Impaired Mitochondrial Energy Metabolism Regulated by p70S6K: A Putative Pathological Feature in Alzheimer’s Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Plasmid Transfection and Cell Treatments
2.3. RNA Extraction, Reverse Transcription, and RT–qPCR
2.4. Protein Extraction and Western Blot
2.5. HPLC Determination of ATP Level
2.6. Flow Cytometry Analysis of Reactive Oxygen Species (ROS)
2.7. Analysis of Activities of Electron Transport Chain (ETC) Complexes
2.8. Laser Confocal Microscopic Detection of Mitochondrial Membrane Potential (MMP)
2.9. Bioinformatics Analysis
2.10. Statistical Analysis
3. Results
3.1. Aβ42 Affects Diverse Pathways in SH–SY5Y Cells and Is Implicated in Multiple Diseases
3.2. Impaired Mitochondrial Energy Metabolism in AD Revealed by Transcriptome Analysis
3.3. Influence of Aβ42 and p70S6K on OXPHOS in SH–SY5Y Cells
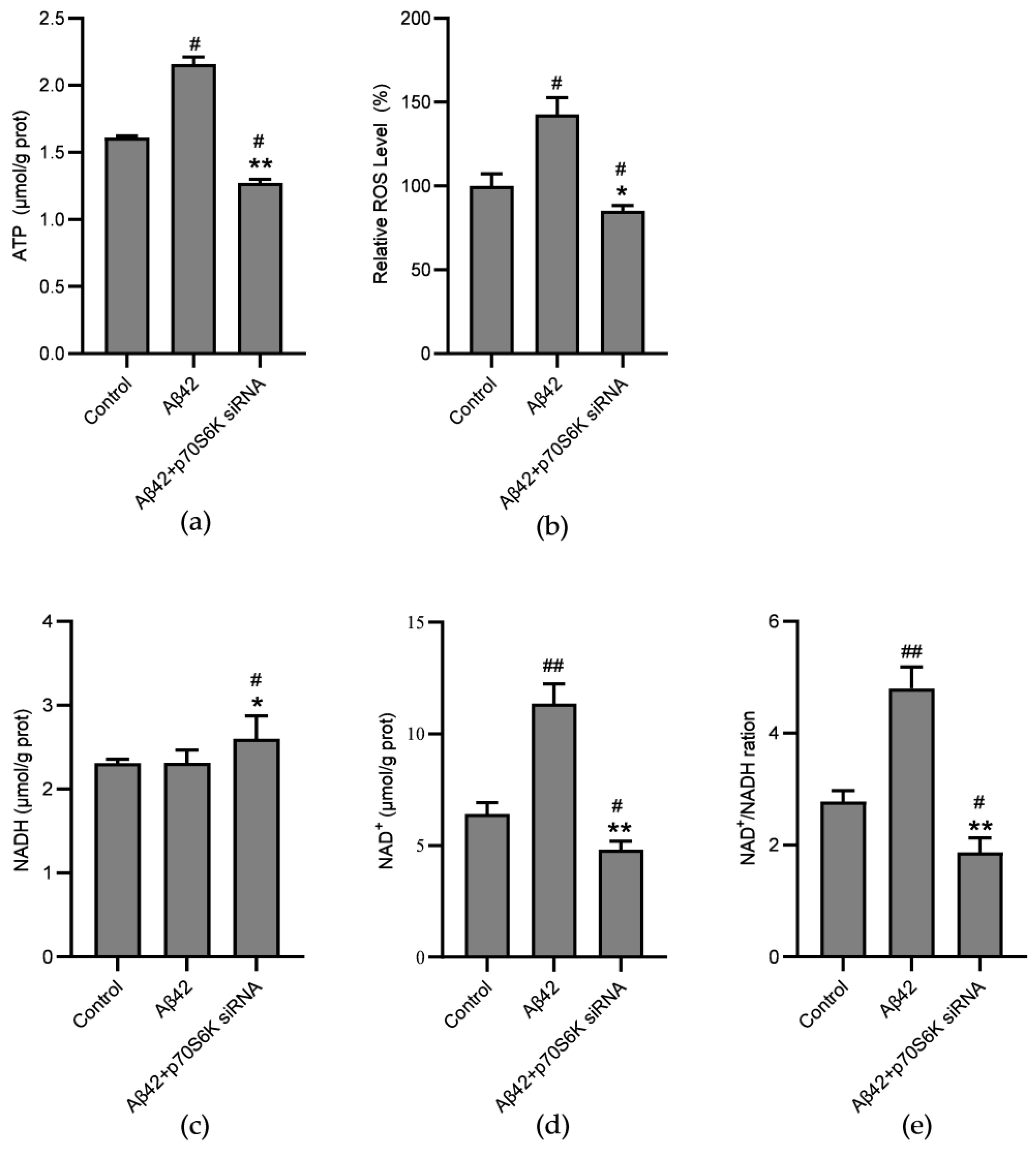
3.4. Influence of Aβ42 and p70S6K on Levels of NAD+, NADH, ATP, and ROS
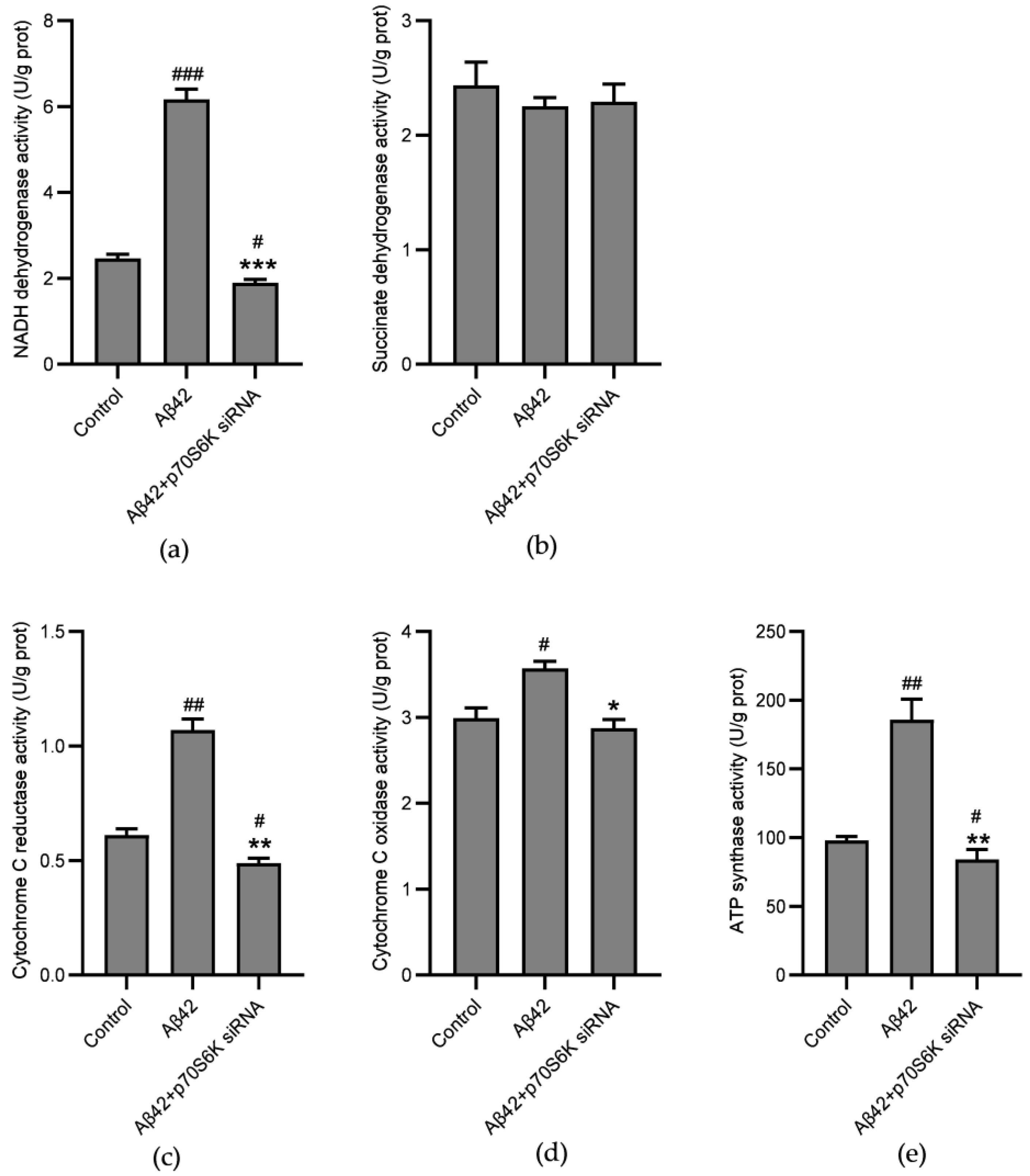
3.5. Influence of Aβ42 and p70S6K on Activities of Mitochondrial ETC Complexes
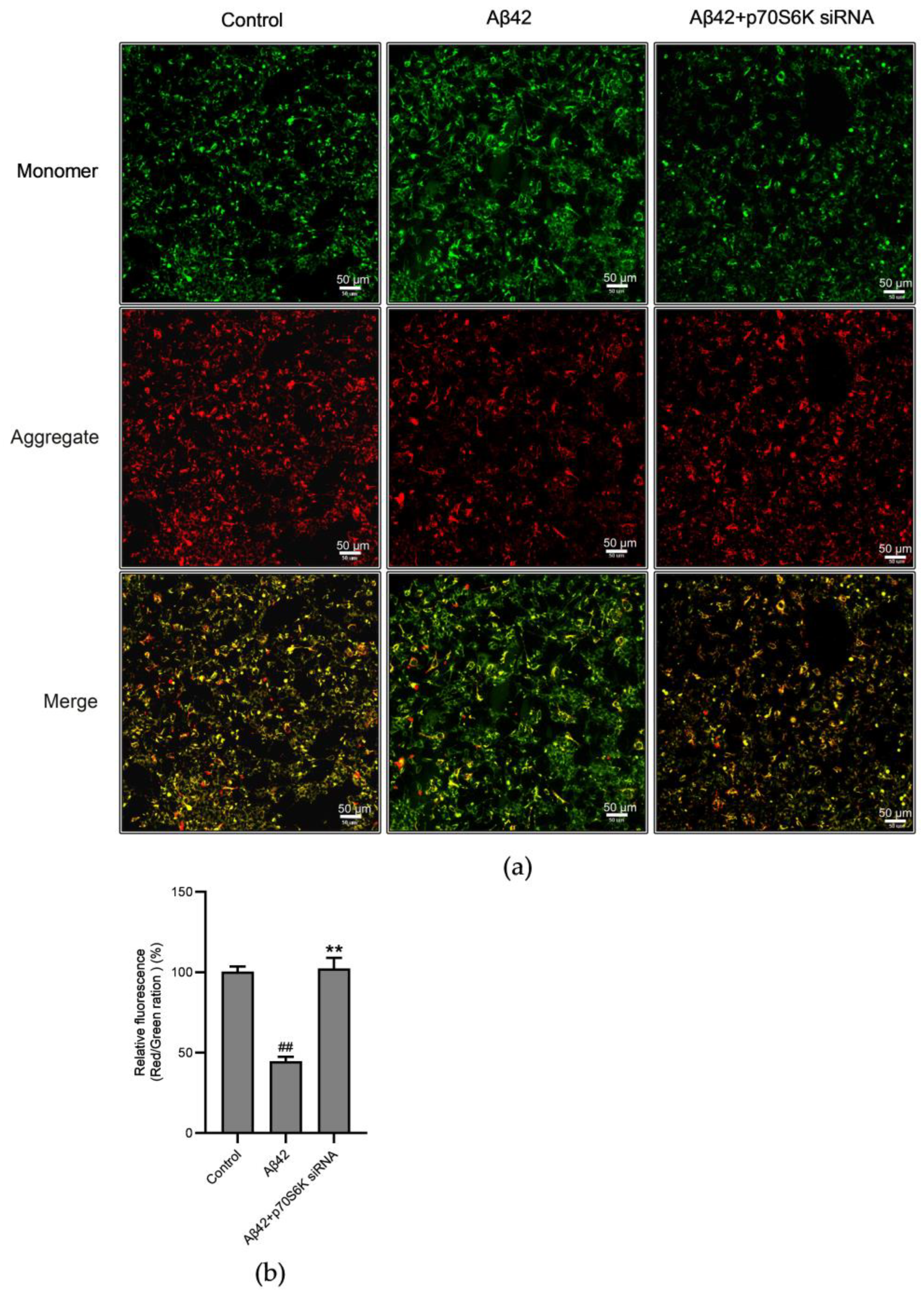
3.6. Influence of Aβ42 and p70S6K on Mitochondrial Membrane Potential (MMP)
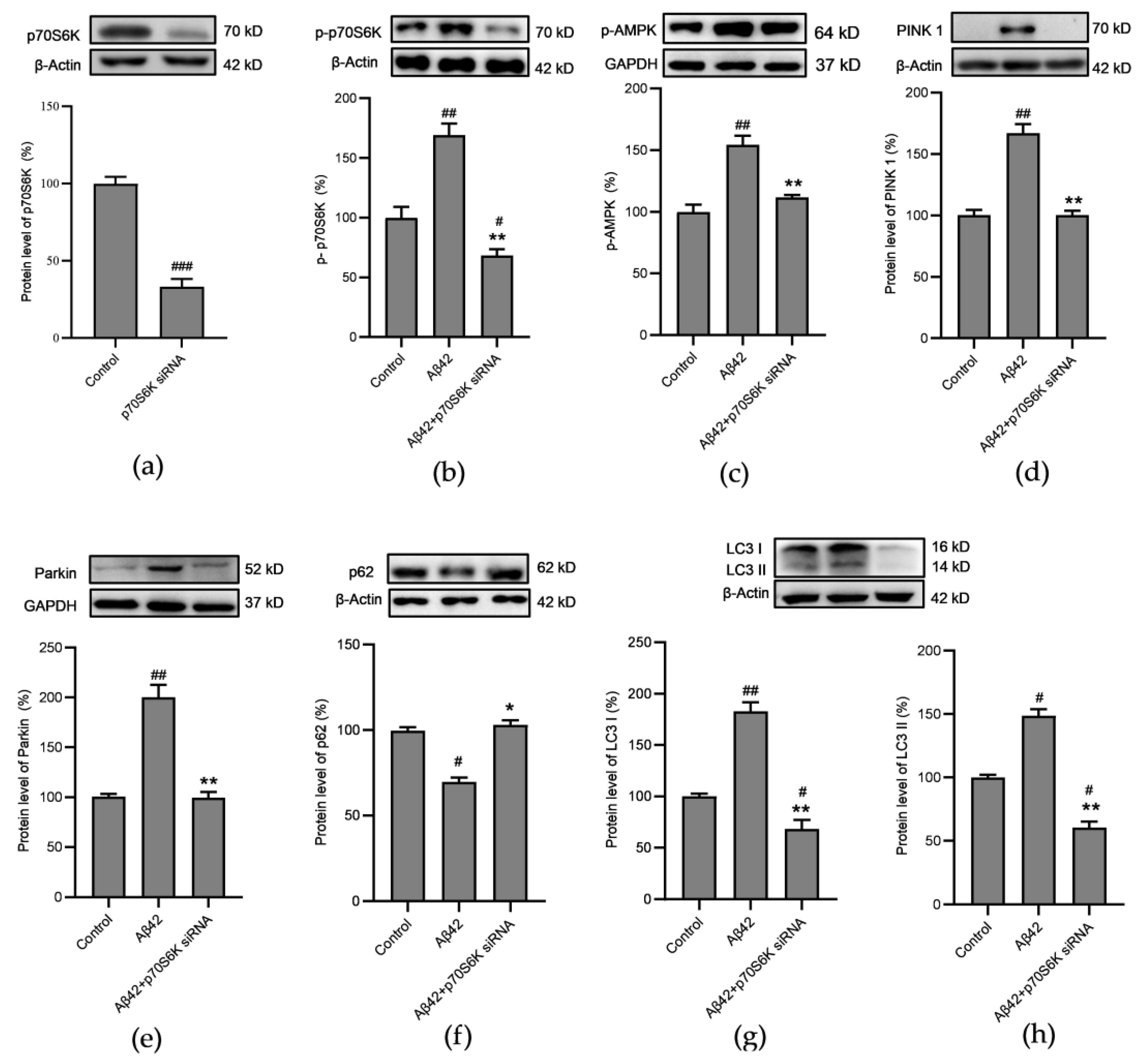
3.7. Influence of Aβ42 and p70S6K on Autophagy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ma, C.; Hong, F.; Yang, S. Amyloidosis in Alzheimer’s Disease: Pathogeny, Etiology, and Related Therapeutic Directions. Molecules 2022, 27, 1210. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Shang, Y.C.; Wang, E.L.; Xu, X.X.; Zhang, Q.Y.; Qian, C.X.; Yang, Z.; Wu, S.; Zhang, T. MST1 mediates neuronal loss and cognitive deficits: A noaavel therapeutic target for Alzheimer’s disease. Prog. Neurobiol. 2022, 214, 102280. [Google Scholar] [CrossRef] [PubMed]
- Tzioras, M.; McGeachan, R.I.; Durrant, C.S.; Spires-Jones, T.L. Synaptic degeneration in Alzheimer disease. Nat. Rev. Neurol. 2023, 19, 19–38. [Google Scholar] [CrossRef] [PubMed]
- Pao, P.C.; Patnaik, D.; Watson, L.A.; Gao, F.; Pan, L.; Wang, J.; Adaikkan, C.; Penney, J.; Cam, H.P.; Huang, W.C.; et al. HDAC1 modulates OGG1-initiated oxidative DNA damage repair in the aging brain and Alzheimer’s disease. Nat. Commun. 2020, 11, 2484. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, D.A.; Halliwell, B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat. Rev. Neurosci. 2019, 20, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zong, S.; Cui, X.; Wang, X.; Wu, S.; Wang, L.; Liu, Y.; Lu, Z. The effects of microglia-associated neuroinflammation on Alzheimer’s disease. Front. Immunol. 2023, 14, 1117172. [Google Scholar] [CrossRef]
- Sharma, C.; Kim, S.; Nam, Y.; Jung, U.J.; Kim, S.R. Mitochondrial Dysfunction as a Driver of Cognitive Impairment in Alzheimer’s Disease. Int. J. Mol. Sci. 2021, 22, 4850. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, R.; Qin, Y.; Wang, T. Brain metabolism in Alzheimer’s disease: Biological mechanisms of exercise. Transl. Neurodegener. 2023, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, R.H. The Alzheimer’s Disease Mitochondrial Cascade Hypothesis: A Current Overview. J. Alzheimers Dis. 2023, 92, 751–768. [Google Scholar] [CrossRef]
- Ashleigh, T.; Swerdlow, R.H.; Beal, M.F. The role of mitochondrial dysfunction in Alzheimer’s disease pathogenesis. Alzheimers Dement. 2023, 19, 333–342. [Google Scholar] [CrossRef]
- Bhatia, V.; Sharma, S. Role of mitochondrial dysfunction, oxidative stress and autophagy in progression of Alzheimer’s disease. J. Neurol. Sci. 2021, 421, 117253. [Google Scholar] [CrossRef]
- Cunnane, S.C.; Trushina, E.; Morland, C.; Prigione, A.; Casadesus, G.; Andrews, Z.B.; Beal, M.F.; Bergersen, L.H.; Brinton, R.D.; de la Monte, S.; et al. Brain energy rescue: An emerging therapeutic concept for neurodegenerative disorders of ageing. Nat. Rev. Drug Discov. 2020, 19, 609–633. [Google Scholar] [CrossRef]
- Chu, X.Y.; Xu, Y.Y.; Tong, X.Y.; Wang, G.; Zhang, H.Y. The Legend of ATP: From Origin of Life to Precision Medicine. Metabolites 2022, 12, 461. [Google Scholar] [CrossRef]
- Yu, L.J.; Jin, J.L.; Xu, Y.; Zhu, X.L. Aberrant energy metabolism in Alzheimer’s disease. J. Transl. Intern. Med. 2022, 10, 197–206. [Google Scholar] [CrossRef]
- Nolfi-Donegan, D.; Braganza, A.; Shiva, S. Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol. 2020, 37, 101674. [Google Scholar] [CrossRef]
- Bai, R.R.; Guo, J.A.; Ye, X.Y.; Xie, Y.Y.; Xie, T. Oxidative stress: The core pathogenesis and mechanism of Alzheimer’s disease. Ageing Res. Rev. 2022, 77, 101619. [Google Scholar] [CrossRef]
- Fang, E.F.; Hou, Y.J.; Palikaras, K.; Adriaanse, B.A.; Kerr, J.S.; Yang, B.M.; Lautrup, S.; Hasan-Olive, M.M.; Caponio, D.; Dan, X.L.; et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 2019, 22, 401–412. [Google Scholar] [CrossRef]
- Fenton, T.R.; Gout, I.T. Functions and regulation of the 70 kDa ribosomal S6 kinases. Int. J. Biochem. Cell Biol. 2011, 43, 47–59. [Google Scholar] [CrossRef]
- Ou, Z.; Kong, X.; Sun, X.; He, X.; Zhang, L.; Gong, Z.; Huang, J.; Xu, B.; Long, D.; Li, J.; et al. Metformin treatment prevents amyloid plaque deposition and memory impairment in APP/PS1 mice. Brain Behav. Immun. 2018, 69, 351–363. [Google Scholar] [CrossRef]
- Wijtenburg, S.A.; Kapogiannis, D.; Korenic, S.A.; Mullins, R.J.; Tran, J.; Gaston, F.E.; Chen, S.; Mustapic, M.; Hong, L.E.; Rowland, L.M. Brain insulin resistance and altered brain glucose are related to memory impairments in schizophrenia. Schizophr. Res. 2019, 208, 324–330. [Google Scholar] [CrossRef]
- Caldwell, A.B.; Anantharaman, B.G.; Ramachandran, S.; Nguyen, P.; Liu, Q.; Trinh, I.; Galasko, D.R.; Desplats, P.A.; Wagner, S.L.; Subramaniam, S. Transcriptomic profiling of sporadic Alzheimer’s disease patients. Mol. Brain 2022, 15, 83. [Google Scholar] [CrossRef]
- Lanning, N.J.; Looyenga, B.D.; Kauffman, A.L.; Niemi, N.M.; Sudderth, J.; DeBerardinis, R.J.; MacKeigan, J.P. A Mitochondrial RNAi Screen Defines Cellular Bioenergetic Determinants and Identifies an Adenylate Kinase as a Key Regulator of ATP Levels. Cell Rep. 2014, 7, 907–917. [Google Scholar] [CrossRef]
- Kong, F.; Binas, B.; Moon, J.H.; Kang, S.S.; Kim, H.J. Differential expression of adenylate kinase 4 in the context of disparate stress response strategies of HEK293 and HepG2 cells. Arch. Biochem. Biophys. 2013, 533, 11–17. [Google Scholar] [CrossRef]
- Li, Z.Q.; Gao, Z.Y.; Sun, T.; Zhang, S.P.; Yang, S.N.; Zheng, M.L.; Shen, H. Meteorin-like/Metrnl, a novel secreted protein implicated in inflammation, immunology, and metabolism: A comprehensive review of preclinical and clinical studies. Front. Immunol. 2023, 14, 1098570. [Google Scholar] [CrossRef]
- Reddy, P.H.; McWeeney, S.; Park, B.S.; Manczak, M.; Gutala, R.V.; Partovi, D.; Jung, Y.; Yau, V.; Searles, R.; Mori, M.; et al. Gene expression profiles of transcripts in amyloid precursor protein transgenic mice: Up-regulation of mitochondrial metabolism and apoptotic genes is an early cellular change in Alzheimer’s disease. Hum. Mol. Genet. 2004, 13, 1225–1240. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Reed, T.; Newman, S.F.; Sultana, R. Roles of amyloid beta-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer’s disease and mild cognitive impairment. Free Radic. Biol. Med. 2007, 43, 658–677. [Google Scholar] [CrossRef]
- Kim, T.; Yi, D.; Byun, M.S.; Ahn, H.; Jung, J.H.; Kong, N.; Kim, M.J.; Jung, G.; Lee, J.Y.; Lee, Y.S.; et al. Synergistic interaction of high blood pressure and cerebral beta-amyloid on tau pathology. Alzheimers Res. Ther. 2022, 14, 193. [Google Scholar] [CrossRef]
- Dong, Z.; Xu, M.; Sun, X.; Wang, X. Mendelian randomization and transcriptomic analysis reveal an inverse causal relationship between Alzheimer’s disease and cancer. J. Transl. Med. 2023, 21, 527. [Google Scholar] [CrossRef]
- Burillo, J.; Marques, P.; Jimenez, B.; Gonzalez-Blanco, C.; Benito, M.; Guillen, C. Insulin Resistance and Diabetes Mellitus in Alzheimer’s Disease. Cells 2021, 10, 1236. [Google Scholar] [CrossRef]
- Dewanjee, S.; Chakraborty, P.; Bhattacharya, H.; Chacko, L.; Singh, B.; Chaudhary, A.; Javvaji, K.; Pradhan, S.R.; Vallamkondu, J.; Dey, A.; et al. Altered glucose metabolism in Alzheimer’s disease: Role of mitochondrial dysfunction and oxidative stress. Free Radic. Biol. Med. 2022, 193, 134–157. [Google Scholar] [CrossRef]
- Chen, Z.; Wang, H.; Wang, Q. Therapeutic potential of NADH: In neurodegenerative diseases characterizde by mitochondrial dysfunction. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi = J. Clin. Otorhinolaryngol. Head Neck Surg. 2024, 38, 57–62. [Google Scholar] [CrossRef]
- Ionescu-Tucker, A.; Cotman, C.W. Emerging roles of oxidative stress in brain aging and Alzheimer’s disease. Neurobiol. Aging 2021, 107, 86–95. [Google Scholar] [CrossRef]
- Zhang, L.; Fang, Y.; Zhao, X.; Zheng, Y.; Ma, Y.; Li, S.; Huang, Z.; Li, L. miR-204 silencing reduces mitochondrial autophagy and ROS production in a murine AD model via the TRPML1-activated STAT3 pathway. Mol. Ther. Nucleic Acids 2021, 24, 822–831. [Google Scholar] [CrossRef]
- Saito, T.; Chiku, T.; Oka, M.; Wada-Kakuda, S.; Nobuhara, M.; Oba, T.; Shinno, K.; Abe, S.; Asada, A.; Sumioka, A.; et al. Disulfide bond formation in microtubule-associated tau protein promotes tau accumulation and toxicity in vivo. Hum. Mol. Genet. 2021, 30, 1955–1967. [Google Scholar] [CrossRef]
- Seledtsov, V.I.; von Delwig, A.A. Therapeutic Stimulation of Glycolytic ATP Production for Treating ROS-Mediated Cellular Senescence. Metabolites 2022, 12, 1160. [Google Scholar] [CrossRef]
- Bang, E.; Kim, D.H.; Chung, H.Y. Protease-activated receptor 2 induces ROS-mediated inflammation through Akt-mediated NF-κB and FoxO6 modulation during skin photoaging. Redox Biol. 2021, 44, 102022. [Google Scholar] [CrossRef]
- Cheignon, C.; Tomas, M.; Bonnefont-Rousselot, D.; Faller, P.; Hureau, C.; Collin, F. Oxidative stress and the amyloid beta peptide in Alzheimer’s disease. Redox Biol. 2018, 14, 450–464. [Google Scholar] [CrossRef]
- Chen, J.X.; Yan, S.D. Amyloid-beta-induced mitochondrial dysfunction. J. Alzheimers Dis. 2007, 12, 177–184. [Google Scholar] [CrossRef]
- Troutwine, B.R.; Strope, T.A.; Franczak, E.; Lysaker, C.R.; Hamid, L.; Mansel, C.; Stopperan, J.A.; Gouvion, C.M.; Haeri, M.; Swerdlow, R.H.; et al. Mitochondrial function and Aβ in Alzheimer’s disease postmortem brain. Neurobiol. Dis. 2022, 171, 105781. [Google Scholar] [CrossRef] [PubMed]
- Pradeepkiran, J.A.; Reddy, P.H. Defective mitophagy in Alzheimer’s disease. Ageing Res. Rev. 2020, 64, 101191. [Google Scholar] [CrossRef] [PubMed]
- Wani, A.; Al Rihani, S.B.; Sharma, A.; Weadick, B.; Govindarajan, R.; Khan, S.U.; Sharma, P.R.; Dogra, A.; Nandi, U.; Reddy, C.N.; et al. Crocetin promotes clearance of amyloid-beta by inducing autophagy via the STK11/LKB1-mediated AMPK pathway. Autophagy 2021, 17, 3813–3832. [Google Scholar] [CrossRef]
- Neurohr, J.M.; Paulson, E.T.; Kinsey, S.T. A higher mitochondrial content is associated with greater oxidative damage, oxidative defenses, protein synthesis and ATP turnover in resting skeletal muscle. J. Exp. Biol. 2021, 224, jeb242462. [Google Scholar] [CrossRef]




| Gene | Sequence |
|---|---|
| SMAD6–F | GTGAATTCTCAGACGCCAGC |
| SMAD6–R | CTGCCCTGAGGTAGGTCGTA |
| AK4–F | CCCTCCTAGCGGAAGGGTAT |
| AK4–R | GCACTCCTCGGCTCTTGTA |
| METRNL–F | GCTGGTTAGGAGGCACAGG |
| METRNL–R | AGCCTCGAACGGCGAAG |
| mt–ND4–F | AAGTCAAAAAGCTATTA |
| mt–ND4–R | CTTACATCCTCATTACTATTC |
| mt–ND4L–F | CTCACACCTCATATCCTCCCTAC |
| mt–ND4L–R | GCTAAGAGGGAGTGGGTGTT |
| mt–ND5–F | ACCGCTAACAACCTATTCCAACTG |
| mt–ND5–R | GATTGCTTGAATGGCTGCTGTG |
| β–Actin–F | TCTTCCAGCCTTCCTTCCTG |
| β–Actin–R | CAATGCCAGGGTACATGGTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gu, W.; Cong, X.; Pei, Y.; Che Ajuyo, N.M.; Min, Y.; Wang, D. Impaired Mitochondrial Energy Metabolism Regulated by p70S6K: A Putative Pathological Feature in Alzheimer’s Disease. Metabolites 2024, 14, 369. https://doi.org/10.3390/metabo14070369
Gu W, Cong X, Pei Y, Che Ajuyo NM, Min Y, Wang D. Impaired Mitochondrial Energy Metabolism Regulated by p70S6K: A Putative Pathological Feature in Alzheimer’s Disease. Metabolites. 2024; 14(7):369. https://doi.org/10.3390/metabo14070369
Chicago/Turabian StyleGu, Wenyu, Xinli Cong, Yechun Pei, Nuela Manka’a Che Ajuyo, Yi Min, and Dayong Wang. 2024. "Impaired Mitochondrial Energy Metabolism Regulated by p70S6K: A Putative Pathological Feature in Alzheimer’s Disease" Metabolites 14, no. 7: 369. https://doi.org/10.3390/metabo14070369
APA StyleGu, W., Cong, X., Pei, Y., Che Ajuyo, N. M., Min, Y., & Wang, D. (2024). Impaired Mitochondrial Energy Metabolism Regulated by p70S6K: A Putative Pathological Feature in Alzheimer’s Disease. Metabolites, 14(7), 369. https://doi.org/10.3390/metabo14070369







