Changes in Metabolite Profiles of Chinese Soy Sauce at Different Time Durations of Fermentation Studied by 1H-NMR-Based Metabolomics
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Preparation
2.3. 1H-NMR Spectroscopic Analysis
2.4. 1H-NMR Data Analysis
2.5. Multivariate Data Analysis
3. Results and Discussion
3.1. 1H-NMR Spectroscopy
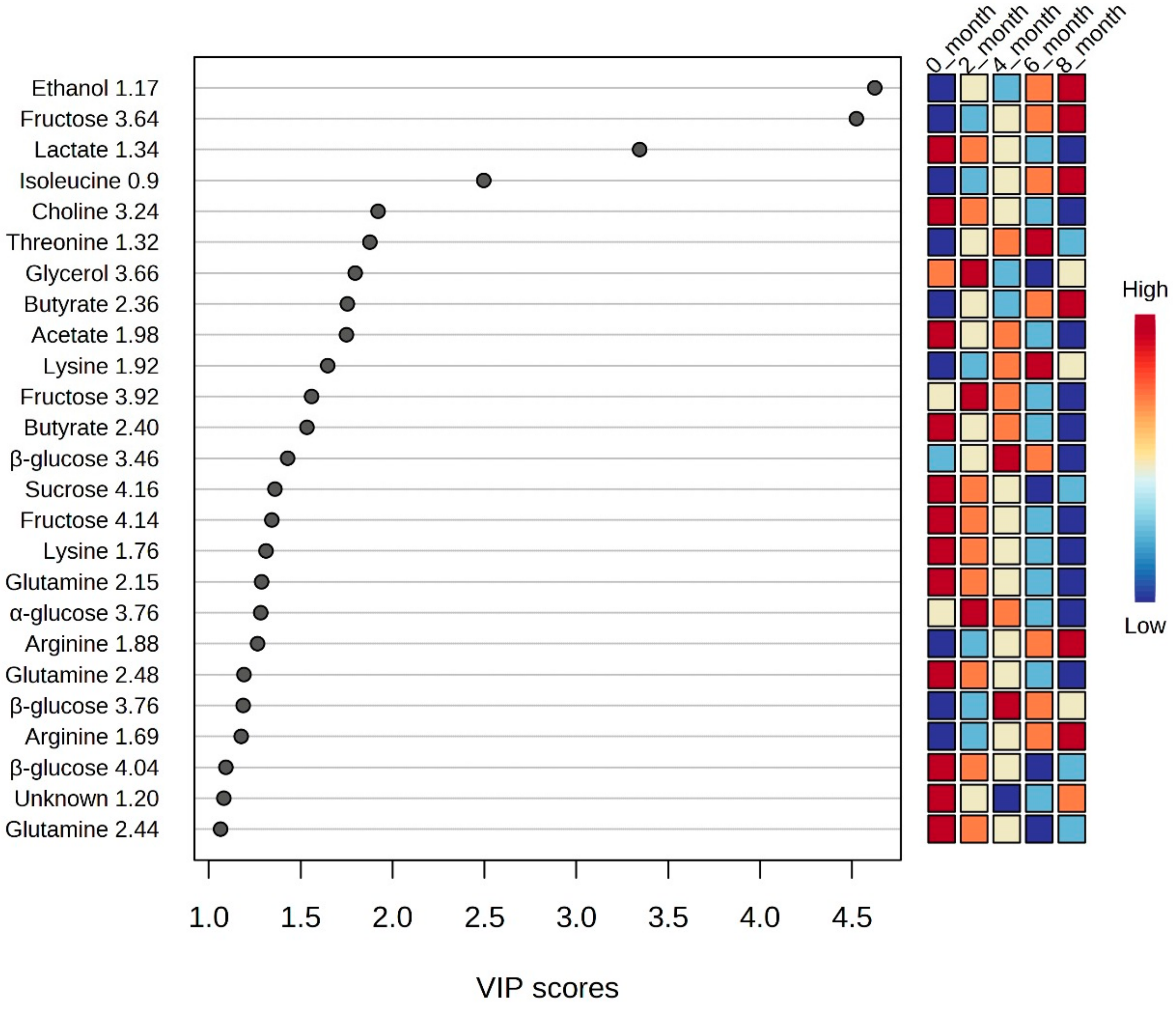
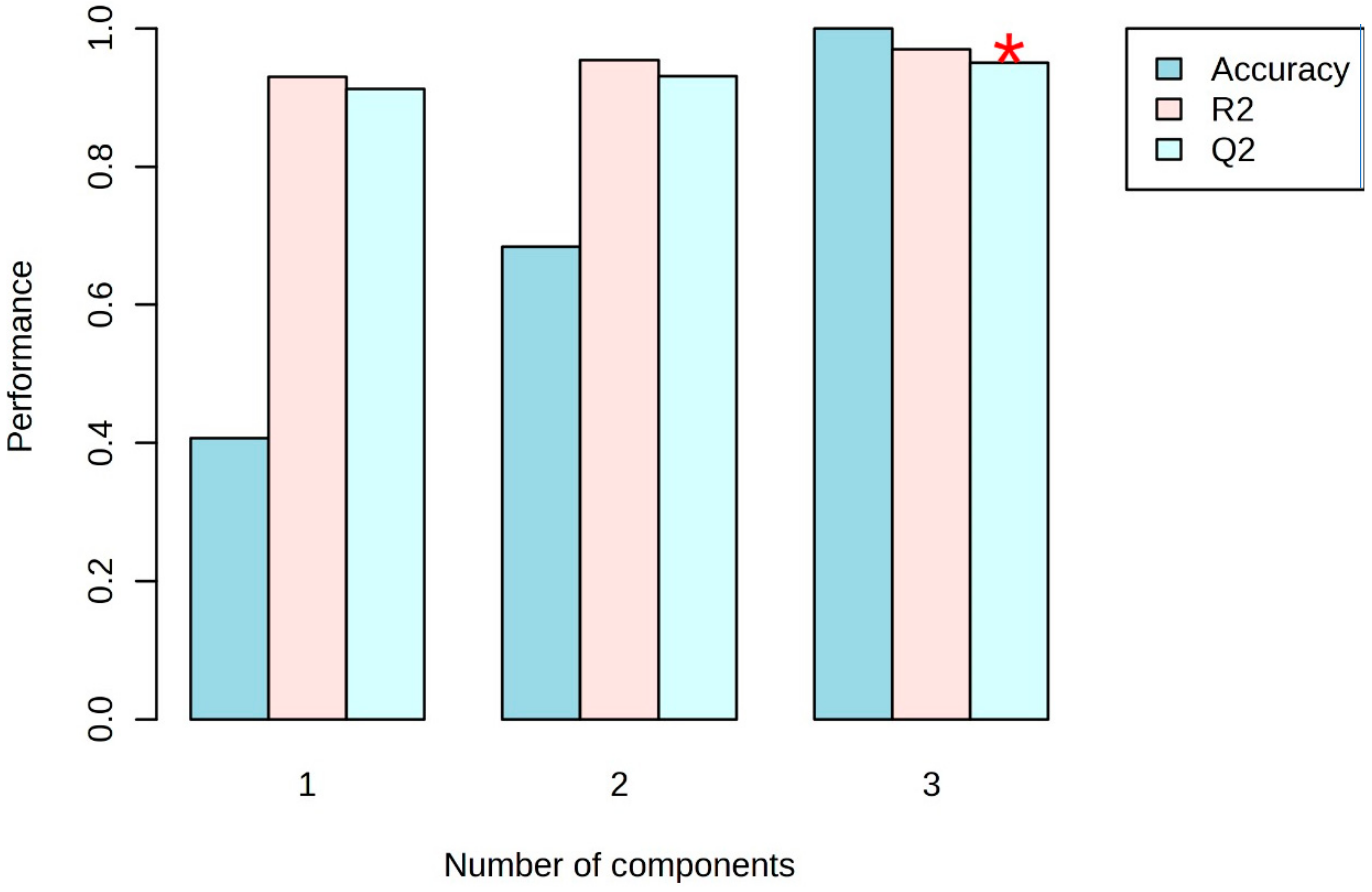
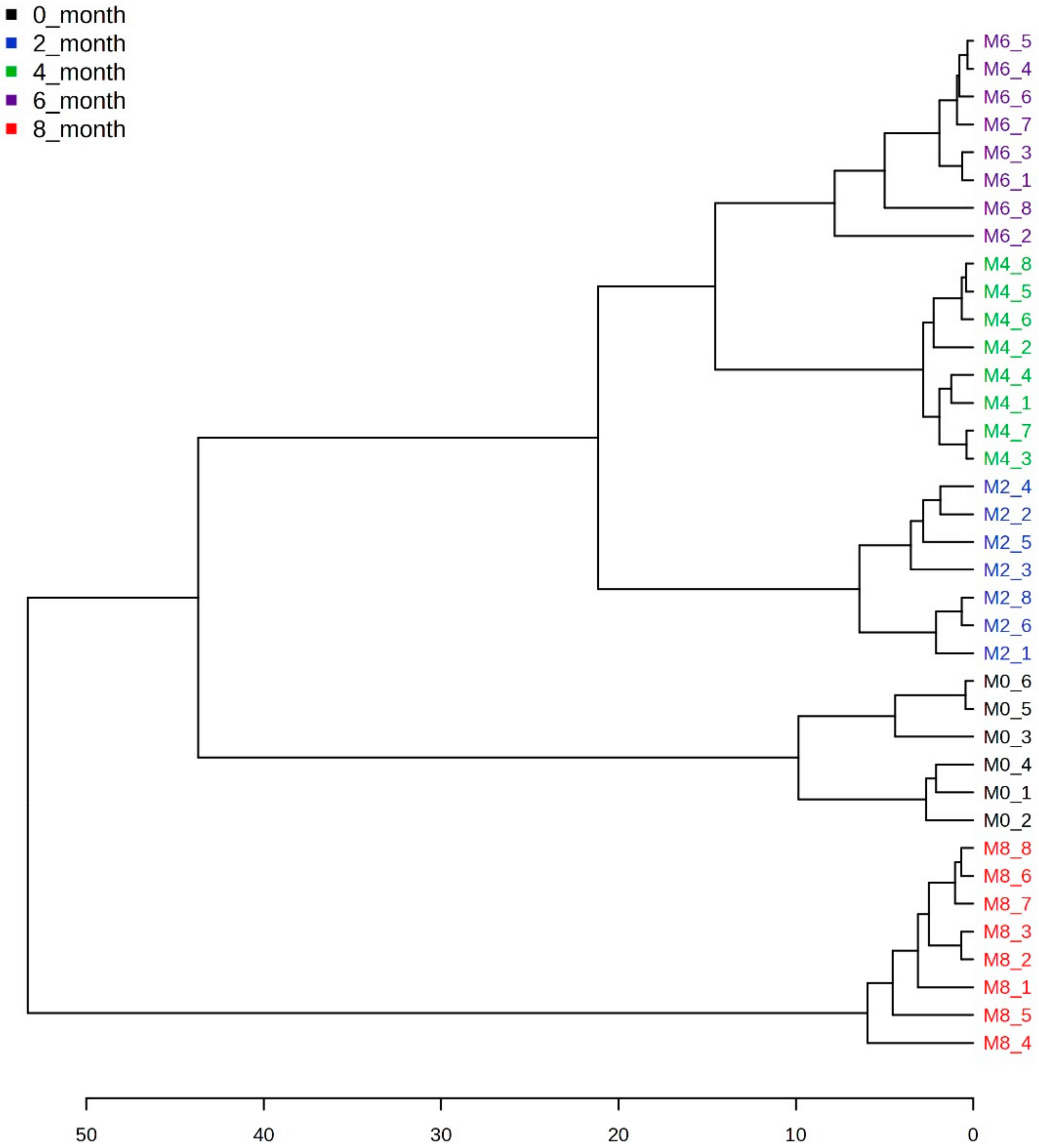
3.2. Multivariate Statistical Analysis
3.3. Complex Carbohydrates to Simple Sugars
3.4. Narrative of Ethanol and Glycerol
3.5. Proteolysis Giving Birth to Amino Acids
3.6. Footsteps of Lactate, Butyrate and Acetate
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, G.; Yao, Y.; Wang, X.; Hou, L.; Wang, C.; Cao, X. Functional properties of soy sauce and metabolism genes of strains for fermentation. Int. J. Food Sci. Technol. 2013, 48, 903–909. [Google Scholar] [CrossRef]
- Gao, X.; Zhao, X.; Hu, F.; Fu, J.; Zhang, Z.; Liu, Z.; Wang, B.; He, R.; Ma, H.; Ho, C.-T. The latest advances on soy sauce research in the past decade: Emphasis on the advances in China. Food Res. Int. 2023, 173, 113407. [Google Scholar] [CrossRef] [PubMed]
- Devanthi, P.V.P.; Gkatzionis, K. Soy sauce fermentation: Microorganisms, aroma formation, and process modification. Food Res. Int. 2019, 120, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Cui, R.; Liu, X.; Zheng, X.; Yao, Y.; Zhao, G. Examining the impact of Tetragenococcus halophilus, Zygosaccharomyces rouxii, and Starmerella etchellsii on the quality of soy sauce: A comprehensive review of microbial population dynamics in fermentation. Crit. Rev. Food Sci. Nutr. 2023. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Komiyama, A.; Sonomoto, K.; Ishizaki, A.; Hall, S.J.; Stanbury, P.F. Two different pathways for D-xylose metabolism and the effect of xylose concentration on the yield coefficient of L-lactate in mixed-acid fermentation by the lactic acid bacterium Lactococcus lactis IO-1. Appl. Microbiol. Biotechnol. 2002, 60, 160–167. [Google Scholar] [CrossRef]
- Feng, Y.; Cai, Y.; Su, G.; Zhao, H.; Wang, C.; Zhao, M. Evaluation of aroma differences between high-salt liquid-state fermentation and low-salt solid-state fermentation soy sauces from China. Food Chem. 2014, 145, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lin, M.; Xu, M.; Yang, S.T. Anaerobic Fermentation for Production of Carboxylic Acids as Bulk Chemicals from Renewable Biomass. Adv. Biochem. Eng. Biotechnol. 2016, 156, 323–361. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Deng, Y.; Jin, Y.; Liu, Y.; Xia, B.; Sun, Q. Dynamics of microbial community during the extremely long-term fermentation process of a traditional soy sauce. J. Sci. Food Agric. 2017, 97, 3220–3227. [Google Scholar] [CrossRef] [PubMed]
- Ross, R.P.; Morgan, S.; Hill, C. Preservation and fermentation: Past, present and future. Int. J. Food Microbiol. 2002, 79, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiong, S.; Du, T.; Xiao, M.; Peng, Z.; Xie, M.; Guan, Q.; Xiong, T. Effects of microbial succession on the dynamics of flavor metabolites and physicochemical properties during soy sauce koji making. Food Biosci. 2023, 53, 102636. [Google Scholar] [CrossRef]
- Chong, S.; Ilham, Z.; Samsudin, N.; Soumaya, S.; Wan-Mohtar, W. Microbial consortia and up-to-date technologies in global soy sauce production: A review. Int. Food Res. J. 2023, 30, 1–24. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, J.; Zhang, L.; Nie, Y.; Xu, Y. Systematic analysis of key fermentation parameters influencing biogenic amines production in spontaneous fermentation of soy sauce. Food Biosci. 2023, 52, 102484. [Google Scholar] [CrossRef]
- Ma, Y.; Gao, Y.; Xu, Y.; Zhou, H.; Zhou, K.; Li, C.; Xu, B. Microbiota dynamics and volatile metabolite generation during sausage fermentation. Food Chem. 2023, 423, 136297. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, D.-S.; Son, Y.; Le, H.-G.; Jo, S.W.; Lee, J.; Song, Y.; Kim, H.-J. Effects of salt treatment time on the metabolites, microbial composition, and quality characteristics of the soy sauce moromi extract. Foods 2021, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, Y.; Shu, L.; He, Y. Study on metabolites of Bacillus producing soy sauce-like aroma in Jiang-flavor Chinese spirits. Food Sci. Nutr. 2020, 8, 97–103. [Google Scholar] [CrossRef]
- Sassi, S.; Ilham, Z.; Jamaludin, N.S.; Halim-Lim, S.A.; Shin Yee, C.; Weng Loen, A.W.; Poh Suan, O.; Ibrahim, M.F.; Wan-Mohtar, W.A.A.Q.I. Critical optimized conditions for gamma-aminobutyric acid (GABA)-Producing Tetragenococcus halophilus strain KBC from a commercial soy sauce moromi in batch fermentation. Fermentation 2022, 8, 409. [Google Scholar] [CrossRef]
- Zhao, G.; Ding, L.-L.; Yao, Y.; Cao, Y.; Pan, Z.-H.; Kong, D.-H. Extracellular proteome analysis and flavor formation during soy sauce fermentation. Front. Microbiol. 2018, 9, 1872. [Google Scholar] [CrossRef] [PubMed]
- Diez-Simon, C.; Eichelsheim, C.; Mumm, R.; Hall, R.D. Chemical and sensory characteristics of soy sauce: A review. J. Agric. Food Chem. 2020, 68, 11612–11630. [Google Scholar] [CrossRef]
- Feng, Y.; Wu, W.; Chen, T.; Huang, M.; Zhao, M. Exploring the core functional microbiota related with flavor compounds in fermented soy sauce from different sources. Food Res. Int. 2023, 173, 113456. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chen, X.; Lu, J.; Wu, D. Evaluation of the differences between low-salt solid-state fermented soy sauce and high-salt diluted-state fermented soy sauce in China: From taste-active compounds and aroma-active compounds to sensory characteristics. J. Sci. Food Agric. 2023, 104, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Byeon, Y.S.; Heo, J.; Park, K.; Chin, Y.-W.; Hong, S.-p.; Lim, S.-D.; Kim, S.S. Consumer Preference of Traditional Korean Soy Sauce (Ganjang) and Its Relationship with Sensory Attributes and Physicochemical Properties. Foods 2023, 12, 2361. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Liu, E.; Zhang, J.; Yang, L.; Huang, Q.; Chen, S.; Ma, H.; Ho, C.T.; Liao, L. Accelerating aroma formation of raw soy sauce using low intensity sonication. Food Chem. 2020, 329, 127118. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; He, S.; Lu, Y.; Gan, J.; Tao, N.; Wang, X.; Jiang, Z.; Hong, Y.; Xu, C. Metabolomics mechanism of traditional soy sauce associated with fermentation time. Food Sci. Hum. Wellness 2022, 11, 297–304. [Google Scholar] [CrossRef]
- Hoang, N.X.; Ferng, S.; Ting, C.-H.; Lu, Y.-C.; Yeh, Y.-F.; Lai, Y.-R.; Chiou, R.Y.-Y.; Hwang, J.-Y.; Hsu, C.-K. Effect of initial 5 days fermentation under low salt condition on the quality of soy sauce. LWT 2018, 92, 234–241. [Google Scholar] [CrossRef]
- Sassi, S.; Wan-Mohtar, W.A.A.Q.I.; Jamaludin, N.S.; Ilham, Z. Recent progress and advances in soy sauce production technologies: A review. J. Food Process. Preserv. 2021, 45, e15799. [Google Scholar] [CrossRef]
- Gao, X.; Cui, C.; Ren, J.; Zhao, H.; Zhao, Q.; Zhao, M. Changes in the chemical composition of traditional Chinese-type soy sauce at different stages of manufacture and its relation to taste. Int. J. Food Sci. Technol. 2011, 46, 243–249. [Google Scholar] [CrossRef]
- Putri, S.P.; Ikram, M.M.M.; Sato, A.; Dahlan, H.A.; Rahmawati, D.; Ohto, Y.; Fukusaki, E. Application of gas chromatography-mass spectrometry-based metabolomics in food science and technology. J. Biosci. Bioeng. 2022, 133, 425–435. [Google Scholar] [CrossRef] [PubMed]
- Truzzi, E.; Marchetti, L.; Fratagnoli, A.; Rossi, M.C.; Bertelli, D. Novel application of 1H-NMR spectroscopy coupled with chemometrics for the authentication of dark chocolate. Food Chem. 2023, 404, 134522. [Google Scholar] [CrossRef] [PubMed]
- Kamal, G.M.; Uddin, J.; Tahir, M.S.; Khalid, M.; Ahmad, S.; Hussain, A.I. Nuclear Magnetic Resonance Spectroscopy in Food Analysis. In Techniques to Measure Food Safety and Quality: Microbial, Chemical, and Sensory; Khan, M.S., Shafiur Rahman, M., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 137–168. [Google Scholar]
- Nurani, L.H.; Rohman, A.; Windarsih, A.; Guntarti, A.; Riswanto, F.D.O.; Lukitaningsih, E.; Fadzillah, N.A.; Rafi, M. Metabolite Fingerprinting Using 1H-NMR Spectroscopy and Chemometrics for Classification of Three Curcuma Species from Different Origins. Molecules 2021, 26, 7626. [Google Scholar] [CrossRef]
- European Union. Commission Regulation (EC) No 889/2008 of 5 September 2008 laying down detailed rules for the implementation of Council Regulation (EC) No 834/2007 on organic production and labelling of organic products with regard to organic production, labelling and control. Off. J. Eur. Union 2008, 8, 173–256. [Google Scholar]
- Wang, X.; Zou, W.; Kamal, G.M.; Wang, J.; Zhou, M.; Chen, L.; Jiang, B.; Khalid, M.; Zhang, X.; Liu, M. An untargeted 13C isotopic evaluation approach for the discrimination of fermented food matrices at natural abundance: Application to vinegar. Talanta 2020, 210, 120679. [Google Scholar] [CrossRef] [PubMed]
- Kamal, G.M.; Wang, X.; Bin, Y.; Wang, J.; Sun, P.; Zhang, X.; Liu, M. Compositional differences among Chinese soy sauce types studied by (13)C NMR spectroscopy coupled with multivariate statistical analysis. Talanta 2016, 158, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Claridge, T. MNova: NMR data processing, analysis, and prediction software. J. Chem. Inf. Model. 2009, 49, 1136–1137. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Psychogios, N.; Young, N.; Wishart, D.S. MetaboAnalyst: A web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009, 37, W652–W660. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Fang, Y.; Ma, H.; Zhang, N.; Li, C. Performance comparison of three scaling algorithms in NMR-based metabolomics analysis. Open Life Sci. 2023, 18, 20220556. [Google Scholar] [CrossRef] [PubMed]
- Duarte, I.; Barros, A.; Belton, P.S.; Righelato, R.; Spraul, M.; Humpfer, E.; Gil, A.M. High-resolution nuclear magnetic resonance spectroscopy and multivariate analysis for the characterization of beer. J. Agric. Food Chem. 2002, 50, 2475–2481. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Shiga, K.; Kodama, Y.; Imamura, M.; Uchida, R.; Obata, A.; Bamba, T.; Fukusaki, E. Analysis of the correlation between dipeptides and taste differences among soy sauces by using metabolomics-based component profiling. J. Biosci. Bioeng. 2014, 118, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Kamal, G.M.; Yuan, B.; Hussain, A.I.; Wang, J.; Jiang, B.; Zhang, X.; Liu, M. 13C-NMR-based metabolomic profiling of typical Asian soy sauces. Molecules 2016, 21, 1168. [Google Scholar] [CrossRef] [PubMed]
- Kamal, G.M.; Uddin, J.; Muhsinah, A.B.; Wang, X.; Noreen, A.; Sabir, A.; Musharraf, S.G. 1H-NMR-Based metabolomics and 13C isotopic ratio evaluation to differentiate conventional and organic soy sauce. Arab. J. Chem. 2022, 15, 103516. [Google Scholar] [CrossRef]
- Islam, S.S.; Ryu, H.-M.; Sohn, S. Tetragenococcus halophilus alleviates intestinal inflammation in mice by altering gut microbiota and regulating dendritic cell activation via CD83. Cells 2022, 11, 1903. [Google Scholar] [CrossRef] [PubMed]
- Golon, A.; Kuhnert, N. Unraveling the chemical composition of caramel. J. Agric. Food Chem. 2012, 60, 3266–3274. [Google Scholar] [CrossRef] [PubMed]
- Alam Shah, S.; Selamat, J.; Haque Akanda, M.J.; Sanny, M.; Khatib, A. Effects of different types of soy sauce on the formation of heterocyclic amines in roasted chicken. Food Addit. Contam. Part A 2018, 35, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Kim, S.B.; Kim, Y.S. Determination of Key Volatile Compounds Related to Long-Term Fermentation of Soy Sauce. J. Food Sci. 2019, 84, 2758–2776. [Google Scholar] [CrossRef] [PubMed]
- Elhalis, H.; Chin, X.H.; Chow, Y. Soybean fermentation: Microbial ecology and starter culture technology. Crit. Rev. Food Sci. Nutr. 2023. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, R.; Bosco, F.; Mizrahi, I.; Bayer, E.A.; Pessione, E. Towards lactic acid bacteria-based biorefineries. Biotechnol. Adv. 2014, 32, 1216–1236. [Google Scholar] [CrossRef] [PubMed]
- Mayo, B.; Aleksandrzak-Piekarczyk, T.; Fernández, M.; Kowalczyk, M.; Álvarez-Martín, P.; Bardowski, J. Updates in the metabolism of lactic acid bacteria. In Biotechnology of Lactic Acid Bacteria: Novel Applications; Wiley: Hoboken, NJ, USA, 2010. [Google Scholar]

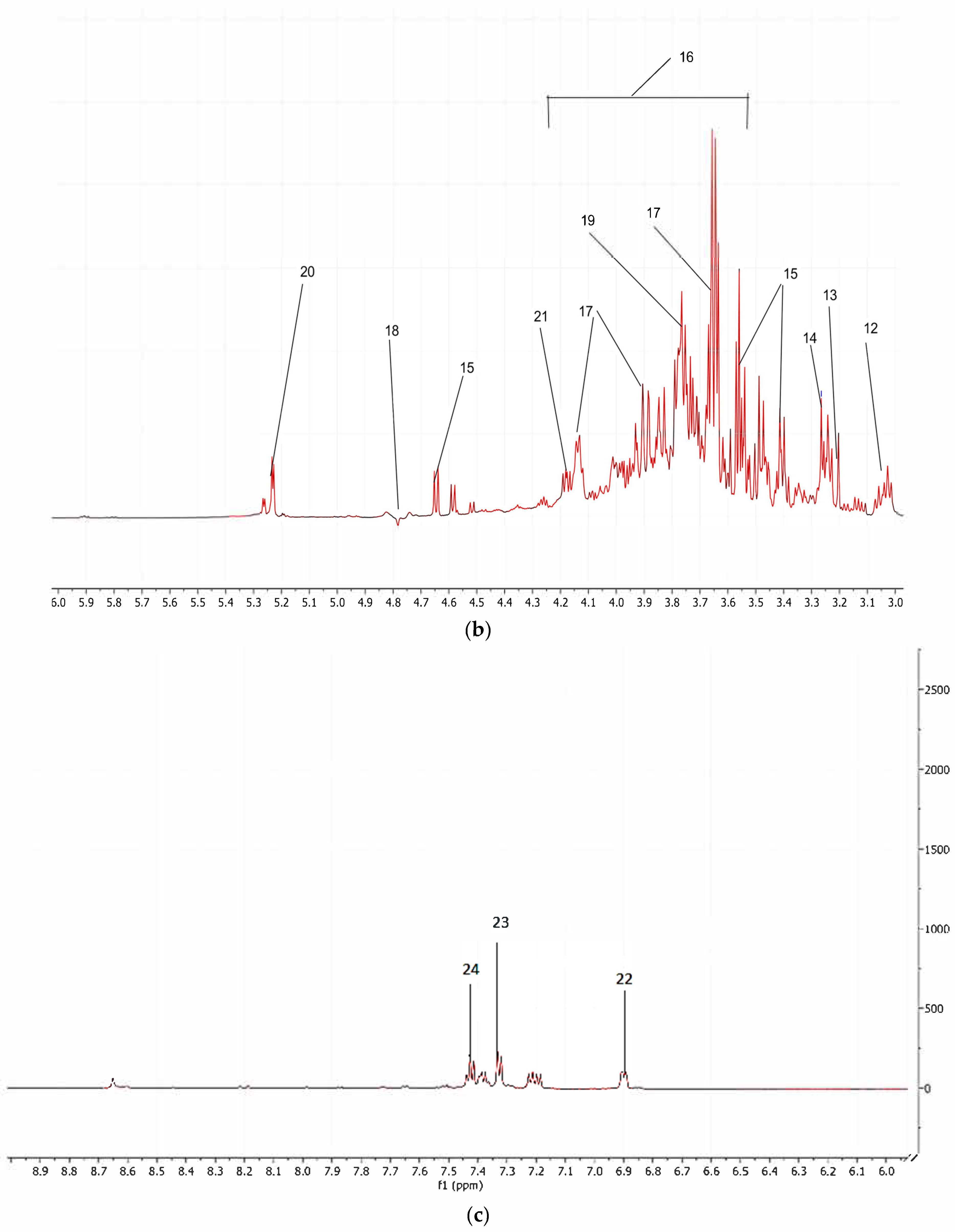



| Sr. No. | Compound | 1H-NMR Chemical Shift |
|---|---|---|
| 1 2 3 4 | TSP Isoleucine Leucine Valine | 0.00 (s) 0.94 (t) 0.98 (d) 0.99 (d) |
| 5 | Ethanol | 1.17 (t) |
| 6 7 | Lactate Alanine | 1.34 (d) 1. 48 (d) |
| 8 | Lysine | 1.73 (m), 1.91 (m) |
| 9 | Acetate | 1.98 (s) |
| 10 | Butyrate | 2.13 (m), 2.39 (m) |
| 11 12 | Methionine γ- amino butyric acid | 2.50 (t) 3.02 (m) |
| 13 14 | Choline Glucose | 3.20 (s) 3.25 (t) |
| 15 16 | β-Glucose Glucose, aliphatic region | 3.40 (t), 3.59 (d), 4.64 (d) 3.50–4.25 |
| 17 18 | Fructose Water (solvent) | 3.64 (m), 3.90 (d), 4.14 (m) 4.78 (s) |
| 19 | Glycerol | 3.76 (m) |
| 20 | α-Glucose | 5.22 (d) |
| 21 22 23 24 | Sucrose Formate Tyrosine Phenylalanine | 4.16 (d) 6.88 (s) 7.30 (d) 7.42 (m) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uddin, J.; Yasmin, S.; Kamal, G.M.; Asmari, M.; Saqib, M.; Chen, H. Changes in Metabolite Profiles of Chinese Soy Sauce at Different Time Durations of Fermentation Studied by 1H-NMR-Based Metabolomics. Metabolites 2024, 14, 285. https://doi.org/10.3390/metabo14050285
Uddin J, Yasmin S, Kamal GM, Asmari M, Saqib M, Chen H. Changes in Metabolite Profiles of Chinese Soy Sauce at Different Time Durations of Fermentation Studied by 1H-NMR-Based Metabolomics. Metabolites. 2024; 14(5):285. https://doi.org/10.3390/metabo14050285
Chicago/Turabian StyleUddin, Jalal, Samra Yasmin, Ghulam Mustafa Kamal, Mufarreh Asmari, Muhammad Saqib, and Heyu Chen. 2024. "Changes in Metabolite Profiles of Chinese Soy Sauce at Different Time Durations of Fermentation Studied by 1H-NMR-Based Metabolomics" Metabolites 14, no. 5: 285. https://doi.org/10.3390/metabo14050285
APA StyleUddin, J., Yasmin, S., Kamal, G. M., Asmari, M., Saqib, M., & Chen, H. (2024). Changes in Metabolite Profiles of Chinese Soy Sauce at Different Time Durations of Fermentation Studied by 1H-NMR-Based Metabolomics. Metabolites, 14(5), 285. https://doi.org/10.3390/metabo14050285







