Metabolome in Tibialis and Soleus Muscles in Wild-Type and Pin1 Knockout Mice through High-Resolution Magic Angle Spinning 1H Nuclear Magnetic Resonance Spectroscopy
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice and Sample Collection
2.2. NMR Spectroscopy Measurements
2.3. NMR Data Processing
3. Results
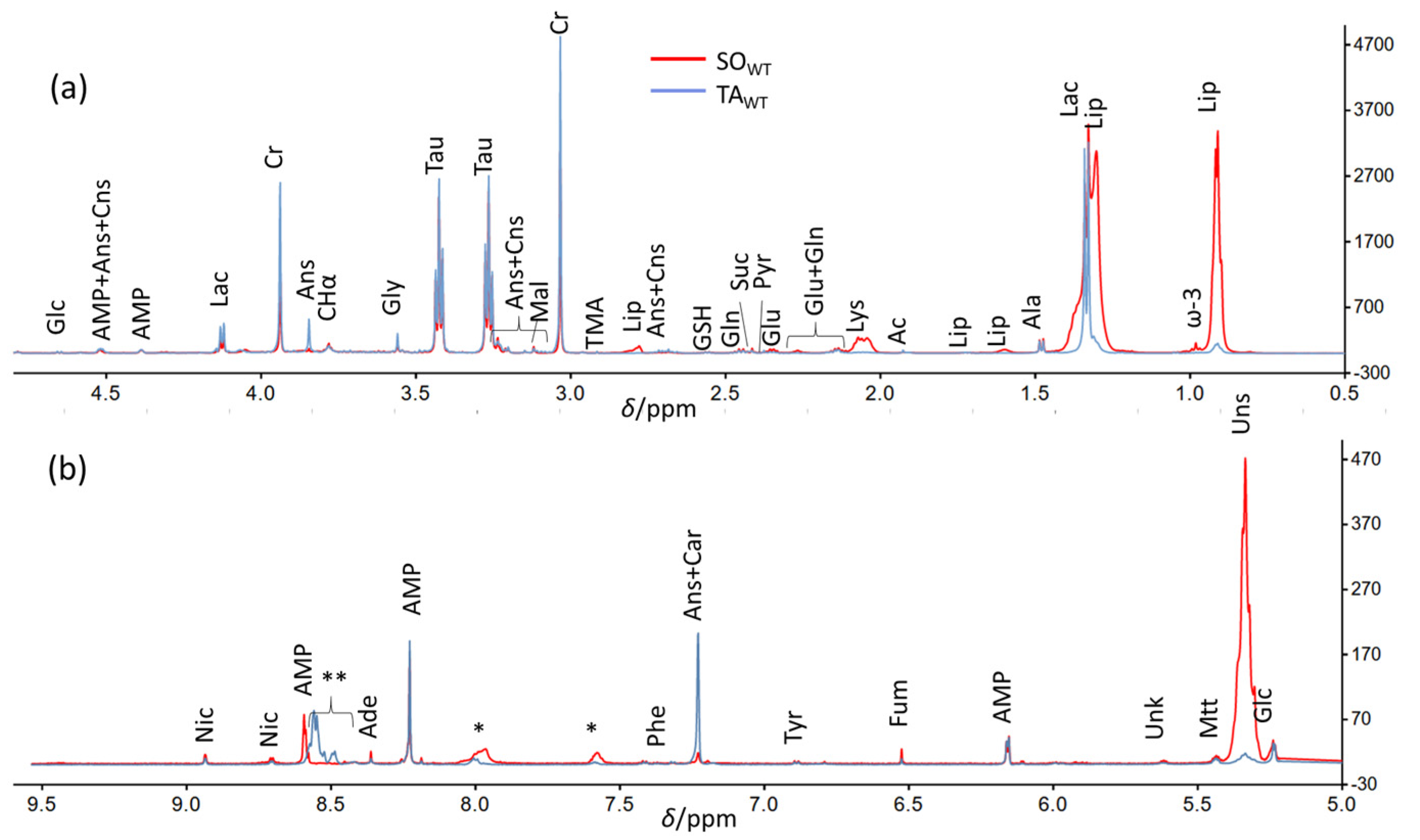
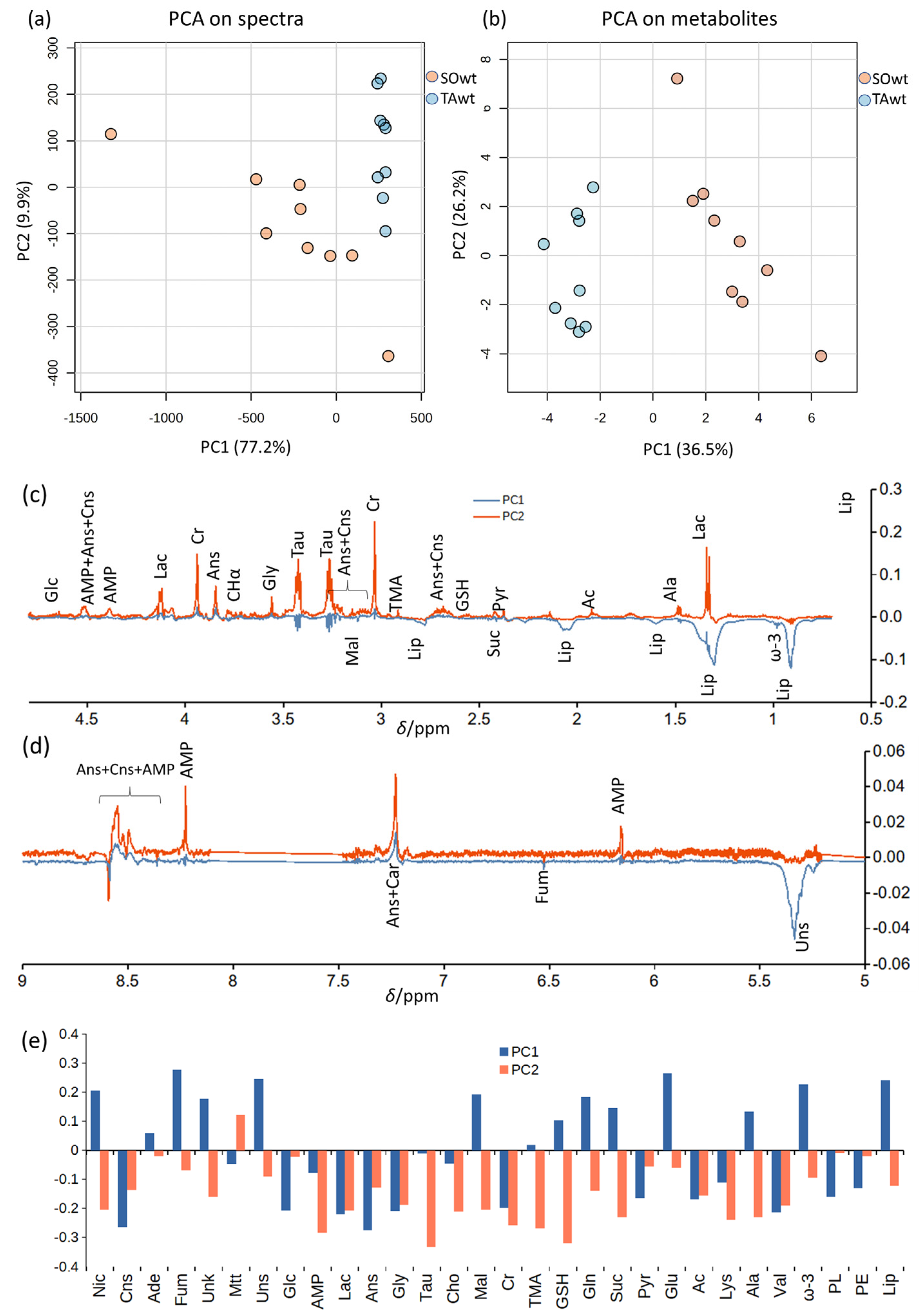
3.1. Detailed Baseline Analysis of WT Mice Muscles
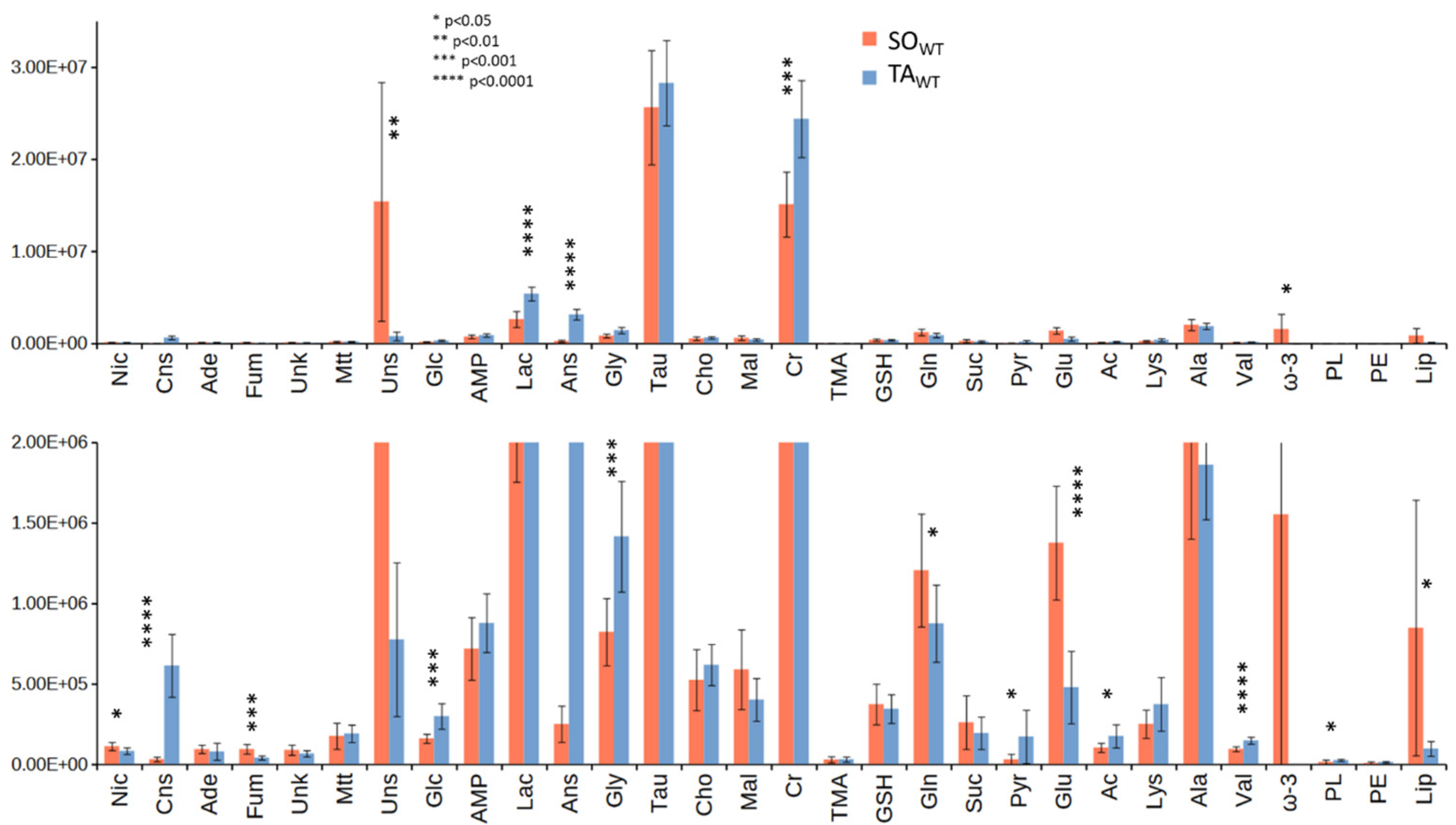
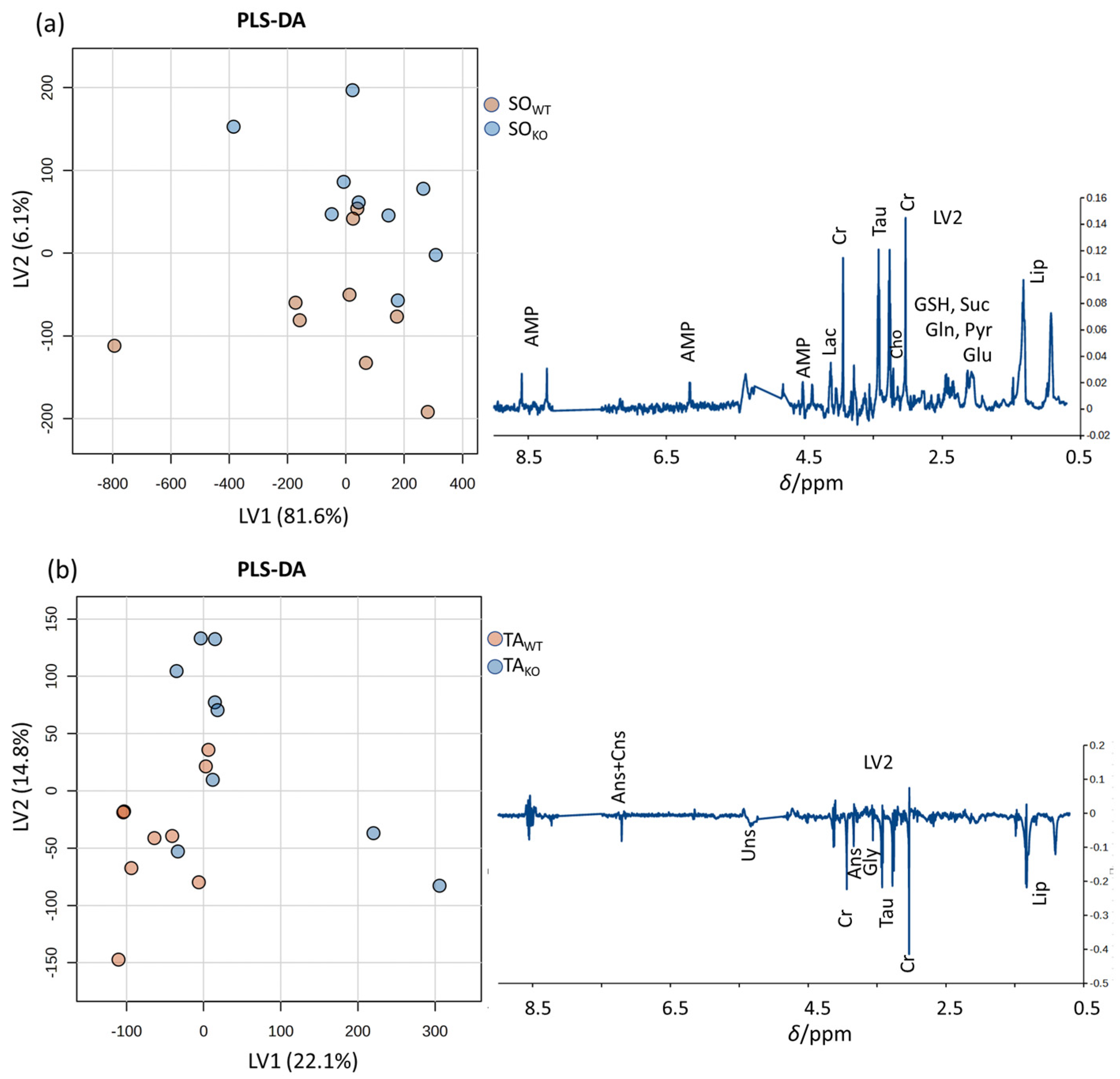
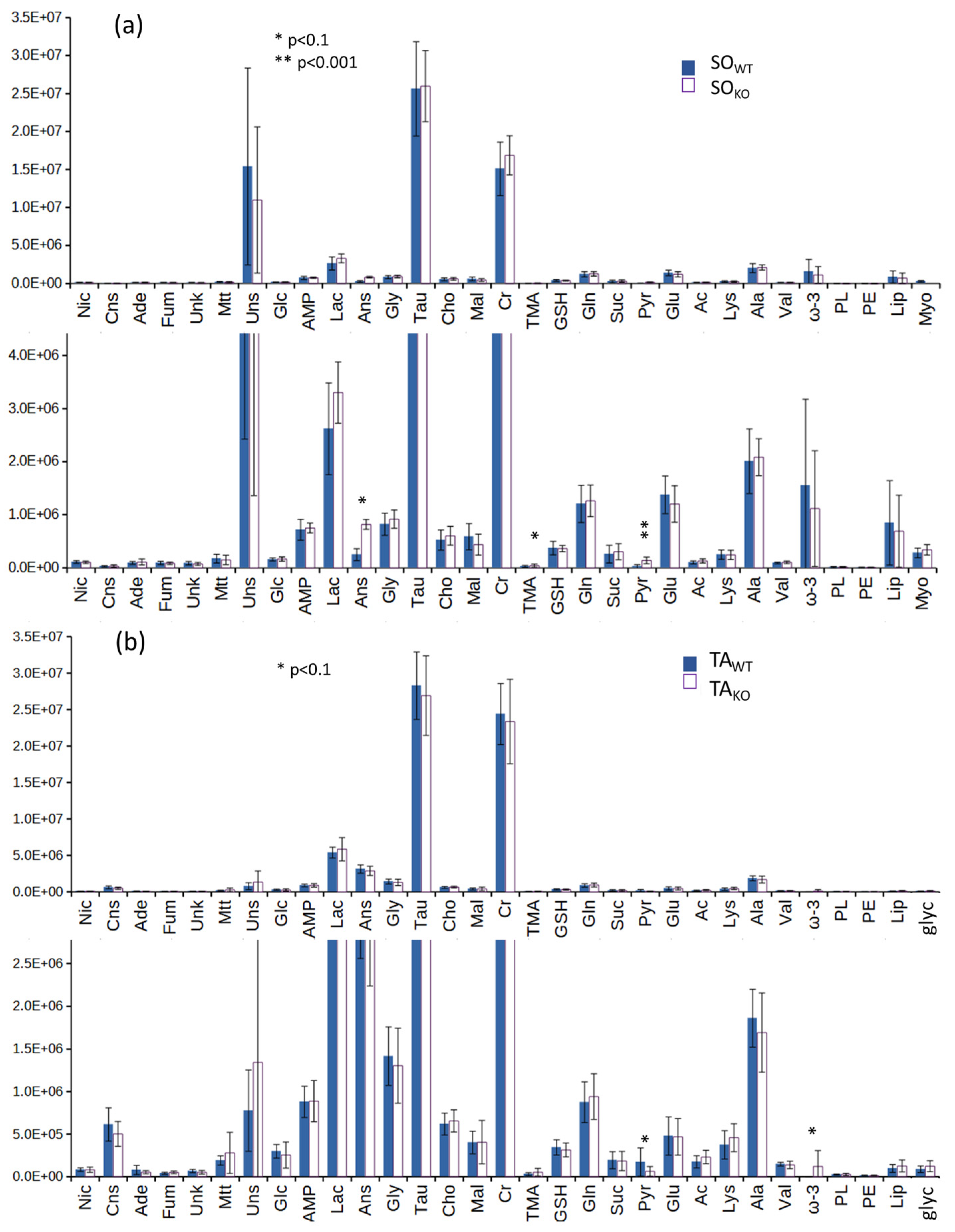
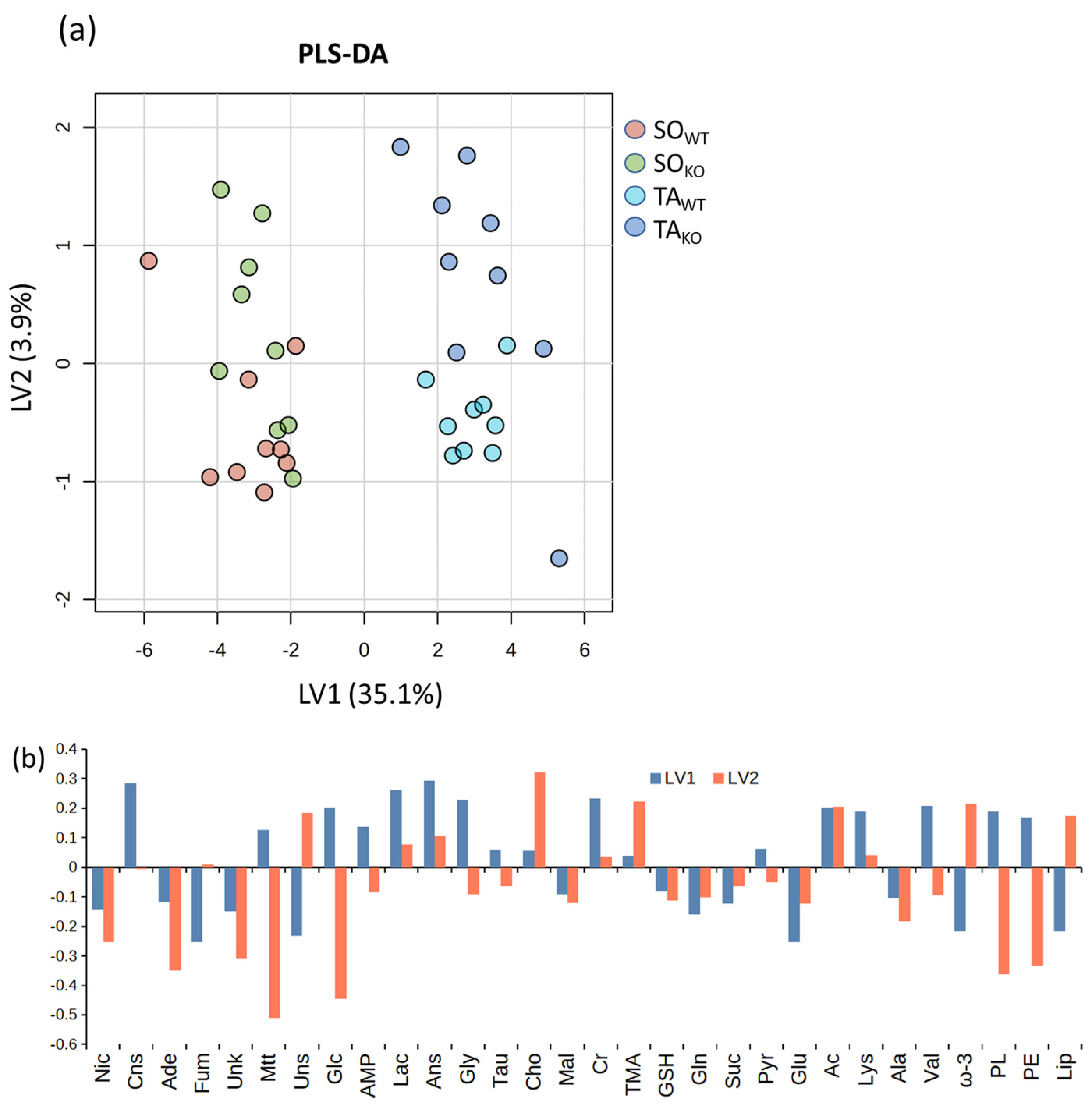
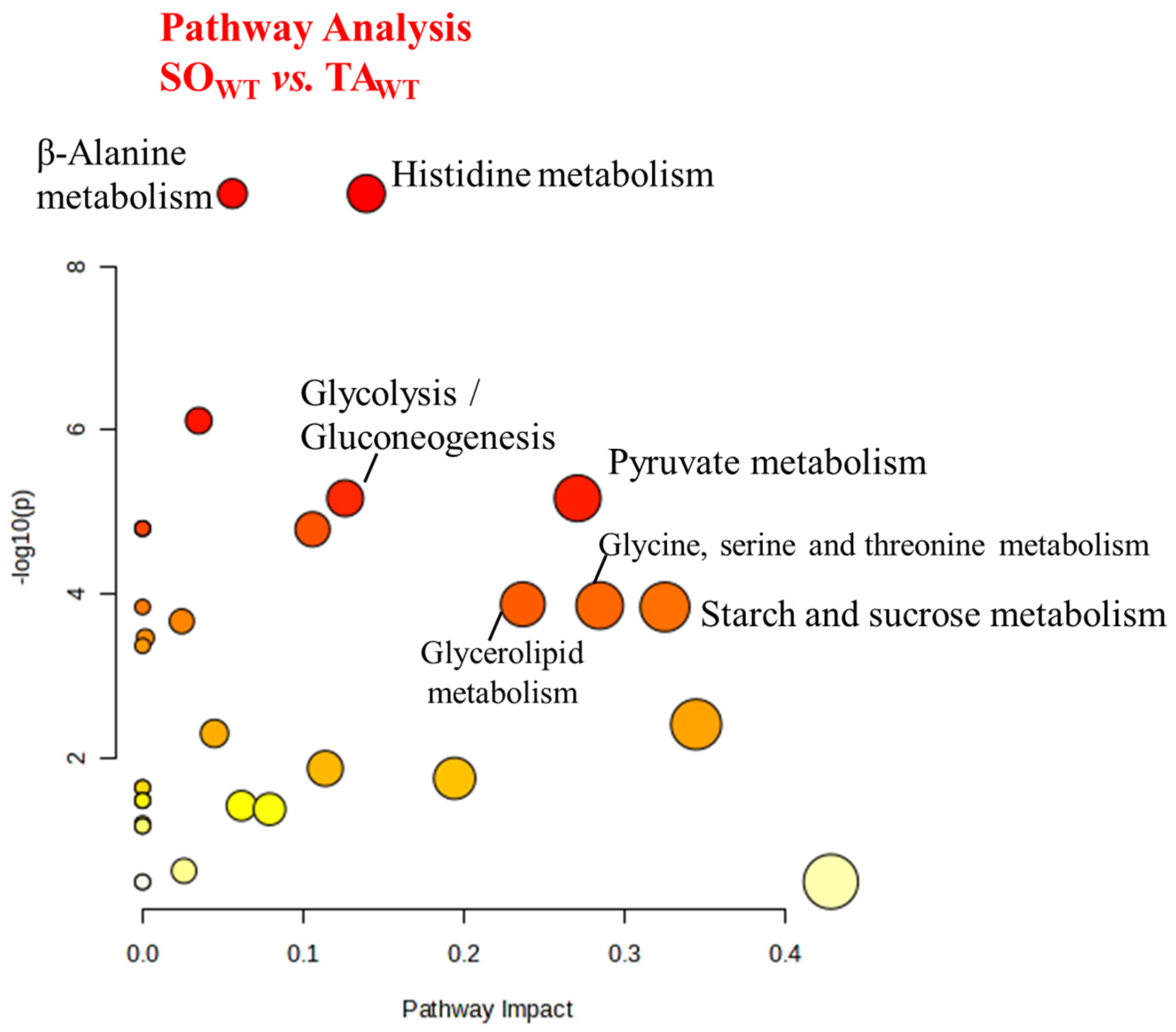
3.2. Comparison between WT and KO Muscles
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schiaffino, S.; Reggiani, C. Fiber types in mammalian skeletal muscles. Physiol. Rev. 2011, 91, 1447–1531. [Google Scholar] [CrossRef] [PubMed]
- Pette, D. Fiber transformation and fiber replacement in chronically stimulated muscle. J. Heart Lung Transplant. 1992, 11, S299–S305. [Google Scholar]
- Miller, J.B.; Crow, M.T.; Stockdale, F.E. Slow and fast myosin heavy chain content defines three types of myotubes in early muscle cell cultures. J. Cell Biol. 1985, 101, 1643–1650. [Google Scholar] [CrossRef] [PubMed]
- Bonen, A.; Tan, M.H.; Watson-Wright, W.M. Insulin binding and glucose uptake differences in rodent skeletal muscles. Diabetes 1981, 30, 702–704. [Google Scholar] [CrossRef] [PubMed]
- Albers, P.H.; Pedersen, A.J.; Birk, J.B.; Kristensen, D.E.; Vind, B.F.; Baba, O.; Nøhr, J.; Højlund, K.; Wojtaszewski, J.F. Human muscle fiber type-specific insulin signaling: Impact of obesity and type 2 diabetes. Diabetes. 2015, 64, 485–497. [Google Scholar] [CrossRef] [PubMed]
- Talbot, J.; Maves, L. Skeletal muscle fiber type: Using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 518–534. [Google Scholar] [CrossRef] [PubMed]
- Liou, Y.C.; Zhou, X.Z.; Lu, K.P. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 2011, 36, 501–514. [Google Scholar] [CrossRef]
- Zhou, X.Z.; Lu, K.P. The isomerase PIN1 controls numerous cancer-driving pathways and is a unique drug target. Nat. Rev. Cancer 2016, 16, 463–478. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Chen, L.F. Pinning Down the Transcription: A Role for Peptidyl-Prolyl cis-trans Isomerase Pin1 in Gene Expression. Front. Cell Dev. Biol. 2020, 8, 179. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, Y.; Matsunaga, Y.; Yamamotoya, T.; Ueda, K.; Inoue, Y.; Mori, K.; Sakoda, H.; Fujishiro, M.; Ono, H.; Kushiyama, A.; et al. Physiological and Pathogenic Roles of Prolyl Isomerase Pin1 in Metabolic Regulations via Multiple Signal Transduction Path-way Modulations. Int. J. Mol. Sci. 2016, 17, 1495. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, Y.; Yamamotoya, T.; Ueda, K.; Ono, H.; Inoue, M.K.; Matsunaga, Y.; Kushiyama, A.; Sakoda, H.; Fujishiro, M.; Matsubara, A.; et al. Prolyl isomerase Pin1 in metabolic reprogramming of cancer cells. Cancer Lett. 2020, 470, 106–114. [Google Scholar] [CrossRef]
- Nakatsu, Y.; Iwashita, M.; Sakoda, H.; Ono, H.; Nagata, K.; Matsunaga, Y.; Fukushima, T.; Fujishiro, M.; Kushiyama, A.; Kamata, H.; et al. Prolyl isomerase Pin1 negatively regulates AMP-activated protein kinase (AMPK) by associating with the CBS domain in the γ subunit. J. Biol. Chem. 2015, 290, 24255–24266. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zheng, Y.; Xia, Y.; Ji, H.; Chen, X.; Guo, F.; Lyssiotis, C.A.; Aldape, K.; Cantley, L.C.; Lu, Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012, 14, 1295–1304, Erratum in Nat. Cell Biol. 2013, 15, 124. [Google Scholar] [CrossRef] [PubMed]
- Ueda, K.; Nakatsu, Y.; Yamamotoya, T.; Ono, H.; Inoue, Y.; Inoue, M.K.; Mizuno, Y.; Matsunaga, Y.; Kushiyama, A.; Sakoda, H.; et al. Prolyl isomerase Pin1 binds to and stabilizes acetyl CoA carboxylase 1 protein, thereby supporting cancer cell proliferation. Oncotarget 2019, 10, 1637–1648. [Google Scholar] [CrossRef]
- Magli, A.; Angelelli, C.; Ganassi, M.; Baruffaldi, F.; Matafora, V.; Battini, R.; Bachi, A.; Messina, G.; Rustighi, A.; Del Sal, G.; et al. Proline isomerase Pin1 represses terminal differentiation and myocyte enhancer factor 2C function in skeletal muscle cells. J. Biol. Chem. 2010, 285, 34518–34527. [Google Scholar] [CrossRef] [PubMed]
- Islam, R.; Yoon, H.; Shin, H.R.; Bae, H.S.; Kim, B.S.; Yoon, W.J.; Woo, K.M.; Baek, J.H.; Lee, Y.S.; Ryoo, H.M. Peptidyl-prolyl cis-trans isomerase NIMA interacting 1 regulates skeletal muscle fusion through structural modification of Smad3 in the linker region. J. Cell Physiol. 2018, 233, 9390–9403. [Google Scholar] [CrossRef]
- Badodi, S.; Baruffaldi, F.; Ganassi, M.; Battini, R.; Molinari, S. Phosphorylation-dependent degradation of MEF2C contributes to regulate G2/M transition. Cell Cycle 2015, 14, 1517–1528. [Google Scholar] [CrossRef] [PubMed]
- Baruffaldi, F.; Montarras, D.; Basile, V.; de Feo, L.; Badodi, S.; Ganassi, M.; Battini, R.; Nicoletti, C.; Imbriano, C.; Musarò, A.; et al. Dynamic phosphorylation of the myocyte enhancer factor 2Cα1 splice variant promotes skeletal muscle regeneration and hypertrophy. Stem Cells 2017, 35, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Fujimori, F.; Takahashi, K.; Uchida, C.; Uchida, T. Mice lacking Pin1 develop normally, but are defective in entering cell cycle from G0arrest. Biochem. Biophys. Res. Commun. 1999, 265, 658–663. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wang, J.; Pan, D.; Wang, X.; Xu, Y.; Yan, J.; Wang, L.; Yang, X.; Yang, M.; Liu, G.P. Applications of multi-omics analysis in human diseases. MedComm 2023, 4, e315. [Google Scholar] [CrossRef]
- Zagatto, A.M.; Bishop, D.J.; Antunes, B.M.; Beck, W.R.; Malta, E.S.; de Poli, R.A.B.; Cavaglieri, C.R.; Chacon-Mikahil, M.P.T.; Castro, A. Impacts of high-intensity exercise on the metabolomics profile of human skeletal muscle tissue. Scand. J. Med. Sci. Sports 2022, 32, 402–413. [Google Scholar] [CrossRef] [PubMed]
- Schenetti, L.; Mucci, A.; Parenti, F.; Cagnoli, R.; Righi, V.; Tosi, M.R.; Tugnoli, V. HR-MAS NMR spectroscopy in the characterization of human tissues: Application to healthy gastric mucosa. Concepts Magn. Reson. Part A 2006, 28A, 430–443. [Google Scholar] [CrossRef]
- MestReNova, ver. 11. 0, 4-18998; Mestrelab Research S. L.: Santiago de Compostela, Spain, 2017.
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Boldyrev, A.A.; Aldini, G.; Derave, W. Physiology and pathophysiology of carnosine. Physiol. Rev. 2013, 93, 1803–1845. [Google Scholar] [CrossRef]
- Kaczmarek, D.; Ochynski, D.; Everaert, I.; Pawlak, M.; Derave, W.; Celichowski, J. Role of histidyl dipeptides in contractile function of fast and slow motor units in rat skeletal muscle. J. Appl. Physiol. 2016, 121, 164–172. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Masuda, T.; Nakano, H.; Miura, H.; Nakaya, S.; Itazawa, S.; Kubokawa, M. In vitro 1H-NMR spectroscopic analysis of metabolites in fast- and slow-twitch muscles of young rats. Magn. Reson. Med. Sci. 2002, 1, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, W.; Peter, J.B. Lipid composition of muscles of nearly homogeneous fiber type. Exp. Neurol. 1973, 39, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, J.; Christensen, A.E.; Nellemann, B.; Christensen, B. Lipid droplet size and location in human skeletal muscle fibers are associated with insulin sensitivity. Am. J. Physiol. Endocrinol. Metab. 2017, 313, E721–E730. [Google Scholar] [CrossRef]
- Leseigneur-Meynier, A.; Gandemer, G. Lipid composition of pork muscle in relation to the metabolic type of the fibres. Meat Sci. 1991, 29, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Naya, F.J.; McKinsey, T.A.; Mercer, B.; Shelton, J.M.; Chin, E.R.; Simard, A.R.; Michel, R.N.; Bassel-Duby, R.; Olson, E.N.; et al. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 2000, 19, 1963–1973. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Righi, V.; Grosso, M.; Battini, R.; Uchida, T.; Gambini, A.; Molinari, S.; Mucci, A. Metabolome in Tibialis and Soleus Muscles in Wild-Type and Pin1 Knockout Mice through High-Resolution Magic Angle Spinning 1H Nuclear Magnetic Resonance Spectroscopy. Metabolites 2024, 14, 262. https://doi.org/10.3390/metabo14050262
Righi V, Grosso M, Battini R, Uchida T, Gambini A, Molinari S, Mucci A. Metabolome in Tibialis and Soleus Muscles in Wild-Type and Pin1 Knockout Mice through High-Resolution Magic Angle Spinning 1H Nuclear Magnetic Resonance Spectroscopy. Metabolites. 2024; 14(5):262. https://doi.org/10.3390/metabo14050262
Chicago/Turabian StyleRighi, Valeria, Martina Grosso, Renata Battini, Takafumi Uchida, Anna Gambini, Susanna Molinari, and Adele Mucci. 2024. "Metabolome in Tibialis and Soleus Muscles in Wild-Type and Pin1 Knockout Mice through High-Resolution Magic Angle Spinning 1H Nuclear Magnetic Resonance Spectroscopy" Metabolites 14, no. 5: 262. https://doi.org/10.3390/metabo14050262
APA StyleRighi, V., Grosso, M., Battini, R., Uchida, T., Gambini, A., Molinari, S., & Mucci, A. (2024). Metabolome in Tibialis and Soleus Muscles in Wild-Type and Pin1 Knockout Mice through High-Resolution Magic Angle Spinning 1H Nuclear Magnetic Resonance Spectroscopy. Metabolites, 14(5), 262. https://doi.org/10.3390/metabo14050262








