Advancing Clinical and Pathophysiological Insights into Pancreatitis Using Lipidomics and Metabolomics
Abstract
1. Introduction
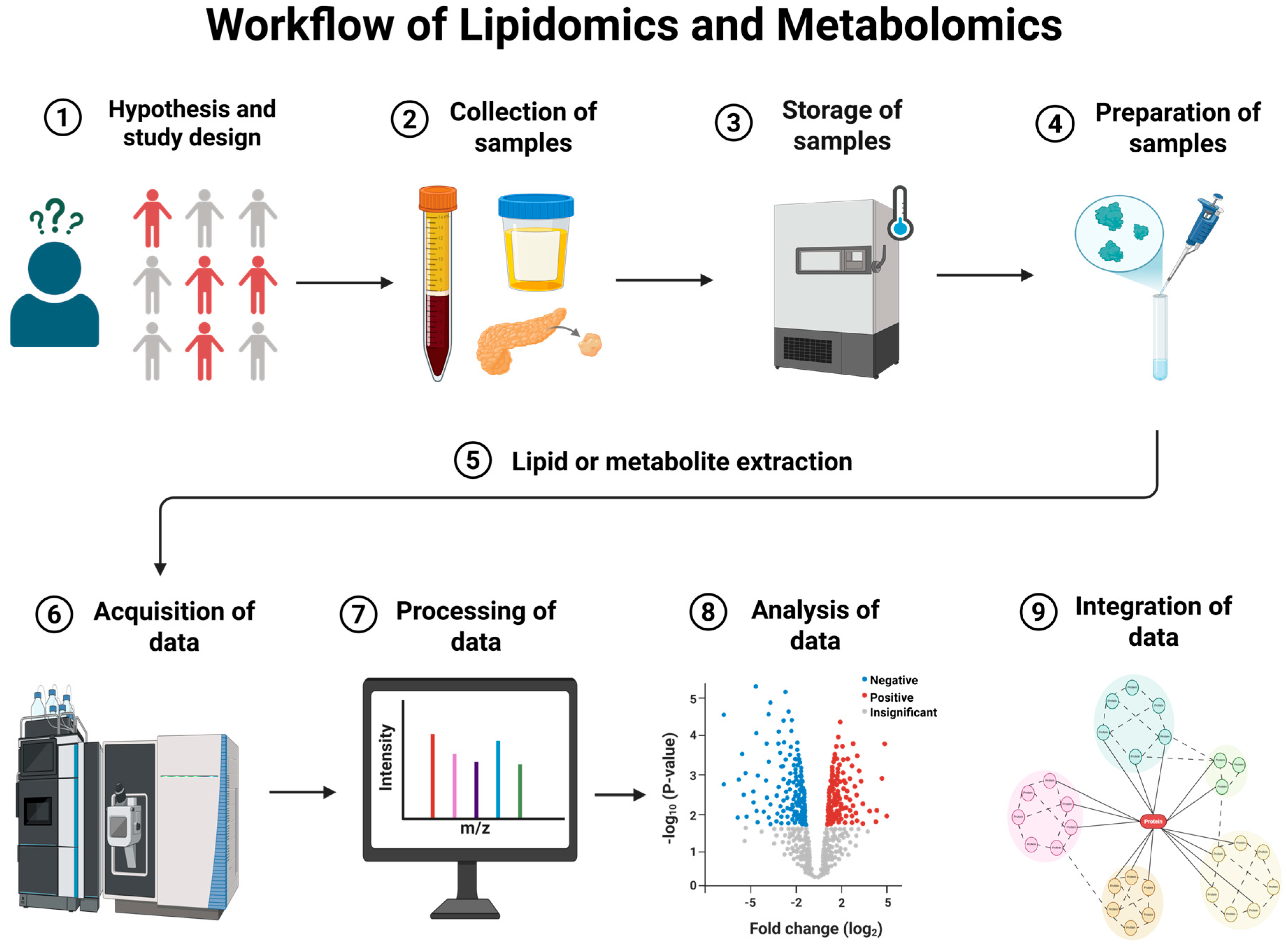
2. Workflow of Lipidomics and Metabolomics
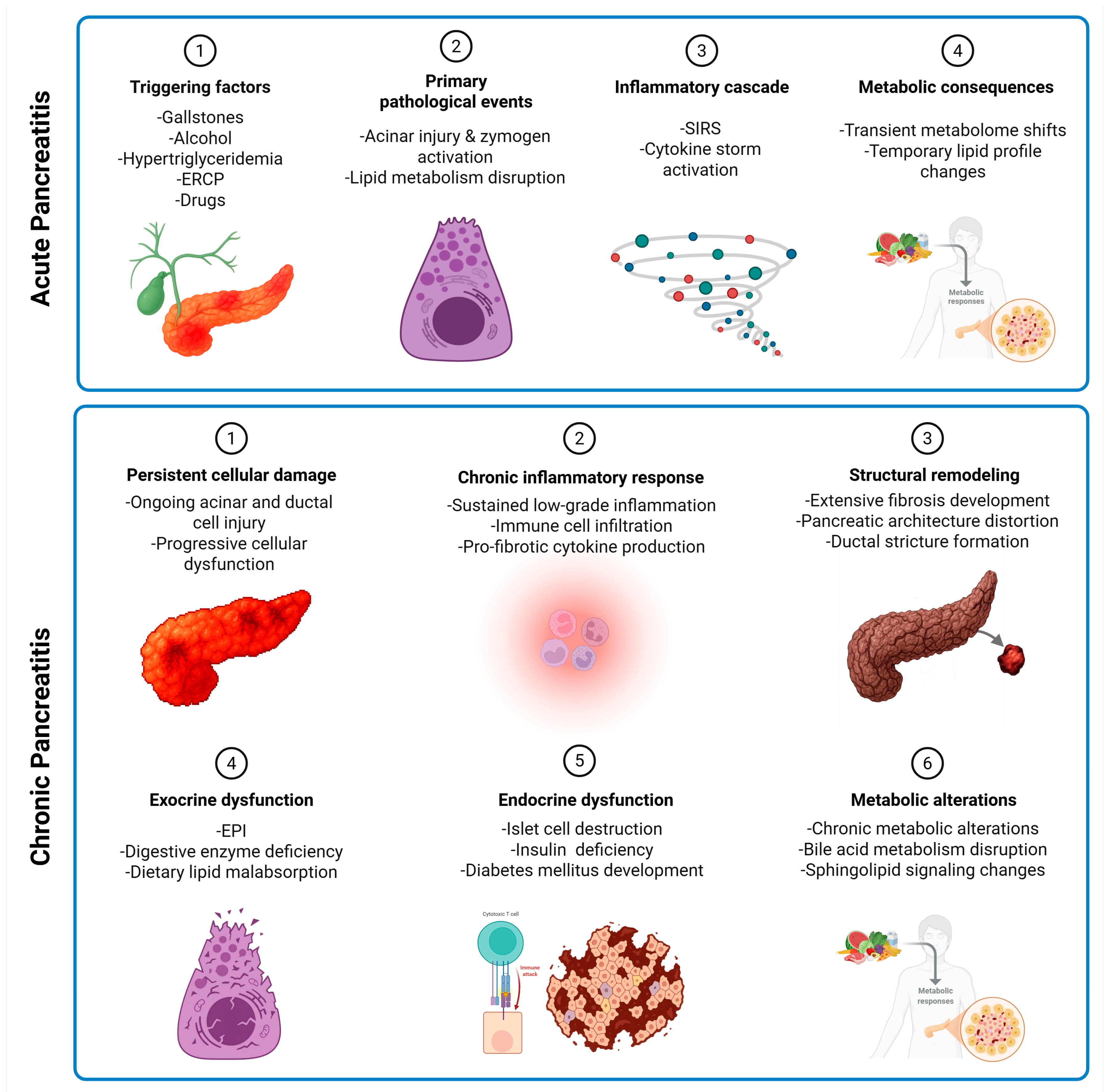
3. Practical Considerations for Sampling and Timing in AP and CP
4. Methods
5. Lipidomics and Metabolomics Studies on Acute Pancreatitis Patients
5.1. Differences in Normal and AP Metabolic States
5.2. Studies Based on Etiology of Acute Pancreatitis
5.3. Studies Based on Severity of Acute Pancreatitis
6. Lipidomics and Metabolomics Studies on Chronic Pancreatitis Patients
7. Altered Metabolic Pathways in Acute and Chronic Pancreatitis
7.1. Alterations in Amino Acid and Energy Metabolism
7.2. Altered Lipid Metabolism
7.3. Etiology-Specific Pathway Alterations
7.4. Alterations in Inflammatory, Oxidative Stress and Disease Progression Pathways
8. Emerging Patterns and Clinical Implications
9. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lee, P.J.; Papachristou, G.I. New insights into acute pancreatitis. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 479–496. [Google Scholar] [CrossRef]
- Peery, A.F.; Crockett, S.D.; Barritt, A.S.; Dellon, E.S.; Eluri, S.; Gangarosa, L.M.; Jensen, E.T.; Lund, J.L.; Pasricha, S.; Runge, T.; et al. Burden of Gastrointestinal, Liver, and Pancreatic Diseases in the United States. Gastroenterology 2015, 149, 1731–1741.e3. [Google Scholar] [CrossRef]
- Yadav, D.; Timmons, L.; Benson, J.T.; Dierkhising, R.A. Incidence, prevalence, and survival of chronic pancreatitis: A population-based study. Am. J. Gastroenterol. 2011, 106, 2192–2199. [Google Scholar] [CrossRef] [PubMed]
- Weiss, F.U.; Laemmerhirt, F.; Lerch, M.M. Etiology and Risk Factors of Acute and Chronic Pancreatitis. Visc. Med. 2019, 35, 73–81. [Google Scholar] [CrossRef]
- Gukovskaya, A.S.; Pandol, S.J.; Gukovsky, I. New insights into the pathways initiating and driving pancreatitis. Curr. Opin. Gastroenterol. 2016, 32, 429–453. [Google Scholar] [CrossRef] [PubMed]
- Testoni, P.A. Acute recurrent pancreatitis: Etiopathogenesis, diagnosis and treatment. World J. Gastroenterol. 2014, 20, 16891–16901. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Snyder, M.P.; Karczewski, K.J.; Snyder, M.P. Integrative omics for health and disease. Nat. Rev. Genet. 2018, 19, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Fahy, E.; Subramaniam, S.; Brown, H.A.; Glass, C.K.; Merrill, A.H.; Murphy, R.C.; Raetz, C.R.; Russell, D.W.; Seyama, Y.; Shaw, W.; et al. A comprehensive classification system for lipids. J. Lipid Res. 2005, 46, 839–861. [Google Scholar] [CrossRef]
- Mayerle, J.; Hoffmeister, A.; Werner, J.; Witt, H.; Lerch, M.M.; Mössner, J. Chronic pancreatitis--definition, etiology, investigation and treatment. Dtsch. Arztebl. Int. 2013, 110, 387. [Google Scholar]
- Peng, Y.; Hong, J.; Raftery, D.; Qing, X.; Dan, D. Metabolomic-based clinical studies and murine models for acute pancreatitis disease: A review. Biochim. Et Biophys. Acta (BBA)—Mol. Basis Dis. 2021, 1867, 166123. [Google Scholar] [CrossRef]
- Vinayavekhin, N.; Saghatelian, A. Untargeted Metabolomics. Curr. Protoc. Mol. Biol. 2010, 90, 30.1.1–30.1.24. [Google Scholar] [CrossRef]
- Roberts, L.D.; Souza, A.L.; Gerszten, R.E.; Clish, C.B. Targeted Metabolomics. Curr. Protoc. Mol. Biol. 2012, 98, 30.2.1–30.2.24. [Google Scholar] [CrossRef]
- Segatto, M.; Pallottini, V. Facts about Fats: New Insights into the Role of Lipids in Metabolism, Disease and Therapy. Int. J. Mol. Sci. 2020, 21, 6651. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.R.; Jeon, B.J.; Park, K.-M.; Lee, E.H.; Hong, S.-C.; Choi, S.J. An Overview of Structured Lipid in Food Science: Synthesis Methods, Applications, and Future Prospects. J. Chem. 2023, 2023, 1222373. [Google Scholar] [CrossRef]
- Cerone, M.; Smith, T.K. A Brief Journey into the History of and Future Sources and Uses of Fatty Acids. Front. Nutr. 2021, 8, 570401. [Google Scholar] [CrossRef]
- Summons, R.E.; Welander, P.V.; Gold, D.A.; Summons, R.E.; Welander, P.V.; Gold, D.A. Lipid biomarkers: Molecular tools for illuminating the history of microbial life. Nat. Rev. Microbiol. 2021, 20, 174–185. [Google Scholar] [CrossRef]
- Wei, J.; Wong, L.C.; Boland, S. Lipids as Emerging Biomarkers in Neurodegenerative Diseases. Int. J. Mol. Sci. 2023, 25, 131. [Google Scholar] [CrossRef]
- Lydic, T.A.; Goo, Y.-H. Lipidomics unveils the complexity of the lipidome in metabolic diseases. Clin. Transl. Med. 2018, 7, 4. [Google Scholar] [CrossRef] [PubMed]
- Hyötyläinen, T.; Orešič, M.; Hyötyläinen, T.; Orešič, M. Optimizing the lipidomics workflow for clinical studies—Practical considerations. Anal. Bioanal. Chem. 2015, 407, 4973–4993. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, J.; Zhang, J.; Rong, J.; Wang, X.; Zhao, C.; Zhang, H.; Shi, H.; Wu, W. UHPLC-MS/MS-based untargeted lipidomics analysis of septic patients. Clin. Chim. Acta 2023, 544, 117136. [Google Scholar] [CrossRef]
- Zhao, X.; Niu, L.; Clerici, C.; Russo, R.; Byrd, M.; Setchell, K.D.R. Data analysis of MS-based clinical lipidomics studies with crossover design: A tutorial mini-review of statistical methods. Clin. Mass Spectrom. 2019, 13, 5–17. [Google Scholar] [CrossRef]
- Köfeler, H.C.; Ahrends, R.; Baker, E.S.; Ekroos, K.; Han, X.; Hoffmann, N.; Holčapek, M.; Wenk, M.R.; Liebisch, G. Recommendations for good practice in MS-based lipidomics. J. Lipid Res. 2021, 62, 100138. [Google Scholar] [CrossRef]
- Yan, M.; Xu, G. Current and future perspectives of functional metabolomics in disease studies–A review. Anal. Chim. Acta 2018, 1037, 41–54. [Google Scholar] [CrossRef]
- Theodoridis, G.; Gika, H.G.; Wilson, I.D. Mass spectrometry-based holistic analytical approaches for metabolite profiling in systems biology studies. Mass Spectrom. Rev. 2011, 30, 884–906. [Google Scholar] [CrossRef]
- Pan, Z.; Raftery, D.; Pan, Z.; Raftery, D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal. Bioanal. Chem. 2006, 387, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, E.-M.; Xu, L.-Y.; Chen, Y.; Li, E.-M.; Xu, L.-Y. Guide to Metabolomics Analysis: A Bioinformatics Workflow. Metabolites 2022, 12, 357. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef]
- Ouyang, D. Metabolomic characterization of human pancreatitis by ¹H-NMR spectroscopy. Hepato-Gastroenterology 2012, 59, 2314–2317. [Google Scholar] [CrossRef]
- Xiao, H.; Huang, J.-h.; Zhang, X.-W.; Ahmed, R.; Xie, Q.-L.; Zhu, Y.-M.; Cai, X.; Peng, Q.-H.; Qin, Y.H. Identification of potential diagnostic biomarkers of acute pancreatitis by serum metabolomic profiles. Pancreatology 2017, 17, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jia, W.; Su, M.; Qiu, Y.; Wang, X. Novel biomarkers of hyperlipidemic acute pancreatitis: Metabolomic identification. Asian Biomed. 2017, 6, 765–769. [Google Scholar] [CrossRef]
- Huang, J.-H.; He, D.; Chen, L.; Dong, C.-Y.; Zhang, S.-H.; Qin, Y.-H.; Yu, R.; Ahmed, R.; Kuang, J.-J.; Zhang, X.-W. GC-MS based metabolomics strategy to distinguish three types of acute pancreatitis. Pancreatology 2019, 19, 630–637. [Google Scholar] [CrossRef]
- Dancu, G.; Tarta, C.; Socaciu, C.; Bende, F.; Danila, M.; Sirli, R.; Sporea, I.; Miutescu, B.; Popescu, A. Unraveling the Metabolic Changes in Acute Pancreatitis: A Metabolomics-Based Approach for Etiological Differentiation and Acute Biomarker Discovery. Biomolecules 2023, 13, 1558. [Google Scholar] [CrossRef]
- Liu, J.; Yan, Q.; Li, S.; Jiao, J.; Hao, Y.; Zhang, G.; Zhang, Q.; Luo, F.; Zhang, Y.; Lv, Q.; et al. Integrative metagenomic and metabolomic analyses reveal the potential of gut microbiota to exacerbate acute pancreatitis. NPJ Biofilms Microbiomes 2024, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lusczek, E.; Paulo, J.; Saltzman, J.; Kadiyala, V.; Banks, P.; Beilman, G.; Conwell, D. Urinary 1H-NMR metabolomics can distinguish pancreatitis patients from healthy controls. J. Pancreas 2013, 14, 161. [Google Scholar]
- Villaseñor, A.; Kinross, J.M.; Li, J.V.; Penney, N.; Barton, R.H.; Nicholson, J.K.; Darzi, A.; Barbas, C.; Holmes, E. 1H NMR Global Metabolic Phenotyping of Acute Pancreatitis in the Emergency Unit. J. Proteome Res. 2014, 13, 5362–5375. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Sun, J.; Chen, Y.Q. Multi-dimensional, comprehensive sample extraction combined with LC-GC/MS analysis for complex biological samples: Application in the metabolomics study of acute pancreatitis. RSC Adv. 2016, 6, 25837–25849. [Google Scholar] [CrossRef]
- Khan, J.; Solakivi, T.; Seppänen, H.; Lappalainen-Lehto, R.; Järvinen, S.; Ronkainen, J.; Sand, J.; Nordback, I. Serum lipid and fatty acid profiles are highly changed in patients with alcohol induced acute pancreatitis. Pancreatology 2012, 12, 44–48. [Google Scholar] [CrossRef]
- Lusczek, E.R.; Colling, K.; Muratore, S.; Conwell, D.; Freeman, M.; Beilman, G. Stereotypical Metabolic Response to Endoscopic Retrograde Cholangiopancreatography Show Alterations in Pancreatic Function Regardless of Post-Procedure Pancreatitis. Clin. Transl. Gastroenterol. 2016, 7, e169. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, L.; Kang, H.; Zhang, J.; Liu, J.; Liu, S. Serum Metabonomics of Mild Acute Pancreatitis. J. Clin. Lab. Anal. 2016, 30, 990–998. [Google Scholar] [CrossRef]
- Skouras, C.; Zheng, X.; Binnie, M.; Homer, N.Z.M.; Murray, T.B.J.; Robertson, D.; Briody, L.; Paterson, F.; Spence, H.; Derr, L.; et al. Increased levels of 3-hydroxykynurenine parallel disease severity in human acute pancreatitis. Sci. Rep. 2016, 6, 33951. [Google Scholar] [CrossRef]
- Lou, D.; Shi, K.; Li, H.-P.; Zhu, Q.; Hu, L.; Luo, J.; Yang, R.; Liu, F. Quantitative metabolic analysis of plasma extracellular vesicles for the diagnosis of severe acute pancreatitis. J. Nanobiotechnol. 2022, 20, 1–10. [Google Scholar] [CrossRef]
- Zhang, L.; Jin, H.; Guo, X.; Yang, Z.; Zhao, L.; Tang, S.; Mo, P.; Wu, K.; Nie, Y.; Pan, Y.; et al. Distinguishing pancreatic cancer from chronic pancreatitis and healthy individuals by (1)H nuclear magnetic resonance-based metabonomic profiles. Clin. Biochem. 2012, 45, 1064–1069. [Google Scholar] [CrossRef] [PubMed]
- Adam, M.G.; Beyer, G.; Christiansen, N.; Kamlage, B.; Pilarsky, C.; Distler, M.; Fahlbusch, T.; Chromik, A.; Klein, F.; Bahra, M.; et al. Identification and validation of a multivariable prediction model based on blood plasma and serum metabolomics for the distinction of chronic pancreatitis subjects from non-pancreas disease control subjects. Gut 2021, 70, 2150–2158. [Google Scholar] [CrossRef] [PubMed]
- Díaz, C.; Jiménez-Luna, C.; Diéguez-Castillo, C.; Martín, A.; Prados, J.; Martín-Ruíz, J.L.; Genilloud, O.; Vicente, F.; Palacio, J.P.d.; Caba, O. Untargeted Metabolomics for the Diagnosis of Exocrine Pancreatic Insufficiency in Chronic Pancreatitis. Medicina 2021, 57, 876. [Google Scholar] [CrossRef]
- Sarkar, S.; Sarkar, P.; Revanth, M.; Hazarika, D.; Prasanna, A.; Pandol, S.J.; Unnisa, M.; Jakkampudi, A.; Bedarkar, A.P.; Dhagudu, N.; et al. Pain, depression, and poor quality of life in chronic pancreatitis: Relationship with altered brain metabolites. Pancreatology 2022, 22, 688–697. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Huang, X.; Ouyang, Q.; Liu, W.; Liu, S.; Huang, Y.; Peng, Y.; Ning, D.; Tan, C. Serum metabolomics study for acute attack of chronic pancreatitis. Clin. Chim. Acta 2023, 541, 117251. [Google Scholar] [CrossRef]
- Lin, Q.; Ye, Z.; Lin, H. Identification of Differential Metabolites Between Type 2 Diabetes and Postchronic Pancreatitis Diabetes (Type 3c) Based on an Untargeted Metabolomics Approach. Lab. Med. 2023, 54, 562–573. [Google Scholar] [CrossRef]
- Ketavarapu, V.; Addipilli, R.; Ragi, N.; Pallera, P.; Simhadri, V.; Manne, S.; Sannapaneni, K.; Aslam, M.; Talukadar, R.; Ch, V.D.; et al. Plasma Metabolite Profiling Identifies Nondiabetic Chronic Pancreatitis Patients with Metabolic Alterations Progressing to Prediabetes Before HbA1c. Clin. Transl. Gastroenterol. 2024, 15, e1. [Google Scholar] [CrossRef]
- Sandstrom, P.; Trulsson, L.; Gasslander, T.; Sundqvist, T.; von, D.U.; Svanvik, J. Serum amino acid profile in patients with acute pancreatitis. Amino Acids 2008, 35, 225–231. [Google Scholar] [CrossRef]
- Zuo, Y.-Y.; Kang, Y.; Yin, W.-H.; Wang, B.; Chen, Y. The association of mean glucose level and glucose variability with intensive care unit mortality in patients with severe acute pancreatitis. J. Crit. Care 2012, 27, 146–152. [Google Scholar] [CrossRef]
- Green, C.D.; Maceyka, M.; Cowart, L.A.; Spiegel, S. Sphingolipids in metabolic disease: The good, the bad, and the unknown. Cell Metab. 2021, 33, 1293–1306. [Google Scholar] [CrossRef]
- Rao, R.P.; Vaidyanathan, N.; Rengasamy, M.; Oommen, A.M.; Somaiya, N.; Jagannath, M.R. Sphingolipid Metabolic Pathway: An Overview of Major Roles Played in Human Diseases. J. Lipids 2013, 2013, 178910. [Google Scholar] [CrossRef]
- vanMeer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. reviews. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Wen, B.; Hou, G.; Lei, L.; Mei, Z.; Jia, X.; Chen, X.; Zhu, W.; Li, J.; Kuang, Y.; et al. Lipidomics profiling reveals the role of glycerophospholipid metabolism in psoriasis. GigaScience 2017, 6, gix087. [Google Scholar] [CrossRef] [PubMed]
- Norris, G.H.; Blesso, C.N.; Norris, G.H.; Blesso, C.N. Dietary and Endogenous Sphingolipid Metabolism in Chronic Inflammation. Nutrients 2017, 9, 1180. [Google Scholar] [CrossRef] [PubMed]
- Leung, K.S.; Galano, J.-M.; Oger, C.; Durand, T.; Lee, J.C.-Y. Enrichment of alpha-linolenic acid in rodent diet reduced oxidative stress and inflammation during myocardial infarction. Free Radic. Biol. Med. 2021, 162, 53–64. [Google Scholar] [CrossRef]
- Sheng, K.; Sheng, K.; Yang, J.; Xu, Y.; Kong, X.; Wang, J.; Wang, Y. Alleviation effects of grape seed proanthocyanidin extract on inflammation and oxidative stress in a D-galactose-induced aging mouse model by modulating the gut microbiota. Food Funct. 2022, 13, 1348–1359. [Google Scholar] [CrossRef]
- Woolbright, B.L.; Dorko, K.; Antoine, D.J.; Clarke, J.I.; Gholami, P.; Li, F.; Kumer, S.C.; Schmitt, T.M.; Forster, J.; Fan, F.; et al. Bile acid-induced necrosis in primary human hepatocytes and in patients with obstructive cholestasis. Toxicol. Appl. Pharmacol. 2015, 283, 168–177. [Google Scholar] [CrossRef]
- Fernstrom, J.D.; Fernstrom, M.H. Tyrosine, Phenylalanine, and Catecholamine Synthesis and Function in the Brain2. J. Nutr. 2007, 137, 1539S–1547S. [Google Scholar] [CrossRef]
- Mierziak, J.; Burgberger, M.; Wojtasik, W.; Wojtasik, W.; Mierziak, J.; Burgberger, M.; Wojtasik, W. 3-Hydroxybutyrate as a Metabolite and a Signal Molecule Regulating Processes of Living Organisms. Biomolecules 2021, 11, 402. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Q.; Yang, Z.; Cai, Y.; Wang, D.; Wu, D. The Role of the Gut Microbiome in the Development of Acute Pancreatitis. Int. J. Mol. Sci. 2024, 25, 1159. [Google Scholar] [CrossRef]
- Cai, Y.; Yang, F.; Huang, X. Oxidative stress and acute pancreatitis (Review). Biomed. Rep. 2024, 21, 1–7. [Google Scholar] [CrossRef]
- Shek, F.W.-T.; Benyon, R.C.; Walker, F.M.; McCrudden, P.R.; Pender, S.L.F.; Williams, E.J.; Johnson, P.A.; Johnson, C.D.; Bateman, A.C.; Fine, D.R.; et al. Expression of Transforming Growth Factor-β1 by Pancreatic Stellate Cells and Its Implications for Matrix Secretion and Turnover in Chronic Pancreatitis. Am. J. Pathol. 2002, 160, 1787–1798. [Google Scholar] [CrossRef]
- Gabr, S.A.; Alghadir, A.H.; Sherif, Y.E.; Ghfar, A.A. Hydroxyproline as a Biomarker in Liver Disease. In Biomarkers in Disease: Methods, Discoveries and Applications; Springer: Dordrecht, The Netherlands, 2016. [Google Scholar]
- Sakai, A.; Nishiumi, S.; Shiomi, Y.; Kobayashi, T.; Izumi, Y.; Kutsumi, H.; Hayakumo, T.; Azuma, T.; Yoshida, M. Metabolomic analysis to discover candidate therapeutic agents against acute pancreatitis. Arch. Biochem. Biophys. 2012, 522, 107–120. [Google Scholar] [CrossRef]
- Teich, N.; Ockenga, J.; Hoffmeister, A.; Manns, M.; Mössner, J.; Keim, V. Chronic pancreatitis associated with an activation peptide mutation that facilitates trypsin activation. Gastroenterology 2000, 119, 461–465. [Google Scholar] [CrossRef]
- Mayers, J.R.; Wu, C.; Clish, C.B.; Kraft, P.; Torrence, M.E.; Fiske, B.P.; Yuan, C.; Bao, Y.; Townsend, M.K.; Tworoger, S.S.; et al. Elevated circulating branched chain amino acids are an early event in pancreatic adenocarcinoma development. Nat. Med. 2014, 20, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Jakkampudi, A.; Sarkar, P.; Unnisa, M.; Patil, A.; Koutarapu, C.; Jaggaiahgari, S.; Naik, P.; Sarkar, S.; Prasanna, A.; Chintaluri, S.; et al. Kynurenine pathway alteration in acute pancreatitis and its role as a biomarker of infected necrosis. Pancreatol. Off. J. Int. Assoc. Pancreatol. (IAP) 2023, 23, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Liu, S.; Zheng, M.; Yan, F.; Xu, D.; Wang, W.; Chen, J.; Shi, C.; Liu, S.; Zheng, M.; et al. Phospholipid and glycerolipid metabolism as potential diagnostic biomarkers for acute pancreatitis. Lipids Health Dis. 2024, 23, 1–11. [Google Scholar] [CrossRef]
- Konończuk, T.; Łukaszuk, B.; Żendzian-Piotrowska, M.; Dąbrowski, A.; Krzyżak, M.; Ostrowska, L.; Kurek, K. Plasma Sphingolipids in Acute Pancreatitis. Int. J. Mol. Sci. 2017, 18, 2606. [Google Scholar] [CrossRef] [PubMed]

| Reference | Sample Types | Groups and Number | Number of Lipids or Metabolites Identified | Lipids/Metabolites Different in AP vs. Other Groups | Prediction Based on Modeling | Pathway or Enrichment Analysis |
|---|---|---|---|---|---|---|
| Ouyang, 2012 [28] | Serum | AP (n = 17), HC (n = 23) | NA | (Up): Isoleucine, acetlyglycine, triglyceride, inosine (Down): 3-hydroxybutyrate, trimethylamine-N-oxide, acetate, acetone | NA | NA |
| Khan et al., 2012 [37] | Serum | At hospitalization: AAP (n = 19), HC (n = 20) | NA | (Up): Palmitic acid C16:0, monounstaurated fatty acids, oleic acid C18:1n9 (Down): Myristic acid, linoleic acid C18:2, gammalinolenic acid C18:3, homogammalinolenic acid C20:3, alphalinolenic acid C18:3, mead acid C20:3n9 | NA | NA |
| At 18–24 months: AAP (n = 16), HC (n = 20) | NA | (Down): Myristic acid, Stearic acid C18:0, homogammalinolenic acid C20:3, alphalinolenic acid C18:3, mead acid C20:3n9 | NA | |||
| Lusczek et al., 2013 [34] | Urine | AP (n = 5), HC (n = 5) | 60 | (Up): Acetone, Ribose | NA | NA |
| Villaseñor et al., 2014 [35] | Plasma | AP (n = 15), non-AP (n = 21) | NA | (Up): Choline, glucose, scyllo-inositol, lipid CH3CH2, lipid (CH2)n, lipid CH2CH═CH, acetone, D-3-hydroxybutyrate, acetoacetic acid (Down): Valine, alanine, | AUC = 0.86 | NA |
| Urine | NA | (Down): Hippurate, creatine, guanine | AUC = 0.91 | |||
| Yang et al., 2016 [36] | Plasma | AP (n = 13), HC (n = 10) | LC-GCMS: 206; GCMS only: 169 | (Up): β-Alanine, inosine, D-sorbitol, D-gluconic acid, L-threitol, D-glucose, D-glucose, arachidonic acid, citric acid, L-glutamine, urea, linolenic acid, myo-inositol, glyceric acid, tetradecanoic acid, cis-9-hexadecenoic acid, L-proline, tyrosine, uric acid, oxalic acid, 2-hydroxypyridine, hexadecanoic acid, glycolic acid, L-tyrosine (Down): L-valine, trans-9-octadecenoic acid, 11-trans-octadecenoic acid, cholesterol, glycylglycine, glycine, dl-isoleucine, L-serine, L-tryptophan, L-isoleucine, L-aspartic acid, phenylalanine, D-fructose, L-proline, L-leucine, D-(−)-lactic acid, L-alanine, L-serine, L-ornithine, 9,12-octadecadienoic acid (Z,Z), L-valine, phosphate, L-leucine, glutamic acid, pyruvic acid | NA | Amino acid metabolism, glucose metabolism, lipid metabolism |
| Xu et al., 2016 [39] | Serum | MAP (n = 38), CHO (n = 26), HC (n = 36) | 432 | MAP vs. HC: (Up): sphinganine, capryloyl choline, glycocholic acid, myristic acid, decanoyl choline, dodecanol, 2-tetradecanone, L-thyronine | AUC = 0.865 | NA |
| MAP vs. CHO: (Up): Sphinganine, capryloyl choline, myristic acid, decanoyl choline, dodecanol, 2-tetradecanone (Down): Glycocholic acid, L-thyronine | ||||||
| Skouras et al., 2016 [40] | Plasma | AP (n = 57), MAP (n = 23), Moderate AP (n = 23), SAP (n = 9) | NA | (Up): SAP vs. others: 3-Hydroxykynurenine (Down): SAP vs. others: Tryptophan | NA | Kynurenine pathway |
| Lusczek et al., 2016 [38] | Serum | AP-post ERCP (n = 9), no AP-post ERCP (n = 18) | 46 | (Up): β-hydroxybutyrate, acetoacetate, glucose | NA | NA |
| Urine | 72 | (Up): β-hydroxybutyrate, acetoacetate | ||||
| Xiao et al., 2017 [29] | Serum | AP (n, identification = 40, validation = 14), HC (n = 37) | 44 | (Up): 3-hydroxybutyric acid, citric acid, D-mannose, D-glucose, D-galactose, hexadecenoic acid, serotonin (Down): Phosphoric acid. Glycerol, Serotonin | AUC = 0.9907 | Galactose metabolism, glycerolipid metabolism, citrate cycle |
| SAP (n = 6), MAP (n = 8) | 3-hydroxybutyric acid, citric acid | |||||
| Zhao et al., 2017 [30] | Serum | HLAP (n = 24), HC (n = 39) | 20 | (Up): Hexadecanoic acid, eicosanoic acid, octadecanoic acid. (Down): Glycine, alanine, citrate, fumaric acid | NA | Tricarboxylic acid cycle (citrate, aconitate), tyrosine metabolism (tyrosine, phenylalanine, tyramine), gut microbiota metabolic activity (p-hydroxyphenylacetate, hippurate) |
| Urine | (Up): Proline, leucine, tyramine, phenylalanine, tyrosine, histidine, octadecanoic acid, hexadecanoic acid. (Down): Glycine, citrate, p-hydroxyphenylacetate, hippurate | |||||
| Huang et al., 2019 [31] | Serum | BAP (n = 27), HC (n = 15) | 32 | (Up): L-lysine (Down): N-acetyl-D-glucosamine, L-lactic acid, L-valine, (R)-3-hydroxybutyric acid, phosphoric acid, glycine, D-galactose, D-glucose, mannitol, L-tyrosine, D-turanose, octadecanoic acid, myo-inositol, oleic acid, cholesterol, glycerol 1-hexadecanoate | AUC= 0.886 | Aminoacyl-tRNA biosynthesis, thiamine metabolism, glycolysis or gluconeogenesis, propanoate metabolism, nitrogen metabolism |
| AAP (n = 20), HC (n = 15) | (Down): L-lactic acid, butyric acid, oxalic acid, (R)-3-hydroxybutyric acid, glycine, L-proline, erythronic acid, L-phenylalanine L-serine, L-threonine, L-glutamine, ornithine, L-tyrosine, octadecanoic acid, hexadecenoic acid, L-tryptophan, linoleic acid, oleic acid, arachidonic acid, cholesterol, glycerol 1-hexadecanoate | AUC = 0.857 | Aminoacyl-tRNA biosynthesis, nitrogen metabolism, glycine, serine and threonine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, fatty acid biosynthesis | |||
| HLAP (n = 29), HC (n = 15) | (Down): N-acetyl-D-glucosamine, L-lactic acid, glycine, D-glucose, mannitol, L-tyrosine, D-turanose, octadecanoic acid, myo-inositol, L-tryptophan, cholesterol, glycerol 1-hexadecanoate | AUC = 0.906 | Nitrogen metabolism, aminoacyl-tRNA biosynthesis, thiamine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, glycolysis or gluconeogenesis | |||
| Lou et al., 2022 [41] | Plasma EVs | SAP (n = 50), HC (n = 50) | 313 | (Up): Propylparaben, N-acetylglucosamine 1-phosphate, N-oleoyl glycine, lysoPC 17:0, glycoursodeoxycholic acid, L-saccharopine, glycochenodeoxycholic acid, proline betaine, L-valine (Down): 2-(methylthio)ethanol, cyclamicacid, methylstearate, diphenylamine, ginkgoic acid, 15-oxoETE, 2-(methylthio)benzothiazol, 4-hydroxy-L-glutamic acid, hyodeoxycholic acid | Discovery: AUC = 1.00; Validation: AUC = 0.886 | NA |
| SAP (n = 50), MAP (n = 50) | 46 | (Up): Hippuric acid, phenylacetyl-L-glutamine, 2-(dimethylamino)guanosine, estrone (Down): L-carnitine, nonadecylic acid, 2-(methylthio)benzothiazole, hexyl acetate | ||||
| Dancu et al., 2023 [32] | Serum | AP (n = 34), HC (n = 26) | 123 | (Up): LPC (20:3), all-trans-Retinyl oleate, DG (37:6), LPE (P-16:0/0:0), PE (30:3), Stearyl linolenate, DG (40:9), TG (57:3), 20:1 Cholesterol ester (Down): Dihydrobiopterin, LPA (20:5), LPC (16:1), LPC (18:0/0:0), (S)-3-hydroxystearic acid | AUC > 0.8 for first 19 metabolites | Glycerophospholipid, sterol lipids, fatty acyls, prenol lipids, glycerolipids, sphingolipids |
| BAP (n = 6), AAP (n = 22) | (Up): Myristyl linolenate, LPC (24:1) (Down): MG (0:0/18:0/0:0), (S)-3-hydroxystearic acid, PC (P-18:0/16:0), all-trans-retinyl oleate, and LPC (O-16:0) | AUC > 0.72 for 3 metabolites | ||||
| Liu et al., 2024 [33] | Serum | AP (n = 45), HC (n = 13) | 705 lipids, 775 metabolites | (Up): Carbohydrates, hematoporphyrin, organic acids, triacylglycerols (Down): Bile acid, glycerophosphocholine, LPA | NA | Arginine biosynthesis, butanoate metabolism, valine, leucine and isoleucine biosynthesis, histidine metabolism, arginine and proline metabolism, alanine, aspartate and glutamate metabolism, phenylalanine metabolism, sphingolipid metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, synthesis and degradation of ketone bodies |
| Reference | Sample Types | Groups and Number | Number of Lipids or Metabolites Identified | Lipid/Metabolites Different in CP vs. Other Groups | Prediction Based on Modeling | Pathway or Enrichment Analysis |
|---|---|---|---|---|---|---|
| Zhang et al., 2012 [42] | Plasma | CP (n = 20), HC (n = 20) | NA | (Up): glucose, lactate, creatine, formate, lipid glyceryls, tyrosine, phenylalanine, lysine, histidine, glutamine, glutamate, alanine (Down): LDL, VLDL, 3-hydroxybutyrate, acetone | NA | NA |
| Lusczek et al., 2013 [34] | Urine | CP (n = 5), HC (n = 5) | 60 | (Up): Adenosine (Down): Citrate | NA | |
| Adam et al., 2021 [43] | Plasma and Serum | Identification (Plasma): CP (n = 80), HC (n = 80) Validation 1 (Plasma): CP (n = 144), HC (n = 204) Validation 2 (Serum): CP (n = 49), HC (n = 56) | Plasma: 620 Serum: 616 | (Up): Mannose, Ceramide (d18:1/ C24:1), Behenic acid (C22:0), N-Acetylcytidine (Down): Beta-carotene, Cryptoxanthin, Indole-3-acetic acid, Hippuric acid | Plasma: AUC = 0.85; Serum: AUC = 0.87 | NA |
| Diaz et al., 2021 [44] | Serum | CP (n = 53), EPI (n = 32), No-EPI (n = 21) | 1262 | (Up): phosphatidylserines (4), phosphatidylcholine (1), Arg-Thr-Pro, pentasine | AUC = 0.79 | NA |
| Sarkar et al., 2022 [45] | Plasma | CP (n = 558), HC (n = 67) | HC: 22; CP: 70 | (Up): benzoic acid (Down): glycine, sarcosine, L-threonine, cholesterol, aminobutanoic acid | NA | NA |
| Wu et al., 2023 [46] | Serum | Exploratory: CP (n = 18), HC (n = 21) Identification: CP (n = 50), HC (n = 17) Validation: CP (n = 23), HC (n = 10) | 239 | (Up): Oleandrin, 1-(9Z-heptadecenoyl)-glycero-3-phosphoserine, 5beta-Cyprinolsulfate, 1-(6Z,9Z,12Z,15Z-octadecatetraenoyl)-glycero-3-phosphate, Fludrocortisone, Atracurium, Gibberellin A51, Lagerstroemine, | AUC > 0.7 | Sphingolipid metabolism, glycerophospholipid metabolism, linoleic acid metabolism, galactose metabolism |
| Qi et al., 2023 [47] | Plasma | T3cDM secondary to CP (n = 16), HC (n = 12) | Positive mode: 2345; Negative mode: 707 | (Up): glycocholate, D- -glucose, glycochenodeoxycholic acid, oleamide (Down): deoxycholic acid, Hippurate, caffeine, indole-3-methyl, γ-linoleic acid, paraxanthine, choline, theophylline, DL-stachydrine | AUC = 0.907 | Bile acid biosynthesis, beta oxidation of very long chain fatty acids, linolenic acid metabolism, fatty acid biosynthesis, sphingolipid metabolism |
| Ketavarapu et al., 2024 [48] | Plasma | Identification: CP (n = 96), HC (n = 7) Validation 1: CP (n = 107), HC (n = 26) Validation 2: CP (n = 43), HC (n = 30) | 57 | (Up): Hexanoyl carnitine, deoxycholic acid, Cer (d18:1/16:0), LPE (16:0), LPC (20:3), PE (38:7), PA (32:0), Cer (d18:1/24:1), LPE (22:6), PE (34:2), PE (36:3), PC(37:6), PC(36:5), PC(32:1), PC(36:5), PC(32:1), PE (34:1), and Cer (d18:2/24:1) (Down): DAG (33:4), PC (O-34:2), PC(36:3), DAG (33:2), PC (34:1), Cer(d18:1/24:0) | AUC for HC and CP = 0.88 for 7 metabolite panel | NA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, F.; Zhao, X.; Setchell, K.D.R.; Abu-El-Haija, M. Advancing Clinical and Pathophysiological Insights into Pancreatitis Using Lipidomics and Metabolomics. Metabolites 2025, 15, 666. https://doi.org/10.3390/metabo15100666
Ahmed F, Zhao X, Setchell KDR, Abu-El-Haija M. Advancing Clinical and Pathophysiological Insights into Pancreatitis Using Lipidomics and Metabolomics. Metabolites. 2025; 15(10):666. https://doi.org/10.3390/metabo15100666
Chicago/Turabian StyleAhmed, Faizan, Xueheng Zhao, Kenneth D. R. Setchell, and Maisam Abu-El-Haija. 2025. "Advancing Clinical and Pathophysiological Insights into Pancreatitis Using Lipidomics and Metabolomics" Metabolites 15, no. 10: 666. https://doi.org/10.3390/metabo15100666
APA StyleAhmed, F., Zhao, X., Setchell, K. D. R., & Abu-El-Haija, M. (2025). Advancing Clinical and Pathophysiological Insights into Pancreatitis Using Lipidomics and Metabolomics. Metabolites, 15(10), 666. https://doi.org/10.3390/metabo15100666





