Lipidomics by Nuclear Magnetic Resonance Spectroscopy and Liquid Chromatography–High-Resolution Mass Spectrometry in Osteosarcoma: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Blood Serum Samples and Lipid Extracts
2.2. NMR Spectra Acquisition
2.3. Statistical Analysis of NMR Data
2.4. LC-MS Analysis of Lipid Extracts
2.5. LC-MS Data Processing and Statistical Analysis
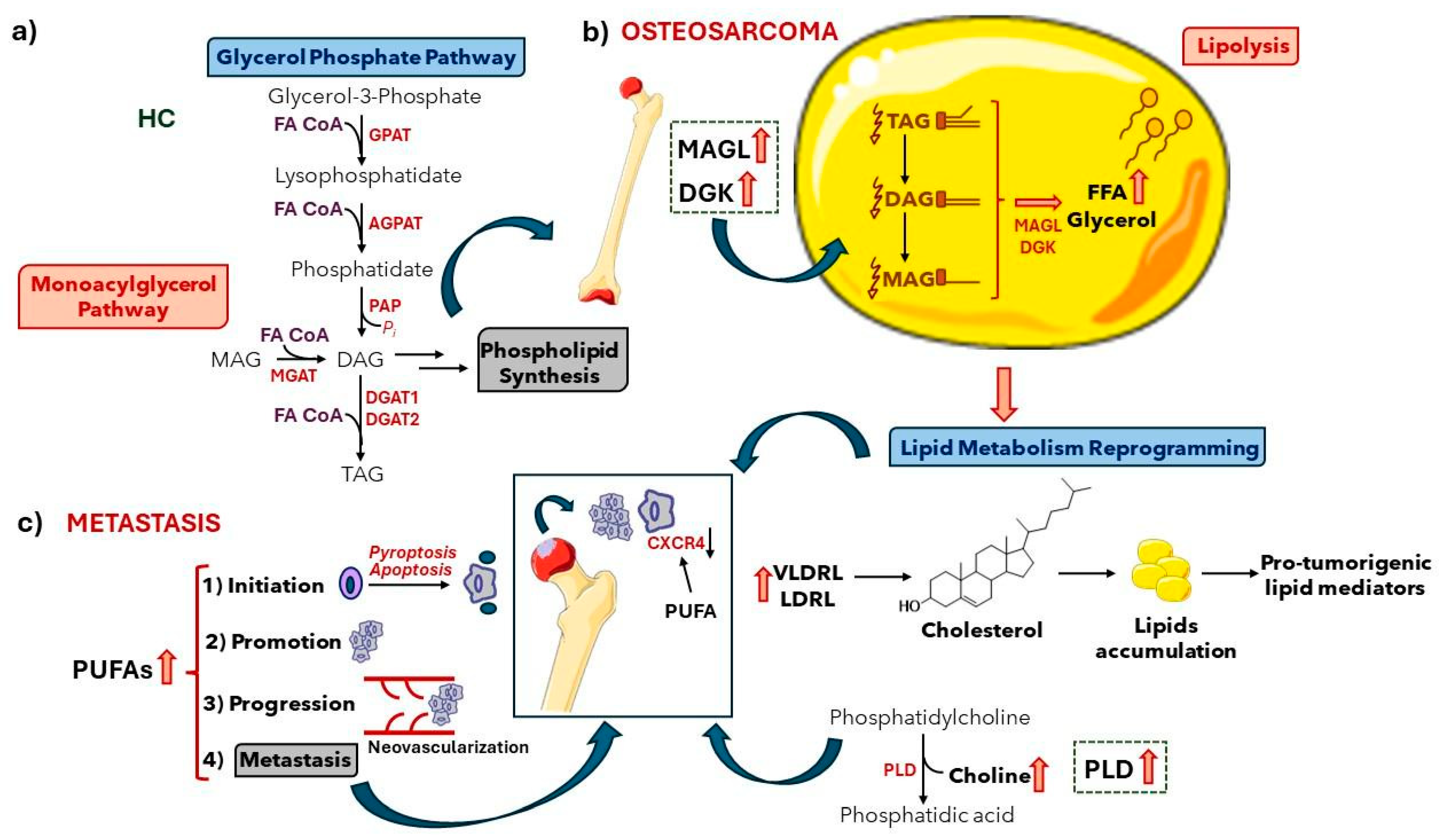
3. Results and Discussion
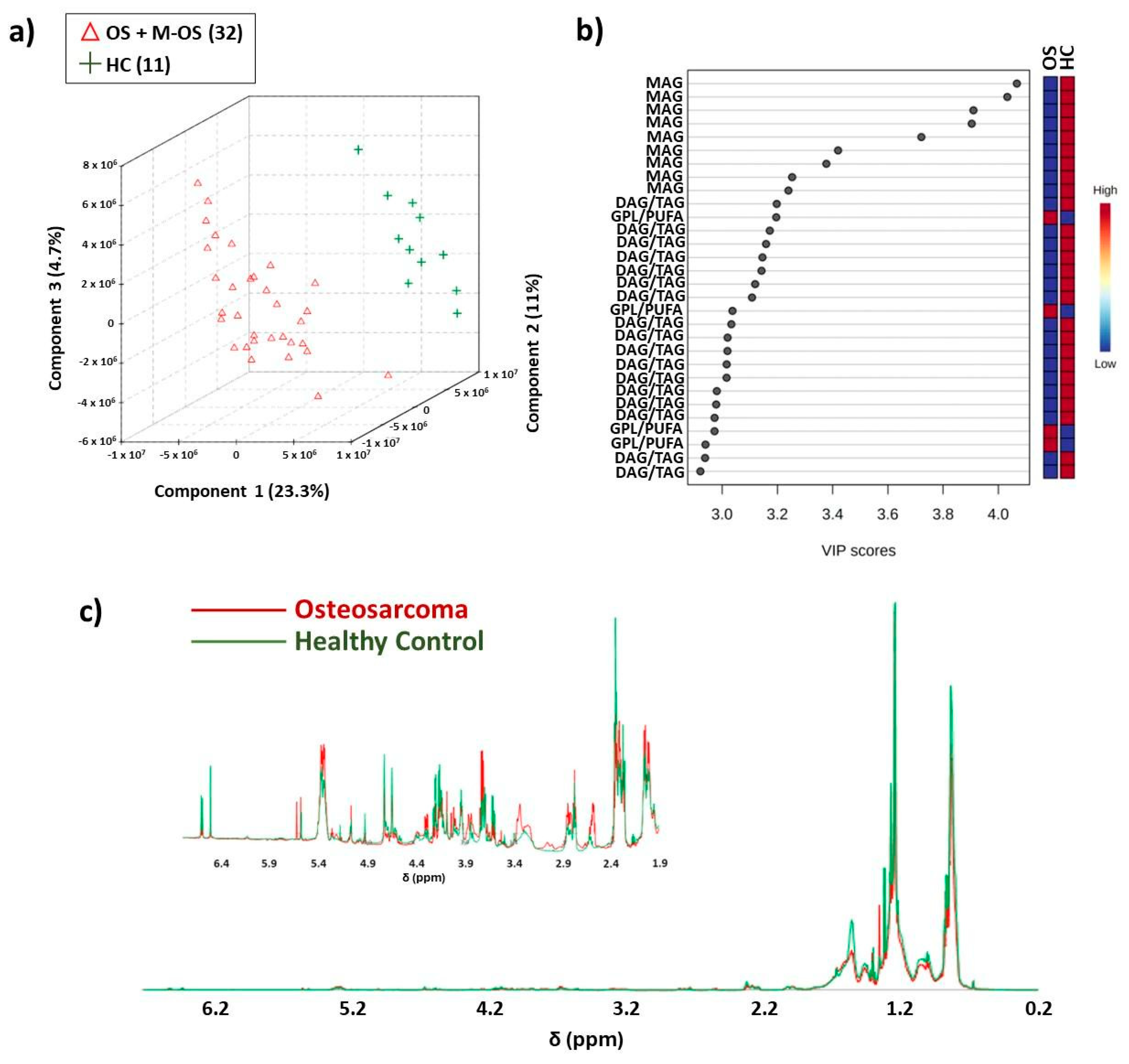
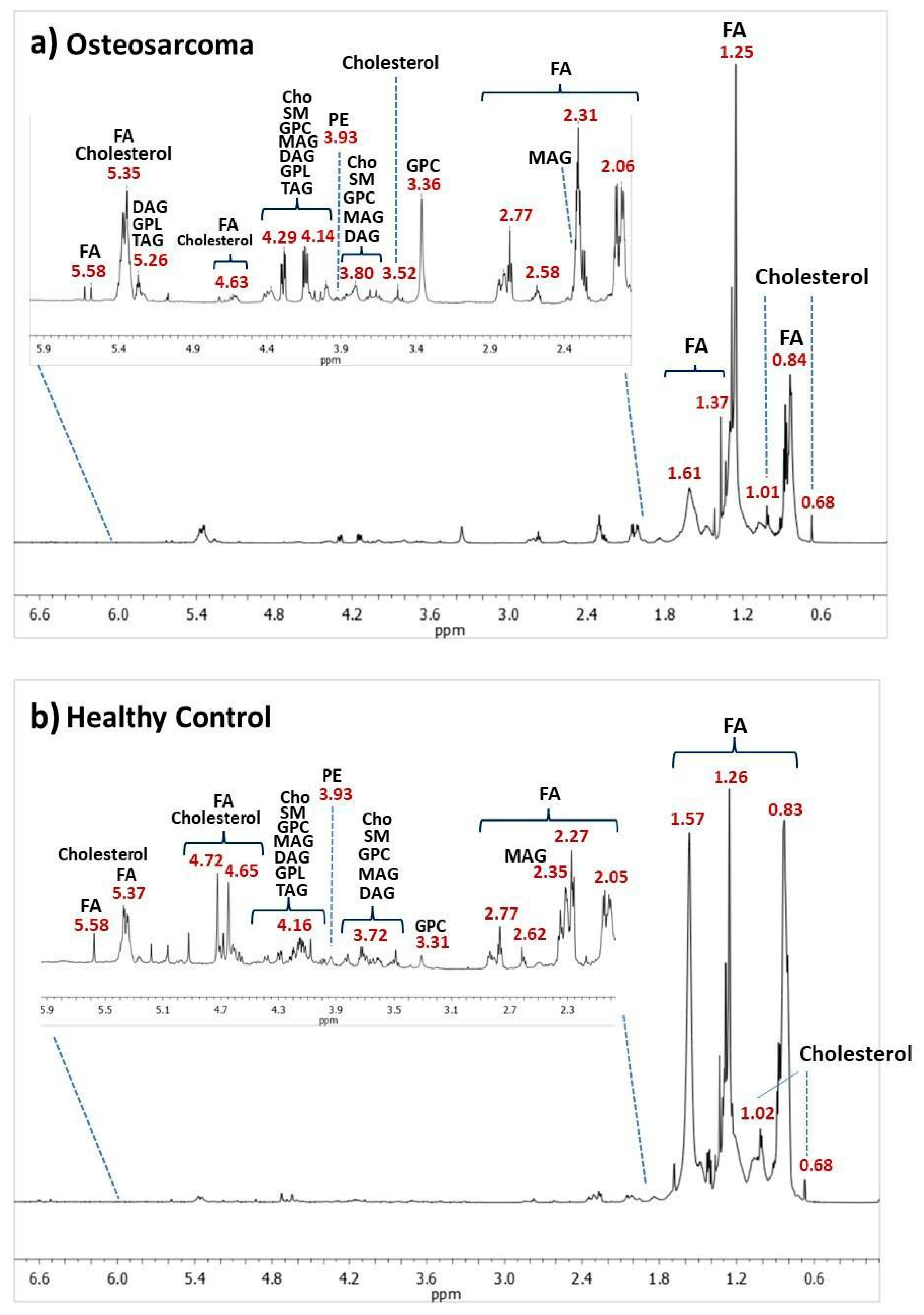
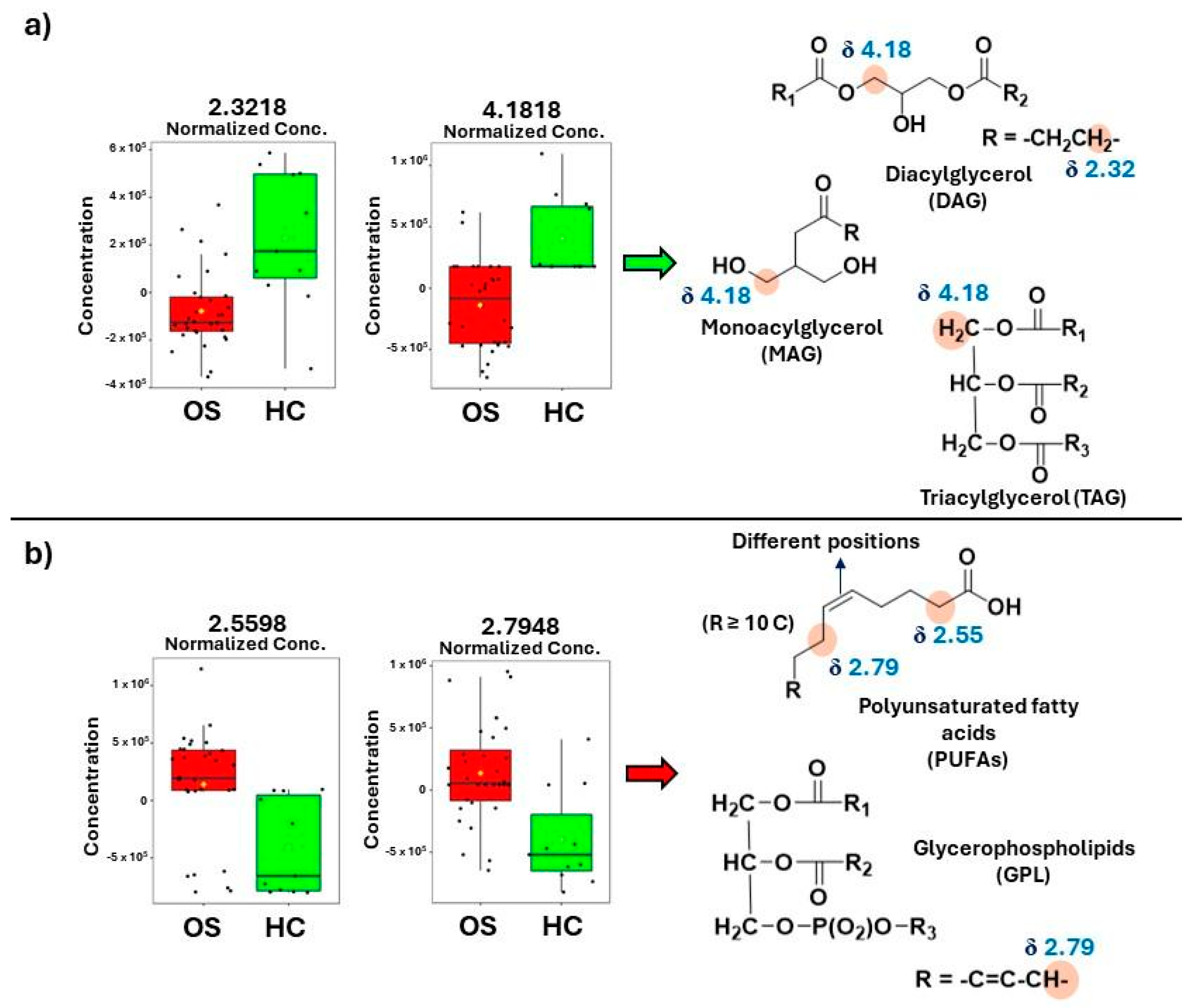
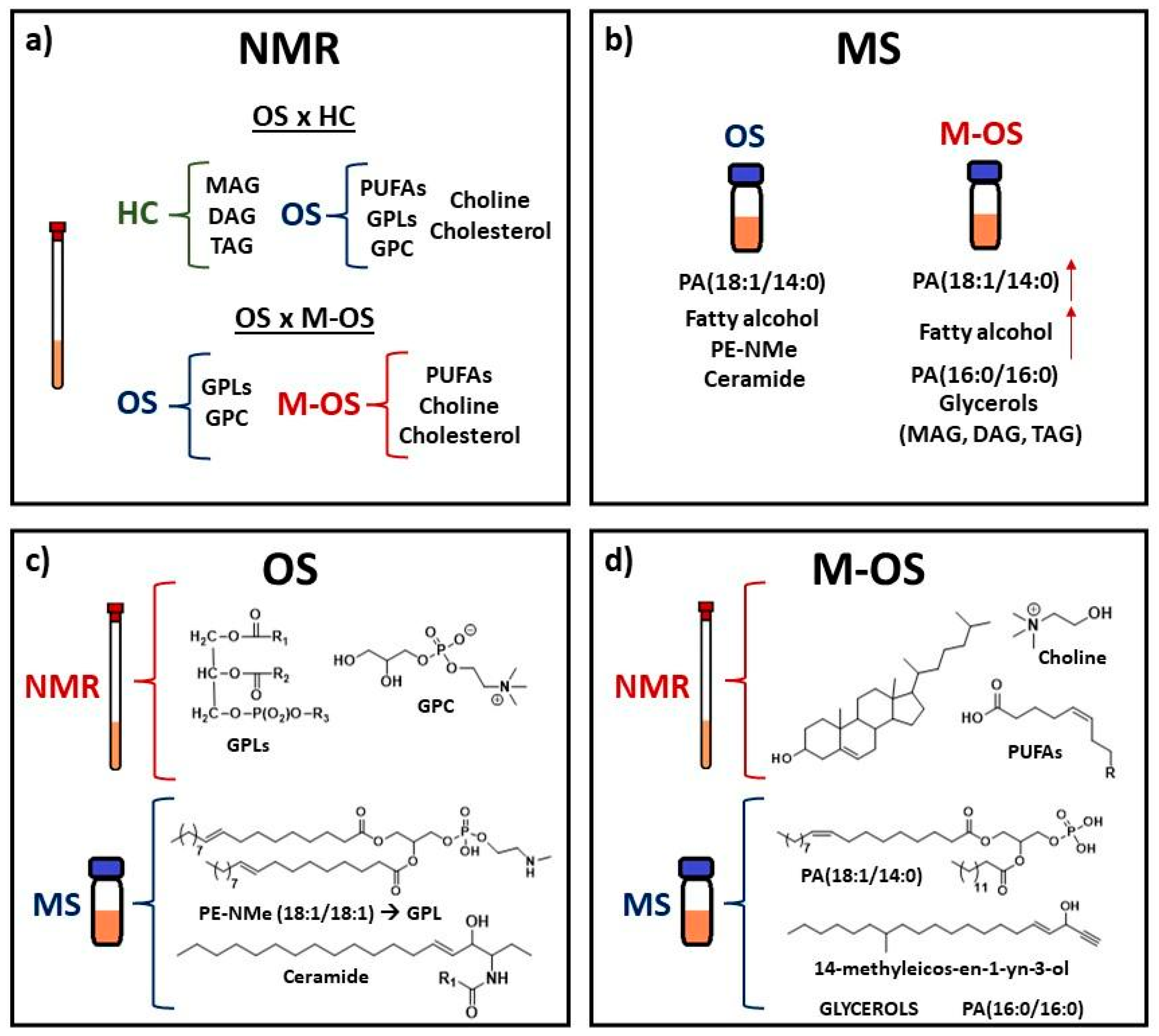
3.1. NMR-Based Lipidomics of Osteosarcoma
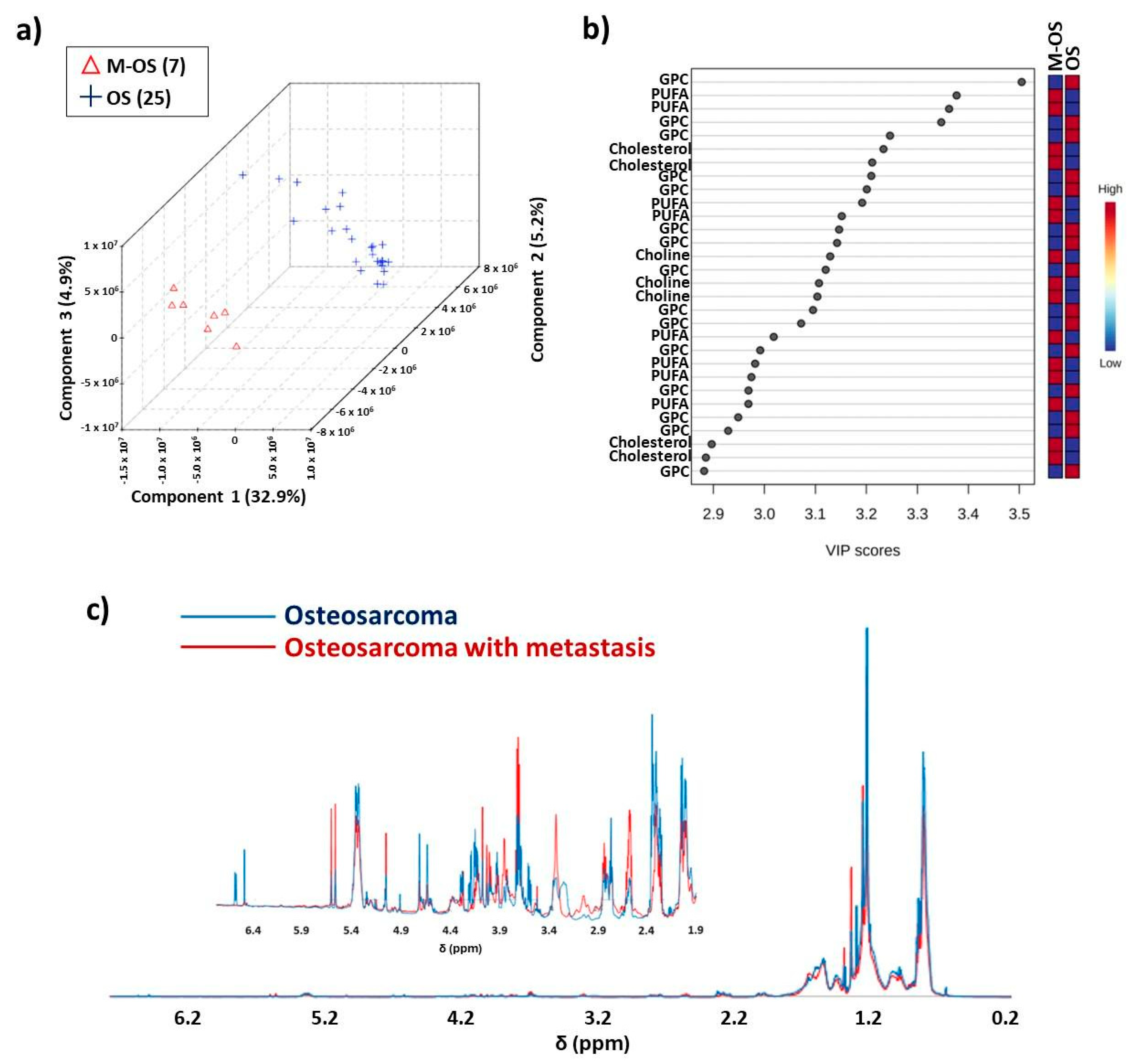
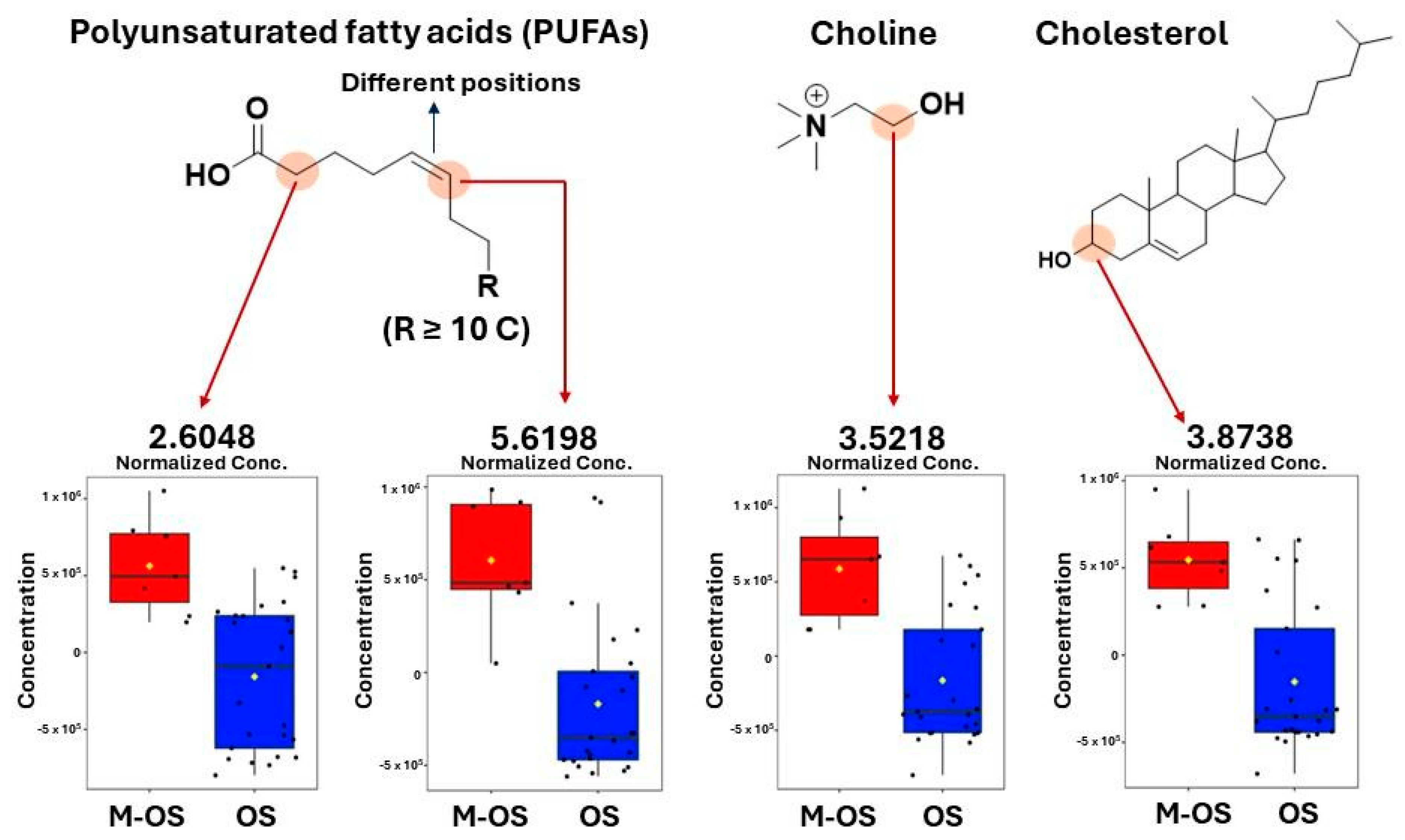
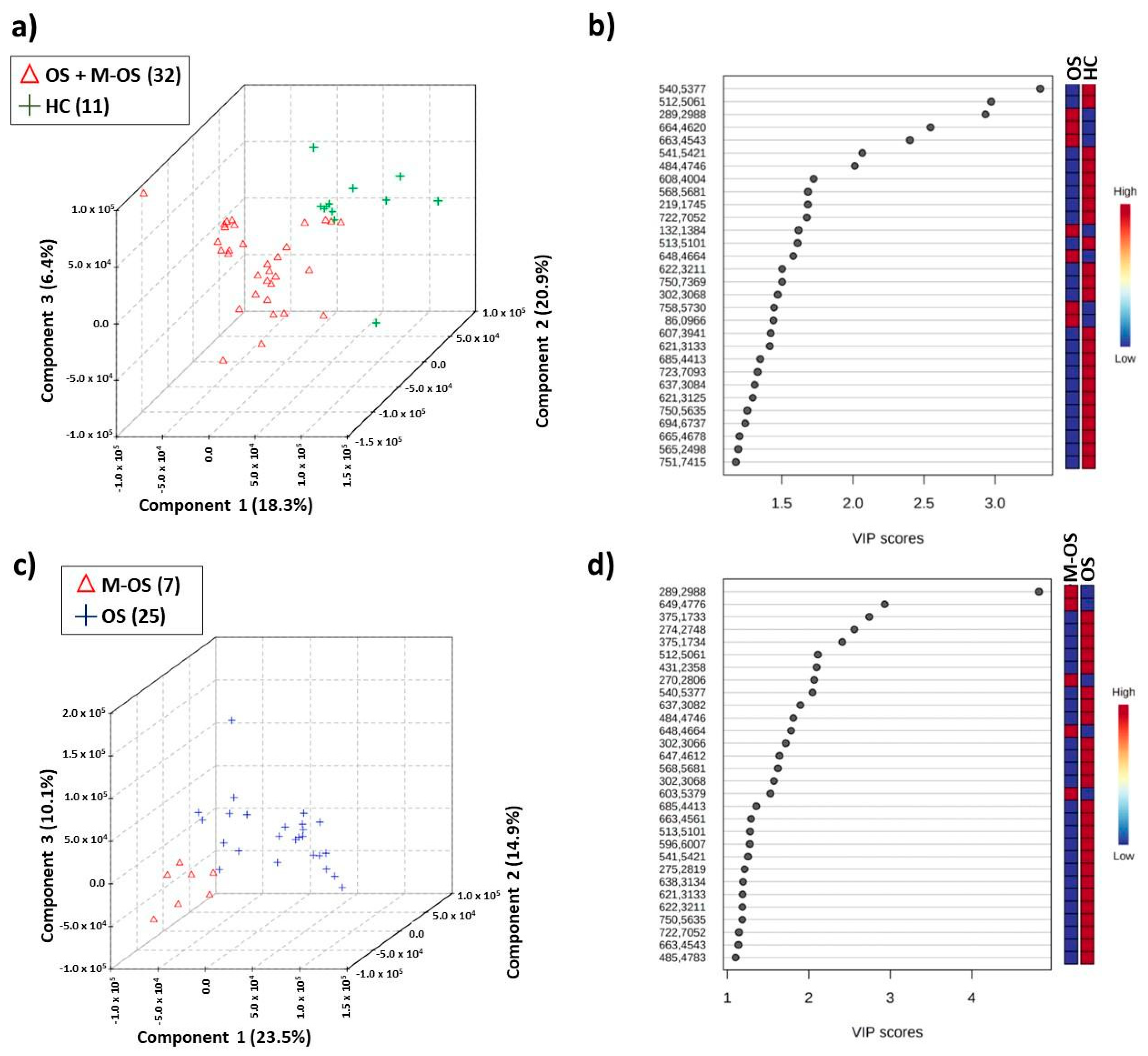
3.2. Differentiation between Osteosarcoma Patients with and without Metastasis
3.3. ESI (+) LC-MS-Based Lipidomics of Osteosarcoma
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Armitage, E.G.; Southam, A.D. Monitoring cancer prognosis, diagnosis and treatment efficacy using metabolomics and lipidomics. Metabolomics 2016, 12, 146. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). Childhood cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer-in-children (accessed on 3 June 2024).
- Guthrie, O.W.; Spankovich, C. Emerging and established therapies for chemotherapy-induced ototoxicity. J. Cancer Surviv. 2023, 17, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.B.; Bjornard, K.L.; Alberts, N.M.; Armstrong, G.T.; Brinkman, T.M.; Chemaitilly, W.; Ehrhardt, M.J.; Fernandez-Pineda, I.; Force, L.M.; Gibson, T.M.; et al. Factors influencing risk-based care of the childhood cancer survivor in the 21st century. CA Cancer J. Clin. 2018, 68, 133–152. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal. Transduct. Target Ther. 2021, 6, 201. [Google Scholar] [CrossRef]
- Escobar, M.Q.; Costa, T.B.B.C.; Martins, L.G.; Costa, S.S.; van Helvoort Lengert, A.; Boldrini, E.; Morini da Silva, S.R.; Lopes, L.F.; Vidal, D.O.; Krepischi, A.C.V.; et al. Insights in osteosarcoma by proton nuclear magnetic resonance serum metabonomics. Front. Oncol. 2020, 10, 506959. [Google Scholar] [CrossRef]
- Yan, F.; Zhao, H.; Zeng, Y. Lipidomics: A promising cancer biomarker. Clin. Transl. Med. 2018, 7, 21. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef] [PubMed]
- Hyötyläinen, T.; Orešič, M. Optimizing the lipidomics workflow for clinical studies–practical considerations. Anal. Bioanal. Chem. 2015, 407, 4973–4993. [Google Scholar] [CrossRef]
- Stromberg, L.R.; Lilley, L.M.; Mukundan, H. Advances in lipidomics for cancer biomarker discovery. In Proteomic and Metabolomic Approaches to Biomarker Discovery; Issaq, H.J., Veenstra, T.D., Eds.; Academic Press: Cambridge, MA, USA, 2020; pp. 421–436. [Google Scholar] [CrossRef]
- Taran, S.J.; Taran, R.; Malipatil, N.B. Pediatric osteosarcoma: An updated review. Indian J. Med. Paediatr. Oncol. 2017, 38, 33–43. [Google Scholar] [CrossRef]
- Chen, K.; Zhu, C.; Cai, M.; Fu, D.; Cheng, B.; Cai, Z.; Li, G.; Liu, J. Integrative metabolome and transcriptome profiling reveals discordant glycolysis process between osteosarcoma and normal osteoblastic cells. J. Cancer Res. Clin. Oncol. 2014, 140, 1715–1721. [Google Scholar] [CrossRef]
- Song, Y.-J.; Xu, Y.; Deng, C.; Zhu, X.; Fu, J.; Chen, H.; Lu, J.; Xu, H.; Song, G.; Tang, Q.; et al. Gene expression classifier reveals prognostic osteosarcoma microenvironment molecular subtypes. Front. Immunol. 2021, 12, 623762. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Jin, X.; Zhang, Y.; Wang, Z.; Zhang, T.; Xu, J.; Shen, J.; Zan, P.; Sun, M.; Wang, C.; et al. Inhibition of sphingolipid metabolism in osteosarcoma protects against CD151-mediated tumorigenicity. Cell. Biosci. 2023, 12, 169. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Meng, Y.; Wang, Y.; Wang, W. NDRG1 regulates osteosarcoma cells via mediating the mitochondrial function and CSCs differentiation. J. Orthop. Surg. Res. 2021, 16, 364. [Google Scholar] [CrossRef] [PubMed]
- Sirikaew, N.; Pruksakorn, D.; Chaiyawat, P.; Chutipongtanate, S. Mass spectrometric-based proteomics for biomarker discovery in osteosarcoma: Current status and future direction. Int. J. Mol. Sci. 2022, 23, 9741. [Google Scholar] [CrossRef] [PubMed]
- Tasic, L.; Lacerda, A.L.T.; Pontes, J.G.M.; Costa, T.B.B.C.; Nani, J.V.; Martins, L.G.; Santos, L.A.; Nunes, M.F.Q.; Adelino, M.P.M.; Pedrini, M.; et al. Peripheral biomarkers allow differential diagnosis between schizophrenia and bipolar disorder. J. Psychiatr. Res. 2019, 119, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Correia, B.S.B.; Pontes, J.G.M.; Nani, J.V.S.; Villalta, F.; Mor, N.C.; Bordini, D.; Brunoni, D.; Brentani, H.; Mari, J.J.; Hayashi, M.A.F.; et al. 1H NMR metabolomics and lipidomics to monitor positive responses in children with autism spectrum disorder following a guided parental intervention: A pilot study. ACS Chem. Neurosci. 2023, 14, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2022, 50, D622–D631. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.R.; Kobayashi, N.; Wedell, J.R.; Baskaran, K.; Iwata, T.; Yokochi, M.; Maziuk, D.; Yao, H.; Fujiwara, T.; Kurusu, G.; et al. BioMagResBank (BMRB) as a resource for structural biology. Methods Mol. Biol. 2020, 2112, 187–218. [Google Scholar] [CrossRef]
- Chambers, M.C.; Maclean, B.; Burke, R.; Amodei, D.; Ruderman, D.L.; Neumann, S.; Gatto, L.; Fischer, B.; Pratt, B.; Egertson, J.; et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 2012, 30, 918–920. [Google Scholar] [CrossRef]
- Grace, S.C.; Hudson, D.A. Processing and visualization of metabolomics data using R. In Metabolomics—Fundamentals and Applications; Prassain, J.K., Ed.; IntechOpen: London, UK, 2016; pp. 67–94. [Google Scholar]
- Partida-Martínez, L.; Winkler, R. Pre-processing and analysis of metabolomics data with XCMS/R and XCMS online. In Processing Metabolomics and Proteomics Data with Open Software: A Practical Guide; Winkler, R., Ed.; Royal Society of Chemistry: London, UK, 2020; pp. 255–280. [Google Scholar] [CrossRef]
- R Development Core Team. Available online: http://www.R-project.org/ (accessed on 3 June 2024).
- Forsberg, E.M.; Huan, T.; Rinehart, D.; Benton, H.P.; Warth, B.; Hilmers, B.; Siuzdak, G. Data processing, multi-omic pathway mapping, and metabolite activity analysis using XCMS online. Nat. Protoc. 2018, 13, 633–651. [Google Scholar] [CrossRef]
- Hobuss, C.B.; da Silva, F.A.; dos Santos, M.A.Z.; Pereira, C.M.P.; Schulz, G.A.S.; Bianchini, D. Synthesis and characterization of monoacylglycerols through glycerolysis of ethyl esters derived from linseed oil by green processes. RSC Adv. 2020, 10, 2327–2336. [Google Scholar] [CrossRef]
- Gong, X.; Zheng, X.; Huang, Y.; Song, W.; Chen, G.; Chen, T. Monoacylglycerol Lipase (MAGL) inhibition impedes the osteosarcoma progression by regulating epithelial mesenchymal transition. Tohoku J. Exp. Med. 2022, 256, 19–26. [Google Scholar] [CrossRef]
- Hu, W.-R.; Lian, Y.-F.; Peng, L.-X.; Lei, J.-J.; Deng, C.-C.; Xu, M.; Feng, Q.-S.; Chen, L.-Z.; Bei, J.-X.; Zeng, Y.-X. Monoacylglycerol lipase promotes metastases in nasopharyngeal carcinoma. Int. J. Clin. Exp. Pathol. 2014, 7, 3704–3713. [Google Scholar] [PubMed]
- Deng, H.; Li, W. Monoacylglycerol lipase inhibitors: Modulators for lipid metabolism in cancer malignancy, neurological and metabolic disorders. Acta Pharm. Sin. B 2020, 10, 582–602. [Google Scholar] [CrossRef]
- Roy, J.; Dibaeinia, P.; Fan, T.M.; Sinha, S.; Das, A. Global analysis of osteosarcoma lipidomes reveal altered lipid profiles in metastatic versus nonmetastatic cells. J. Lipid Res. 2019, 60, 376–387. [Google Scholar] [CrossRef]
- Mika, A.; Kaczynski, Z.; Stepnowski, P.; Kaczor, M.; Proczko-Stepaniak, M.; Kaska, L.; Sledzinski, T. Potential application of 1H NMR for routine serum lipidome analysis-evaluation of effects of bariatric surgery. Sci. Rep. 2017, 7, 15530. [Google Scholar] [CrossRef]
- Hatzakis, E.; Agiomyrgianaki, A.; Kostidis, S.; Dais, P. High-resolution NMR spectroscopy: An alternative fast tool for qualitative and quantitative analysis of diacylglycerol (DAG) oil. J. Am. Oil Chem. Soc. 2011, 88, 1695–1708. [Google Scholar] [CrossRef]
- Yu, W.; Tang, L.; Lin, F.; Yao, Y.; Shen, Z. DGKZ acts as a potential oncogene in osteosarcoma proliferation through its possible interaction with ERK1/2 and MYC pathway. Front. Oncol. 2019, 8, 655. [Google Scholar] [CrossRef]
- Lehmann, M. Diverse roles of phosphatidate phosphatases in insect development and metabolism. Insect Biochem. Mol. Biol. 2021, 133, 103469. [Google Scholar] [CrossRef]
- Duarte, I.F.; Marques, J.; Ladeirinha, A.F.; Rocha, C.; Lamego, I.; Calheiros, R.; Silva, T.M.; Marques, M.P.M.; Melo, J.B.; Carreira, I.M.; et al. Analytical approaches toward successful human cell metabolome studies by NMR spectroscopy. Anal. Chem. 2009, 81, 5023–5032. [Google Scholar] [CrossRef]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef]
- Santini, M.T.; Romano, R.; Rainaldi, G.; Indovina, P.; Ferrante, A.; Motta, A.; Indovina, P.L. Temporal dynamics of 1H-NMR-visible metabolites during radiation-induced apoptosis in MG-63 human osteosarcoma spheroids. Radiat. Res. 2006, 166, 734–745. [Google Scholar] [CrossRef]
- Skorupa, A.; Poński, M.; Ciszek, M.; Cichoń, B.; Klimek, M.; Witek, A.; Pakulo, S.; Boguszewicz, S.; Sokół, M. Grading of endometrial cancer using 1H HR-MAS NMR-based metabolomics. Sci. Rep. 2021, 11, 18160. [Google Scholar] [CrossRef]
- Misra, D.; Bajpai, U. Metabolite characterization in serum samples from normal healthy human subjects by 1H and 13C NMR spectroscopy. Bull. Chem. Soc. Ethiop. 2009, 23, 211–221. [Google Scholar] [CrossRef]
- Wu, X.; Cao, H.; Zhao, L.; Song, J.; She, Y.; Feng, Y. Metabolomic analysis of glycerophospholipid signatures of inflammation treated with non-steroidal anti-inflammatory drugs-induced-RAW264.7 cells using 1H NMR and U-HPLC/Q-TOF-MS. J. Chromatogr. B 2016, 1028, 199–215. [Google Scholar] [CrossRef]
- Lei, T.; Qian, H.; Lei, P.; Hu, Y. Ferroptosis-related gene signature associates with immunity and predicts prognosis accurately in patients with osteosarcoma. Cancer Sci. 2021, 112, 4785–4798. [Google Scholar] [CrossRef]
- Yang, S.; Tian, Z.; Feng, Y.; Zhang, K.; Pan, Y.; Li, Y.; Wang, Z.; Wei, W.; Qiao, X.; Zhou, R.; et al. Transcriptomics and metabolomics reveal changes in the regulatory mechanisms of osteosarcoma under different culture methods in vitro. BMC Med. Genom. 2022, 15, 265. [Google Scholar] [CrossRef]
- Quiroz-Acosta, T.; Flores-Martinez, Y.M.; Becerra-Martínez, E.; Pérez-Hernández, E.; Pérez-Hernández, N.; Bañuelos-Hernández, A.E. Aberrant sphingomyelin 31P-NMR signatures in giant cell tumour of bone. Biochem. Cell. Biol. 2021, 99, 717–724. [Google Scholar] [CrossRef]
- Sonnino, S.; Aureli, M.; Mauri, L.; Ciampa, M.G.; Prinetti, A. Membrane lipid domains in the nervous system. Front. Biosci. 2015, 20, 280–302. [Google Scholar] [CrossRef]
- Xu, F.; Yan, J.; Peng, Z.; Liu, J.; Li, Z. Comprehensive analysis of a glycolysis and cholesterol synthesis-related genes signature for predicting prognosis and immune landscape in osteosarcoma. Front. Immunol. 2022, 13, 1096009. [Google Scholar] [CrossRef]
- Sheng, G.; Gao, Y.; Yang, Y.; Wu, H. Osteosarcoma and Metastasis. Front. Oncol. 2021, 11, 780264. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, X.; Xu, Y.; Han, X.; Cai, J.; Wang, X.; Wang, G. Lung metastases at the initial diagnosis of high-grade osteosarcoma: Prevalence, risk factors and prognostic factors. A large population-based cohort study. Sao Paulo Med. J. 2019, 137, 423–429. [Google Scholar] [CrossRef]
- Jafari, F.; Javdansirat, S.; Sanaie, S.; Naseri, A.; Shamekh, A.; Rostamzadeh, D.; Dolati, S. Osteosarcoma: A comprehensive review of management and treatment strategies. Ann. Diagn. Pathol. 2020, 49, 151654. [Google Scholar] [CrossRef]
- Casati, S.; Giannasi, C.; Minoli, M.; Niada, S.; Ravelli, A.; Angeli, I.; Mergenthaler, V.; Ottria, R.; Ciuffreda, P.; Orioli, M.; et al. Quantitative lipidomic analysis of osteosarcoma cell-derived products by UHPLC-MS/MS. Biomolecules 2020, 10, 1302. [Google Scholar] [CrossRef] [PubMed]
- Putra, R.N.; Rahardjo, P.; Setiawati, R. The detection of elevated choline metabolite in magnetic resonance spectroscopy to differentiate between benign and malignant bone tumor. Int. J. Res. 2020, 60, 36–44. [Google Scholar] [CrossRef]
- Setiawati, R.; Lay, E.S.; Testini, V.; Rahardjo, P.; Edward, M.; Mustokoweni, S.; Guglielmi, G. Advance MR evaluation of synchronous multifocal osteosarcoma with pathologic fracture. BJR Case Rep. 2021, 7, 20210015. [Google Scholar] [CrossRef]
- Abdallah, D.; Skafi, N.; Hamade, E.; Borel, M.; Reibel, S.; Vitale, N.; El Jamal, A.; Bougault, C.; Laroche, N.; Vico, L.; et al. Effects of phospholipase D during cultured osteoblast mineralization and bone formation. J. Cell. Biochem. 2019, 120, 5923–5935. [Google Scholar] [CrossRef]
- Haas, E.; Stanley, D.W. Phospholipase D. In xPharm: The Comprehensive Pharmacology Reference; Enna, S.J., Bylund, D.B., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–4. [Google Scholar] [CrossRef]
- Thyssel, E.; Surowiec, I.; Hörnberg, E.; Crnalic, S.; Widmark, A.; Johansson, A.I.; Stattin, P.; Bergh, A.; Moritz, T.; Antti, H.; et al. Metabolomic characterization of human prostate cancer bone metastases reveals increased levels of cholesterol. PLoS ONE 2010, 5, e14175. [Google Scholar] [CrossRef]
- Dean, D.C.; Shen, S.; Hornicek, F.J.; Duan, Z. From genomics to metabolomics: Emerging metastatic biomarkers in osteosarcoma. Cancer Metastasis Rev. 2018, 37, 719–731. [Google Scholar] [CrossRef]
- Hua, Y.; Qiu, Y.; Zhao, A.; Wang, X.; Chen, T.; Zhang, Z.; Chi, Y.; Li, Q.; Sun, W.; Li, G.; et al. Dynamic metabolic transformation in tumor invasion and metastasis in mice with LM-8 osteosarcoma cell transplantation. J. Proteome Res. 2011, 10, 3513–3521. [Google Scholar] [CrossRef]
- Pang, X.; Yin, P.; Han, J.; Wang, Z.; Zheng, F.; Chen, X. cPLA2a correlates with metastasis and poor prognosis of osteosarcoma by facilitating epithelial-mesenchymal transition. Pathol. Res. Pract. 2019, 215, 152398. [Google Scholar] [CrossRef]
- Luo, X.; Cheng, C.; Tan, Z.; Li, N.; Tang, M.; Yang, L.; Cao, Y. Emerging roles of lipid metabolism in cancer metastasis. Mol. Cancer 2017, 16, 76. [Google Scholar] [CrossRef]
- Munem, M.; Zaar, O.; Nilsson, K.D.; Neittaanmäki, N.; Paoli, J.; Fletcher, J.S. Chemical imaging of aggressive basal cell carcinoma using time-of-flight secondary ion mass spectrometry. Biointerphases 2018, 13, 03B402. [Google Scholar] [CrossRef]
- Kim, K.-M.; Park, T.-S.; Shim, S.-M. Optimization and validation of HRLC-MS method to identify and quantify triacylglycerol molecular species in human milk. Anal. Methods 2015, 7, 4362–4370. [Google Scholar] [CrossRef]
- Liu, G.Y.; Han, F.; Yang, Y.; Xie, Y.; Jiang, H.; Mao, Y.Y.; Wang, H.; Wang, M.; Chen, R.; Yang, J.; et al. Evaluation of sphingolipid metabolism in the renal cortex of rats with streptozotocin-induced diabetes and the effects of rapamycin. Nephrol. Dial. Transplant. 2011, 26, 1493–1502. [Google Scholar] [CrossRef]
- Wegner, M.-S.; Gruber, L.; Mattjus, P.; Geisslinger, G.; Grösch, S. The UDP-glucose ceramide glycosyltransferase (UGCG) and the link to multidrug resistance protein 1 (MDR1). BMC Cancer 2018, 18, 153. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, Z.; Chen, X.; Zhang, G.; Wang, Y.; Pan, L.; Yan, C.; Yang, G.; Zhao, L.; Han, J.; et al. Plasma lipidomics profiling reveals biomarkers for papillary thyroid cancer diagnosis. Front. Cell. Dev. Biol. 2021, 9, 682269. [Google Scholar] [CrossRef]
- Calzada, E.; Onguka, O.; Claypool, S.M. Phosphatidylethanolamine metabolism in health and disease. Int. Rev. Cell. Mol. Biol. 2016, 321, 29–88. [Google Scholar] [CrossRef]
- Barbeau, L.M.O.; Keulers, T.G.H.; Rouschop, K.M.A. Tumors responsive to autophagy-inhibition: Identification and biomarkers. Cancers 2020, 12, 2463. [Google Scholar] [CrossRef] [PubMed]
- Mowers, E.E.; Sharifi, M.N.; Macleod, K.F. Autophagy in cancer metastasis. Oncogene 2017, 36, 1619–1630. [Google Scholar] [CrossRef]
- Dacheux, M.A.; Norman, D.D.; Tigyi, G.J.; Lee, S.C. Emerging roles of lysophosphatidic acid receptor subtype 5 (LPAR5) in inflammatory diseases and cancer. Pharmacol. Ther. 2023, 245, 108414. [Google Scholar] [CrossRef]
- Peyruchaud, O.; Karin, N.J. Lysophosphatidic acid: Role in bone and bone cancer. In Bone and Cancer, Topics in Bone Biology (Topics in Bone Biology 5); Bronner, F., Farach-Carson, M.C., Eds.; Springer: London, UK, 2010; pp. 73–88. [Google Scholar] [CrossRef]
- Yen, C.-L.E.; Stone, S.J.; Koliwad, S.; Harris, C.; Farese, R.V., Jr. DGAT enzymes and triacylglycerol biosynthesis. J. Lipid Res. 2008, 49, 2283–2301. [Google Scholar] [CrossRef]
- Liput, K.P.; Lepczyński, A.; Ogłuszka, M.; Nawrocka, A.; Polawska, E.; Grzesiak, A.; Slaska, B.; Pareek, C.S.; Czarnik, U.; Pierzchała, M. Effects of dietary n–3 and n–6 polyunsaturated fatty acids in inflammation and cancerogenesis. Int. J. Mol. Sci. 2021, 22, 6965. [Google Scholar] [CrossRef]
- Broadfield, L.A.; Pane, A.A.; Talebi, A.; Swinnen, J.V.; Fendt, S.-M. Lipid metabolism in cancer: New perspectives and emerging mechanisms. Dev. Cell. 2021, 56, 1363–1393. [Google Scholar] [CrossRef]
- Purkayastha, K.; Dhar, R.; Pethusamy, K.; Srivastava, T.; Shankar, A.; Rath, G.K.; Karmakar, S. The issues and challenges with cancer biomarkers. J. Cancer Res. Ther. 2023, 19, S20–S35. [Google Scholar] [CrossRef]
- Semplonatto, J.R.; Lasalde-Ramírez, J.A.; Mahato, K.; Wang, J.; Gao, W. Wearable chemical sensors for biomarker discovery in the omics era. Nat. Rev. Chem. 2022, 6, 899–915. [Google Scholar] [CrossRef]
| Entry | Experimental m/z and Class | Theoretical m/z | Ions | Lipid Assignments | Proposed Formula | Reference/HMDB * ID LipidMaps ID |
|---|---|---|---|---|---|---|
| 1 | 289.2923 (OS and M-OS) | 289.2890 | [M+H-H2O]+ | 14-methylic-en-1- yn-3-ol | C21H38O | LMFA05000766 |
| 2 | 603.5379 (M-OS) | 603.5352 | [M+H-H2O]+ (DAG) or [M-RCOO]+ (TAG) | Glycerols | C39H72O5 | HMDB0007030, HMDB0007109, HMDB0007137, HMDB0007161, HMDB0007218 [60,61] |
| 3 | 664.4620 (OS) | 664.46 | [M+H2O+H]+ | Cer(d18:2/24:1)-Ceramide | C42H79NO3 | HMDB0240680 [62] |
| 4 | 648.4664 (OS and M-OS) | 648.4646 | [M+H+1]+ | PA (18:1/14:0) | C35H67O8P | HMDB0114921 LMGP10010882 |
| 5 | 649.4776 (M-OS) | 649.4803 | [M+H]+ | PA (16:0/16:0) | C35H69O8P | LMGP10010027 |
| 6 | 758.5730 (OS) | 758.5674 | [M+H]+ | PE-NMe (18:1/18:1) | C42H80NO8P | HMDB0010565 LMGP02010338 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pontes, J.G.d.M.; Jadranin, M.; Assalin, M.R.; Quintero Escobar, M.; Stanisic, D.; Costa, T.B.B.C.; van Helvoort Lengert, A.; Boldrini, É.; Morini da Silva, S.R.; Vidal, D.O.; et al. Lipidomics by Nuclear Magnetic Resonance Spectroscopy and Liquid Chromatography–High-Resolution Mass Spectrometry in Osteosarcoma: A Pilot Study. Metabolites 2024, 14, 416. https://doi.org/10.3390/metabo14080416
Pontes JGdM, Jadranin M, Assalin MR, Quintero Escobar M, Stanisic D, Costa TBBC, van Helvoort Lengert A, Boldrini É, Morini da Silva SR, Vidal DO, et al. Lipidomics by Nuclear Magnetic Resonance Spectroscopy and Liquid Chromatography–High-Resolution Mass Spectrometry in Osteosarcoma: A Pilot Study. Metabolites. 2024; 14(8):416. https://doi.org/10.3390/metabo14080416
Chicago/Turabian StylePontes, João Guilherme de Moraes, Milka Jadranin, Márcia Regina Assalin, Melissa Quintero Escobar, Danijela Stanisic, Tássia Brena Barroso Carneiro Costa, André van Helvoort Lengert, Érica Boldrini, Sandra Regina Morini da Silva, Daniel Onofre Vidal, and et al. 2024. "Lipidomics by Nuclear Magnetic Resonance Spectroscopy and Liquid Chromatography–High-Resolution Mass Spectrometry in Osteosarcoma: A Pilot Study" Metabolites 14, no. 8: 416. https://doi.org/10.3390/metabo14080416
APA StylePontes, J. G. d. M., Jadranin, M., Assalin, M. R., Quintero Escobar, M., Stanisic, D., Costa, T. B. B. C., van Helvoort Lengert, A., Boldrini, É., Morini da Silva, S. R., Vidal, D. O., Liu, L. H. B., Maschietto, M., & Tasic, L. (2024). Lipidomics by Nuclear Magnetic Resonance Spectroscopy and Liquid Chromatography–High-Resolution Mass Spectrometry in Osteosarcoma: A Pilot Study. Metabolites, 14(8), 416. https://doi.org/10.3390/metabo14080416








