Advancing Glucose Conjugated Gibberellins Discovery: A Structure–Oriented Screening and Identification Method for Unraveling Gibberellin Metabolites in Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Collection of Plant Samples
2.3. Extraction and Derivatization of Glc–GAs in Plant Samples
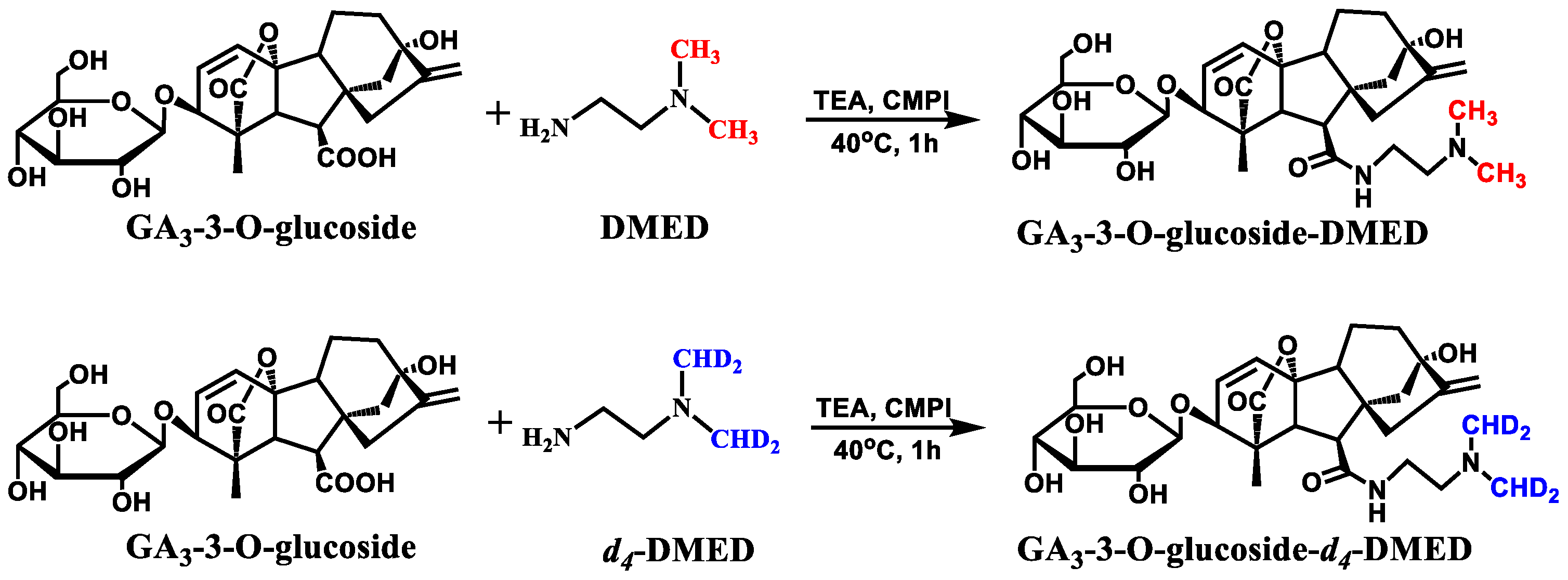
2.4. Synthesis of Glc–GA3
2.5. Instrumentation and Analytical Conditions
3. Results and Discussion
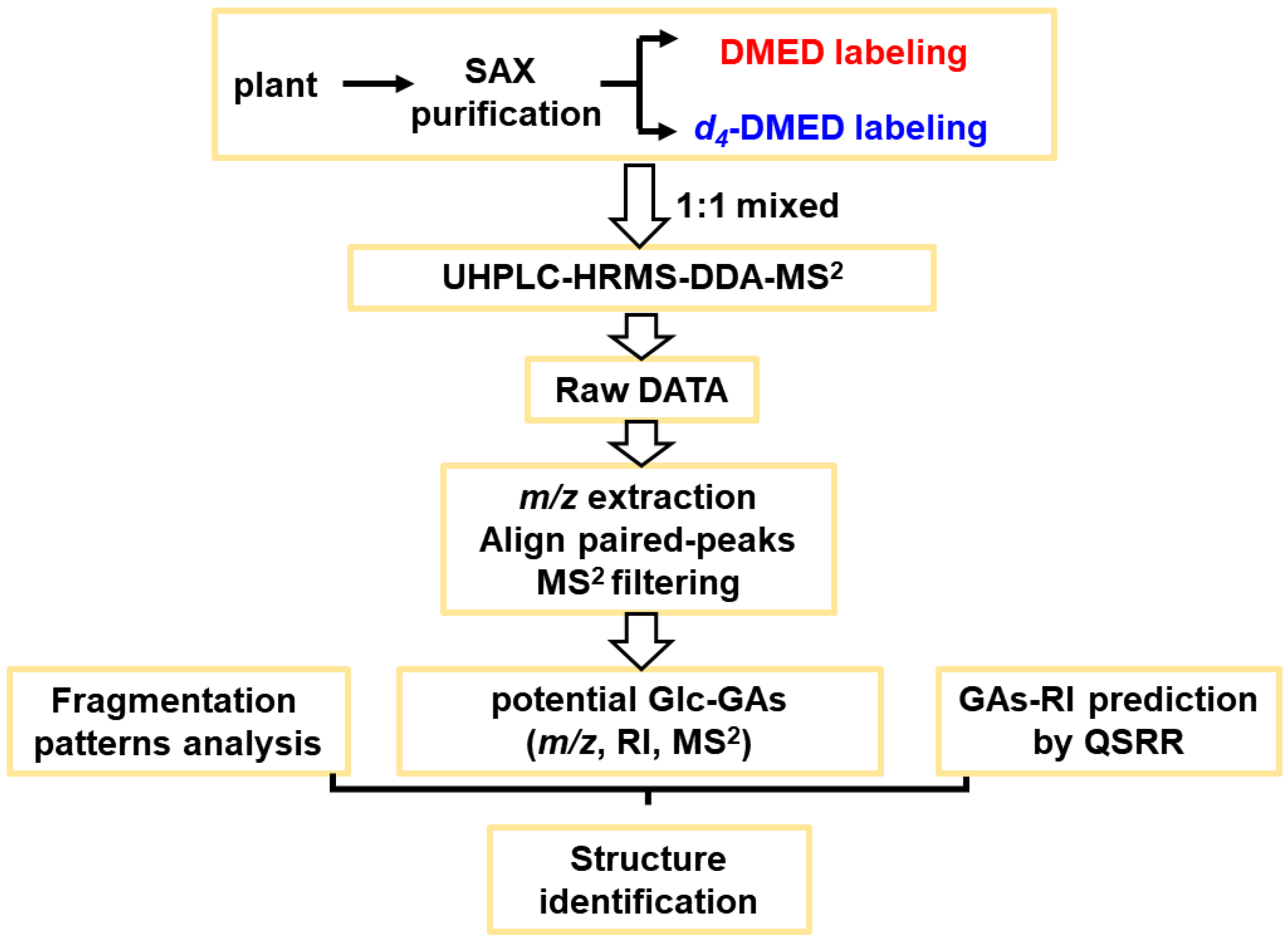
3.1. Strategies for Screening and Identifying Glc–GAs
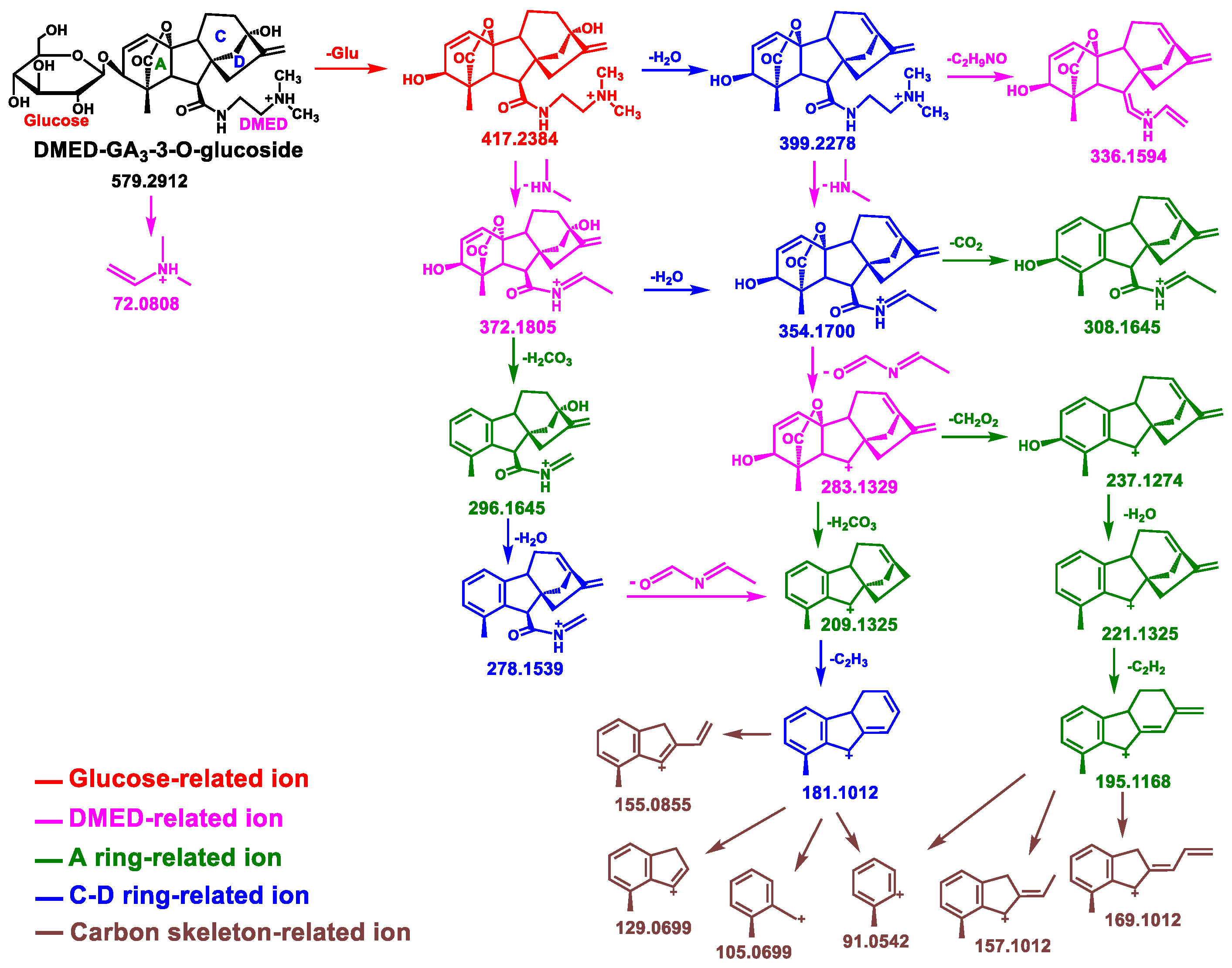
3.2. Analysis of Mass Fragmentation Behavior of Glc–GAs–Labeled Products
3.3. Establishment of Quantitative Structure–Retention Relationship (QSRR) Models
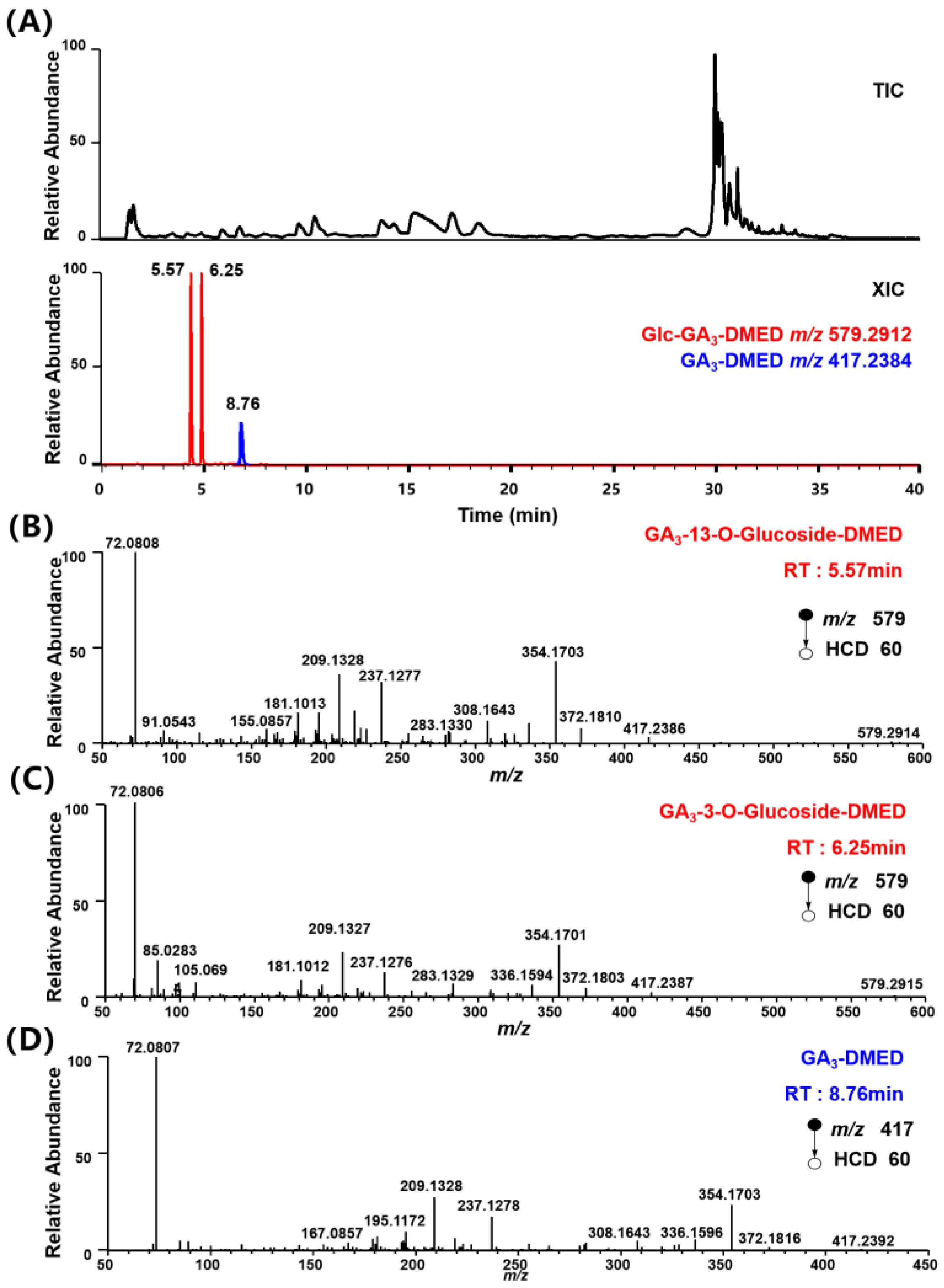
3.4. Screening Potential Glc–GAs in Plant Samples
3.5. Identification of Potential Glc–GAs in Plant Samples
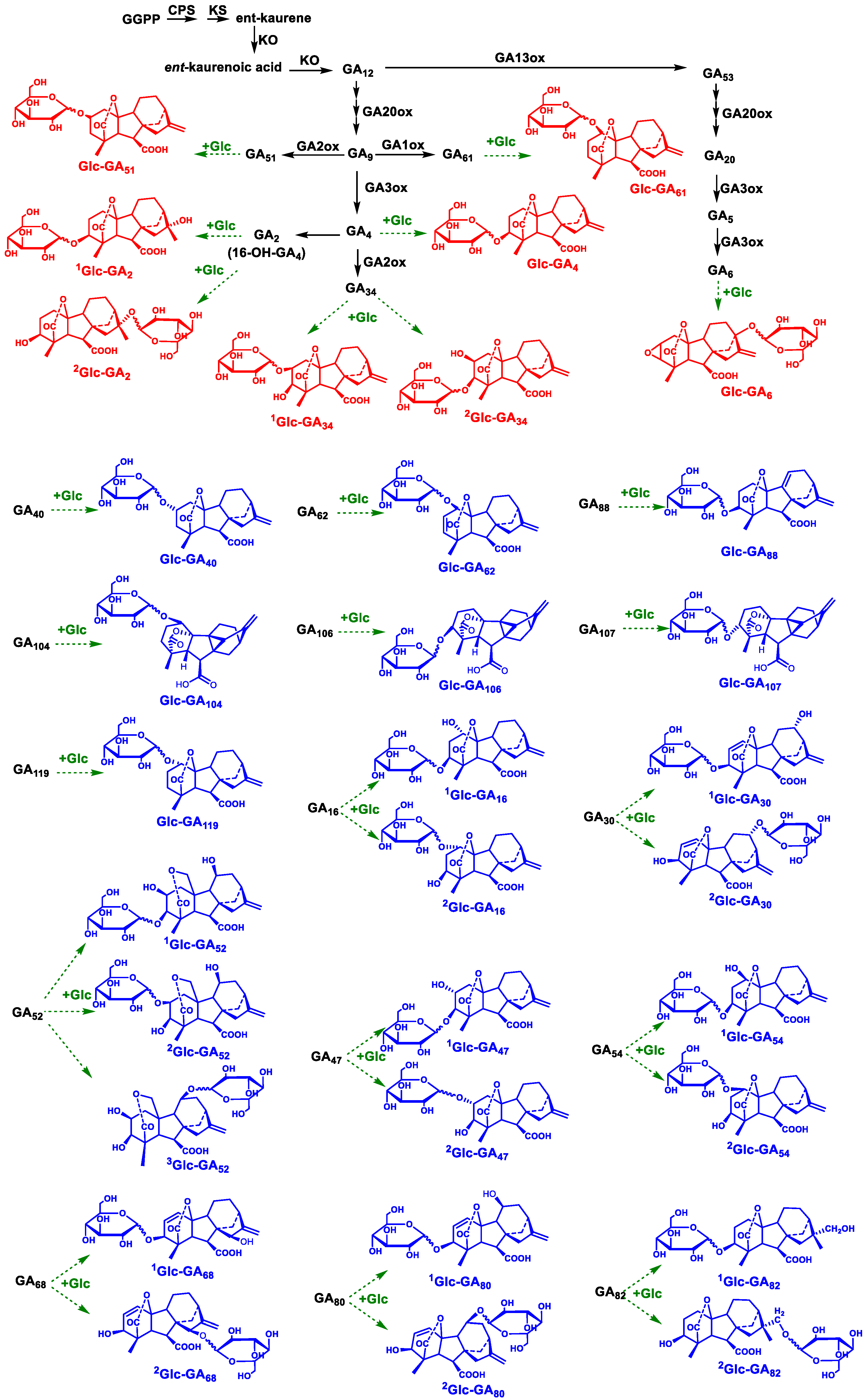
3.6. Possible Locations of Potential Glc–GAs in the Metabolic Pathway of GAs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hedden, P.; Sponsel, V. A Century of Gibberellin Research. J. Plant Growth Regul. 2015, 34, 740–760. [Google Scholar] [CrossRef] [PubMed]
- Plackett, A.R.G.; Wilson, Z.A. Gibberellins and Plant Reproduction. In Annual Plant Reviews; Hedden, P., Thomas, S.G., Eds.; Wiley: Hoboken, NJ, USA, 2016; Volume 49, pp. 323–358. ISBN 978-1-119-21042-9. [Google Scholar]
- Sembdner, G.; Atzorn, R. Plant Hormone Conjugation. Plant Mol. Biol. 1994, 26, 1459–1481. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Schliemann, W. Gibberellin Conjugates: An Overview. Plant Growth Regul. 1994, 15, 247–260. [Google Scholar] [CrossRef]
- Yokota, T.; Murofushi, N.; Takahashi, N.; Katsumi, M. Biological Activities of Gibberellins and Their Glycosides in Pharbitis Nil. Phytochemistry 1971, 10, 2943–2949. [Google Scholar] [CrossRef]
- Von Schirach–Szmigiel, L. Alterations in Endogenous Levels of Gibberellin–like Substances during Germination of Phaseolus vulgaris Seeds. Physiol. Plant. 1979, 46, 54–57. [Google Scholar] [CrossRef]
- Piotrowska, A.; Bajguz, A. Conjugates of Abscisic Acid, Brassinosteroids, Ethylene, Gibberellins, and Jasmonates. Phytochemistry 2011, 72, 2097–2112. [Google Scholar] [CrossRef]
- Knöfel, H.-D.; Schwarzkopf, E.; Müller, P.; Sembdner, G. Enzymic Glycosylation of Gibberellins. J. Plant Growth Regul. 1984, 3, 127–140. [Google Scholar] [CrossRef]
- Sakamoto, T.; Miura, K.; Itoh, H.; Tatsumi, T.; Ueguchi-Tanaka, M.; Ishiyama, K.; Kobayashi, M.; Agrawal, G.K.; Takeda, S.; Abe, K.; et al. An Overview of Gibberellin Metabolism Enzyme Genes and Their Related Mutants in Rice. Plant Physiol. 2004, 134, 1642–1653. [Google Scholar] [CrossRef] [PubMed]
- Hiraga, K.; Yamane, H.; Takahashi, N. Biological Activity of Some Synthetic Gibberellin Glycosyl Esters. Phytochemistry 1974, 13, 2371–2376. [Google Scholar] [CrossRef]
- Yamane, H.; Yamaguchi, I.; Yokota, T.; Murofushi, N.; Takahashi, N.; Katsumi, M. Biological Activities of New Gibberellins A30–A35 and A35 Glycoside. Phytochemistry 1973, 12, 255–261. [Google Scholar] [CrossRef]
- Baidsx, G.W.M. Formation of Bound Gibberellins. Planta 1971, 99, 290–301. [Google Scholar] [CrossRef]
- Sembdner, G.; Schliemann, W.; Schneider, G. Biochemical and Physiological Aspects of Gibberellin Conjugation. In Gibberellins; Takahashi, N., Phinney, B.O., MacMillan, J., Eds.; Springer: New York, NY, USA, 1991; pp. 249–263. ISBN 978-1-4612-7754-5. [Google Scholar]
- Stephan, M.; Bangerth, F.; Schneider, G. Transport and Metabolism of Exogenously Applied Gibberellins to Malus Domestica Borkh. Cv. Jonagold. Plant Growth Regul. 2001, 33, 77–85. [Google Scholar] [CrossRef]
- Ostrowski, M.; Jakubowska, A. Udp–Glycosyltransferases of Plant Hormones. Adv. Cell Biol. 2014, 4, 43–60. [Google Scholar] [CrossRef]
- Piccoli, P.; Lucangeli, C.D.; Schneider, G.; Bottini, R. Hydrolysis of [17,17–2H2] Gibberellin A20–Glycoside and [17,17–2H2]Gibberellin A20–Glycosyl Ester by Azospirillum Lipoferum Cultured in a Nitrogen–Free Biotin–Based Chemically–Defined Medium. Plant Growth Regul. 1997, 23, 179–182. [Google Scholar] [CrossRef]
- Moritz, T.; Monteiro, A.M. Analysis of Endogenous Gibberellins and Gibberellin Metabolites from Dalbergia dolichopetMaby Gas Chromatography–Mass Spectrometry and High–Performance Liquid Chromatography–Mass Spectrometry. Planta 1994, 193, 1–8. [Google Scholar] [CrossRef]
- Rivier, L.; Gaskin, P.; Albone, K.S.; MacMillan, J. GC–MS Identification of Endogenous Gibberellins and Gibberellin Conjugates as Their Permethylated Derivatives. Phytochemistry 1981, 20, 687–692. [Google Scholar] [CrossRef]
- Binks, R.; MacMillan, J.; Pryce, R.J. Plant Hormones—VIII: Combined Gas Chromatography–Mass Spectrometry of the Methyl Esters of Gibberellins A1 to A24 and Their Trimethylsilyl Ethers. Phytochemistry 1969, 8, 271–284. [Google Scholar] [CrossRef]
- Schneider, G.; Sembdner, G.; Jensen, E.; Bernhard, U.; Wagenbreth, D. GC–MS Identification of Native Gibberellin–O–Glycosides in Pea Seeds. J. Plant Growth Regul. 1992, 11, 15–18. [Google Scholar] [CrossRef]
- Schneider, G.; Schmidt, J.; Phinney, B.O. GC–MS Identification of GA20–13–O–Glycoside Formed from GA20 in Normal Plants and Dwarf–1 Mutants of Zea mays L. J. Plant Growth Regul. 1987, 5, 217–223. [Google Scholar] [CrossRef]
- Senns, B.; Fuchs, P.; Schneider, G. GC–MS Quantification of Gibberellin A20–13–O–Glycoside and Gibberellin A8–2–O–Glycoside in Developing Barley Caryopses. Phytochemistry 1998, 48, 1275–1280. [Google Scholar] [CrossRef]
- Moritz, T. The Use of Combined Capillary Liquid Chromatography/Mass Spectrometry for the Identification of a Gibberellin Glycosyl Conjugate. Phytochem. Anal. 1992, 3, 32–37. [Google Scholar] [CrossRef]
- Cai, W.-J.; Zeng, C.; Zhang, X.-Y.; Ye, T.; Feng, Y.-Q. A Structure–Guided Screening Strategy for the Discovery and Identification of Potential Gibberellins from Plant Samples Using Liquid Chromatography–Mass Spectrometry Assisted by Chemical Isotope Labeling. Anal. Chim. Acta 2021, 1163, 338505. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.-H.; Zhang, Z.; Wang, L.; Liu, C.; Lei, A.-W.; Yuan, B.-F.; Feng, Y.-Q. Stable Isotope Labeling Assisted Liquid Chromatography–Electrospray Tandem Mass Spectrometry for Quantitative Analysis of Endogenous Gibberellins. Talanta 2015, 144, 341–348. [Google Scholar] [CrossRef]
- Zhu, Q.-F.; Yan, J.-W.; Gao, Y.; Zhang, J.-W.; Yuan, B.-F.; Feng, Y.-Q. Highly Sensitive Determination of Fatty Acid Esters of Hydroxyl Fatty Acids by Liquid Chromatography–Mass Spectrometry. J. Chromatogr. B 2017, 1061–1062, 34–40. [Google Scholar] [CrossRef]
- Schneider, G.; Sembdner, G.; Schreiber, K. ChemInform Abstract: GIBBERELLINE 26. MITT. SYNTHESE VON GIBBERELLIN–A3–BETA–D–GLYCOPYRANOSIDEN. Chem. Inf. 1975, 6, 226. [Google Scholar] [CrossRef]
- Noma, M.; Huber, J.; Ernst, D.; Pharis, R.P. Quantitation of Gibberellins and the Metabolism of [3H]Gibberellin A1 during Somatic Embryogenesis in Carrot and Anise Cell Cultures. Planta 1982, 155, 369–376. [Google Scholar] [CrossRef]
- Schliemann, W.; Schaller, B.; Jensen, E.; Schneider, G. Native Gibberellin-O-Glucosides from Mature Seeds of Phaseolus Coccineus. Phytochemistry 1993, 35, 35–38. [Google Scholar] [CrossRef]
- Schneider, G.; Fuchs, P.; Schmidt, J. Evidence for the Direct 2β- and 3β-hydroxylation of [2H2]GA20-13-O-[6′-2H2]Glucoside in Seedlings of Phaseolus coccineus. Physiologia Plantarum 2002, 116, 144–147. [Google Scholar] [CrossRef] [PubMed]
- Tamura, S.; Takahashi, N.; Yokota, T.; Murofushi, N.; Ogawa, Y. Isolation of Water-Soluble Gibberellins from Immature Seeds of Pharbitis Nil. Planta 1967, 78, 208–212. [Google Scholar] [CrossRef]
- Yamaguchi, I.; Yokota, T.; Yoshida, S.; Takahashi, N. High Pressure Liquid Chromatography of Conjugated Gibberellins. Phytochemistry 1979, 18, 1699–1702. [Google Scholar] [CrossRef]
- Hasegawa, M.; Nakajima, M.; Takeda, K.; Yamaguchi, I.; Murofushi, N. A Novel Gibberellin Glucoside 16α,` 17-Dihydroxy-16,17-Dihydrogibberellin A4-17-O-β-d-Glucopyranoside, from Rice Anthers. Phytochemistry 1994, 37, 629–634. [Google Scholar] [CrossRef]
- Hiraga, K.; Kawabe, S.; Yokota, T.; Murofushi, N.; Takahashi, N. Isolation and Characterization of Plant Growth Substances in Immature Seeds and Etiolated Seedlings of Phaseolus vulgaris. Agric. Biol. Chem. 1974, 38, 2521–2527. [Google Scholar] [CrossRef]
- Yokota, T.; Takahashi, N.; Murofushi, N.; Tamura, S. Isolation of Gibberellins A26 and A27 and Their Glucosides from Immature Seeds of Pharbitis Nil. Planta 1969, 87, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Yokota, T.; Takahashi, N.; Murofushi, N.; Tamura, S. Structures of new gibberellin glucosides in immature seeds of pharbitis nil. Tetrahedron Lett. 1969, 10, 2081–2084. [Google Scholar] [CrossRef]
- Yamane, H.; Yamaguchi, I.; Murofushi, N.; Takahashi, N. Isolation and Structures of Gibberellin A35 and Its Glucoside from Immature Seed of Cytisus scoparius. J. Agric. Chem. Soc. Jpn. 1974, 38, 649–655. [Google Scholar] [CrossRef]
- Hiraga, K.; Yokota, T.; Murofushi, N.; Takahashi, N. Isolation and Characterization of a Free Gibberellin and Glucosyl Esters of Gibberellins in Mature Seeds of Phaseolus vulgaris. Agric. Biol. Chem. 1972, 36, 345–347. [Google Scholar] [CrossRef]
- Yamaguchi, I.; Kobayashi, M.; Takahashi, N. Isolation and Characterization of Glucosyl Esters of Gibberellin A 5 and A 44 from Immature Seeds of Pharbitis purpurea. Agric. Biol. Chem. 1980, 44, 1975–1977. [Google Scholar] [CrossRef]
- Lorenzi, R.; Horgan, R.; Heald, J.K.; Lorenzi, R.; Horgan, R.; Heald, J.K. Gibberellins in Picea Sitchensis Carriere: Seasonal Variation and Partial Characterization. Planta 1975, 126, 75–82. [Google Scholar] [CrossRef]
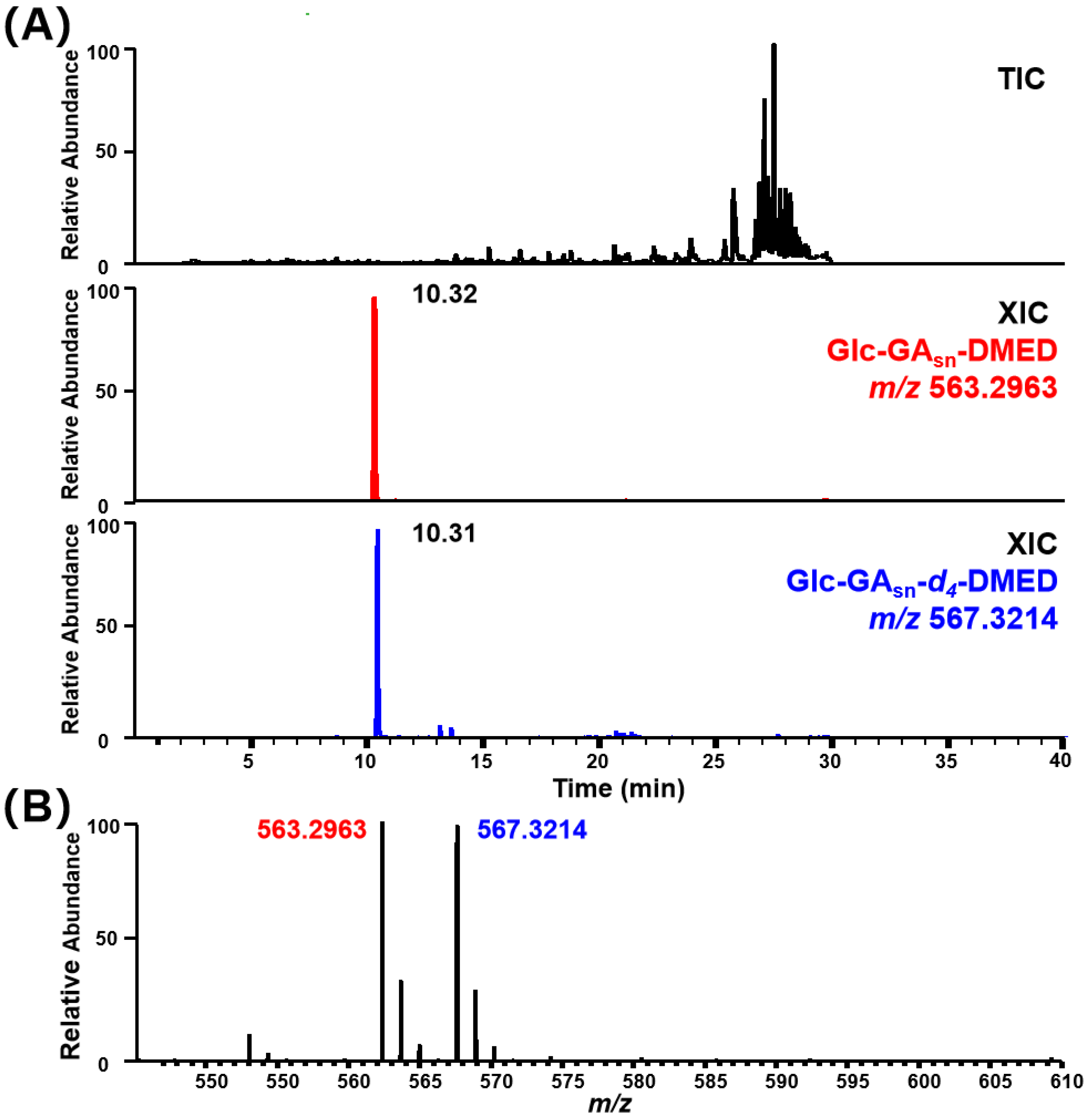
| ID | Candidates | Theoretical m/z | Derivatized Formula | Native Formula | RT | RI | Identification | |
|---|---|---|---|---|---|---|---|---|
| DMED –Labeled | d4–DMED –Labeled | |||||||
| 1 | Glc–GA5, 31, 95, 96, 108, 109, 117, 121, 122,7, 11, 62, 88, 104, 105, 106, 107 | 563.2963 | 567.3214 | C29H44O9N2 | C25H32O10 | 10.32 | 718.97 | Glc–GA62, 88, 104,106, 107 a,b |
| 2 | 12.10 | 760.66 | Glc–GA62, 88, 104,106, 107 c | |||||
| 3 | Glc–GA20, 69, 70, 84, 4, 40, 51, 61, 119, 45 | 565.3119 | 569.3370 | C29H46O9N2 | C25H34O10 | 8.70 | 682.35 | Glc–GA4, 40, 51, 61, 119 c |
| 4 | 10.79 | 729.98 | Glc–GA4, 40, 51, 61, 119 d | |||||
| 5 | 13.01 | 781.97 | Glc–GA4, 40, 51, 61, 119 a,b,c,e | |||||
| 6 | Glc–GA126, 3, 6, 30, 80, 92, 94, 22, 68 | 579.2912 | 583.3163 | C29H44O10N2 | C25H32O11 | 7.47 | 655.56 | Glc–GA6, 30, 80, 68 a |
| 7 | 8.20 | 671.46 | Glc–GA6, 30, 80, 68 e | |||||
| 8 | 8.94 | 687.58 | Glc–GA6, 30 e | |||||
| 9 | 11.12 | 737.70 | Glc–GA6, 30 f | |||||
| 10 | Glc–GA77, 130, 136, 1, 29, 35, 58, 60, 71, 81, 118, 16, 34, 47, 54, 90, 63, 67, 131 | 581.3068 | 585.3320 | C29H46O10N2 | C25H34O11 | 10.77 | 729.51 | Glc–GA16, 34, 47, 54 a |
| 11 | Glc–GA2, 82 | 583.3225 | 587.3476 | C29H48O10N2 | C25H36O11 | 8.70 | 682.35 | Glc–GA2, 82 e |
| 12 | Glc–GA125, 129, 23, 52, 13, 17, 46, 66, 99, 102 | 611.3174 | 615.3425 | C30H48O11N2 | C26H36O12 | 9.55 | 700.94 | Glc–GA52 e |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, C.; Cai, W.-J.; Jiang, L.-C.; Ye, T.; Feng, Y.-Q. Advancing Glucose Conjugated Gibberellins Discovery: A Structure–Oriented Screening and Identification Method for Unraveling Gibberellin Metabolites in Plants. Metabolites 2024, 14, 96. https://doi.org/10.3390/metabo14020096
Zeng C, Cai W-J, Jiang L-C, Ye T, Feng Y-Q. Advancing Glucose Conjugated Gibberellins Discovery: A Structure–Oriented Screening and Identification Method for Unraveling Gibberellin Metabolites in Plants. Metabolites. 2024; 14(2):96. https://doi.org/10.3390/metabo14020096
Chicago/Turabian StyleZeng, Chen, Wen-Jing Cai, Liu-Cheng Jiang, Tiantian Ye, and Yu-Qi Feng. 2024. "Advancing Glucose Conjugated Gibberellins Discovery: A Structure–Oriented Screening and Identification Method for Unraveling Gibberellin Metabolites in Plants" Metabolites 14, no. 2: 96. https://doi.org/10.3390/metabo14020096
APA StyleZeng, C., Cai, W.-J., Jiang, L.-C., Ye, T., & Feng, Y.-Q. (2024). Advancing Glucose Conjugated Gibberellins Discovery: A Structure–Oriented Screening and Identification Method for Unraveling Gibberellin Metabolites in Plants. Metabolites, 14(2), 96. https://doi.org/10.3390/metabo14020096







