 5 | qed | 282 |
| 6 | SMR_VSA10 | 278 |
| 7 | VSA_EState2 | 272 |
| 8 | SlogP_VSA6 | 257 |
| 9 | FractionCSP3 | 257 |
| 10 | VSA_EState6 | 251 |
| 11 | BCUT2D_MWLOW | 248 |
| 12 | EState_VSA7 | 233 |
| 13 | MinAbsEStateIndex | 231 |
| 14 | Chi2n | 228 |
| 15 | VSA_EState8 | 223 |
| 16 | PEOE_VSA9 | 213 |
| Compound | Chemical Formula | SMILES | ML-System Prediction |
|---|---|---|---|
| Epicatechin | C15H14O6 | O[C@H]1CC2=C(O)C=C(O)C=C2O[C@H]1C1=CC(O)=C(O)C=C1 | Active |
| Apigenin-7-O-glucoside | C21H20O10 | OCC1OC(OC2=CC(O)=C3C(=O)C=C(OC3=C2)C2=CC=C(O)C=C2)C(O)C(O)C1O | Active |
| Sesquiterpenoid | C15H18O3 | C[C@@H]1[C@@H]2CC[C@]3(C)C=CC(=O)C(C)=C3[C@@H]2OC1=O | Not Active |
| Sapogenin | C30H50O3 | C[C@H]1CC[C@@]2([C@H]([C@H]3[C@@H](O2)C[C@@H]4[C@@]3(CC[C@H]5[C@H]4CCC6[C@@]5(CCCC6)C)C)C)OC1 | Active |
| Salidroside | C14H20O7 | OCC1OC(OCCC2=CC=C(O)C=C2)C(O)C(O)C1O | Active |
| Quercetin-3-glucuronide | C21H18O13 | OC1C(O)C(OC2=C(OC3=CC(O)=CC(O)=C3C2=O)C2=CC=C(O)C(O)=C2)OC(C1O)C(O)=O | Active |
| Procyanidin dimer | C30H26O12 | [H][C@]1([C@@H](O)[C@H](OC2=CC(O)=CC(O)=C12)C1=CC(O)=C(O)C=C1)C1=C2O[C@@H]([C@H](O)[C@@]([H])(C3=C(O)C=C(O)C4=C3O[C@@H]([C@H](O)C4)C3=CC(O)=C(O)C=C3)C2=C(O)C=C1O)C1=CC(O)=C(O)C=C1 | Active |
| p-Coumaroyl-ester | C16H14O5 | O[C@@H]1C[C@](O)(C[C@@H](OC(=O)\C=C/C2=CC=C(O)C(O)=C2)[C@@H]1O)C(O)=O | Active |
| Malic acid | C4H6O5 | OC(CC(O)=O)C(O)=O | Not Active |
| L-ascorbic acid | C6H8O6 | OCC(O)C1OC(=O)C(O)=C1O | Active |
| Kaempferol | C15H12O6 | OC1=CC=C(C=C1)C1=C(O)C(=O)C2=C(O1)C=C(O)C=C2O | Active |
| Kaempferol acetylhexose | C23H22O12 | OCC1OC(OC2C(O)C(O)C(CO)OC2OC2=C(OC3=CC(OC4OC(C(O)C(O)C4O)C(O)=O)=CC(O)=C3C2=O)C2=CC=C(O)C=C2)C(O)C(O)C1O | Active |
| Isorhamnetin hexose | C22H20O12 | COC1=C(OC2OC(CO)C(O)C(O)C2O)C=CC(=C1)C1=C(O)C(=O)C2=C(O)C=C(O)C=C2O1 | Active |
| Galloylquinic acid | C16H12O10 | OS(=O)(=O)OC(=O)C1=CC=CC=C1 | Active |
| Gallic acid monohydrate | C7H6O5 | C(O)(=O)C1=CC(O)=C(O)C(O)=C1.[H]O[H] | Active |
| Ellagic acid deoxyhexose | C20H16O12 | OC1=CC=C(\C=C\C2=CC(O)=CC(O)=C2)C=C1 | Active |
| Ellagic acid | C14H6O8 | OC1=C(O)C2=C3C(=C1)C(=O)OC1=C3C(=CC(O)=C1O)C(=O)O2 | Active |
| Dicaffeoylquinic acid | C25H24O12 | O[C@H]1[C@H](OC(=O)\C=C\C2=CC=C(O)C(O)=C2)C[C@@](O)(C[C@H]1OC(=O)\C=C\C1=CC=C(O)C(O)=C1)C(=O)O | Active |
| Dehydroascorbic acid | C6H6O6 | [H][C@@]1(OC(=O)C(=O)C1=O)[C@@H](O)CO | Not Active |
| Coumaroylquinic acid | C16H16O9 | O[C@@H]1C[C@@](O)(C[C@@H](OC(=O)\C=C\C2=CC=C(O)C=C2)[C@H]1O)C(O)=O | Active |
| Citric acid | C6H8O7 | OC(=O)CC(O)(CC(O)=O)C(O)=O | Not Active |
| Catechin | C15H14O6 | O[C@H]1CC2=C(O)C=C(O)C=C2O[C@@H]1C1=CC(O)=C(O)C=C1 | Active |
| Caffeic acid | C9H8O4 | OC(=O)\C=C\C1=CC(O)=C(O)C=C1 | Active |
| Tryptophan | C11H12N2O2 | NC(CC1=CNC2=C1C=CC=C2)C(O)=O | Active |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ladika, G.; Christodoulou, P.; Kritsi, E.; Tsiaka, T.; Sotiroudis, G.; Cavouras, D.; Sinanoglou, V.J. Exploring Postharvest Metabolic Shifts and NOX2 Inhibitory Potential in Strawberry Fruits and Leaves via Untargeted LC-MS/MS and Chemometric Analysis. Metabolites 2025, 15, 321. https://doi.org/10.3390/metabo15050321
Ladika G, Christodoulou P, Kritsi E, Tsiaka T, Sotiroudis G, Cavouras D, Sinanoglou VJ. Exploring Postharvest Metabolic Shifts and NOX2 Inhibitory Potential in Strawberry Fruits and Leaves via Untargeted LC-MS/MS and Chemometric Analysis. Metabolites. 2025; 15(5):321. https://doi.org/10.3390/metabo15050321
Chicago/Turabian StyleLadika, Georgia, Paris Christodoulou, Eftichia Kritsi, Thalia Tsiaka, Georgios Sotiroudis, Dionisis Cavouras, and Vassilia J. Sinanoglou. 2025. "Exploring Postharvest Metabolic Shifts and NOX2 Inhibitory Potential in Strawberry Fruits and Leaves via Untargeted LC-MS/MS and Chemometric Analysis" Metabolites 15, no. 5: 321. https://doi.org/10.3390/metabo15050321
APA StyleLadika, G., Christodoulou, P., Kritsi, E., Tsiaka, T., Sotiroudis, G., Cavouras, D., & Sinanoglou, V. J. (2025). Exploring Postharvest Metabolic Shifts and NOX2 Inhibitory Potential in Strawberry Fruits and Leaves via Untargeted LC-MS/MS and Chemometric Analysis. Metabolites, 15(5), 321. https://doi.org/10.3390/metabo15050321









