Integrated Transcriptomics and Metabolomics Analysis Reveals the Effects of Cutting on the Synthesis of Flavonoids and Saponins in Chinese Herbal Medicine Astragalus mongholious
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Measurement of Biomass
2.3. Measuring the Contents of Flavonoids and Saponins
2.4. Sequencing Data Analysis
2.5. Quantitative Real-Time PCR Analysis
2.6. Metabolome Analysis
3. Results
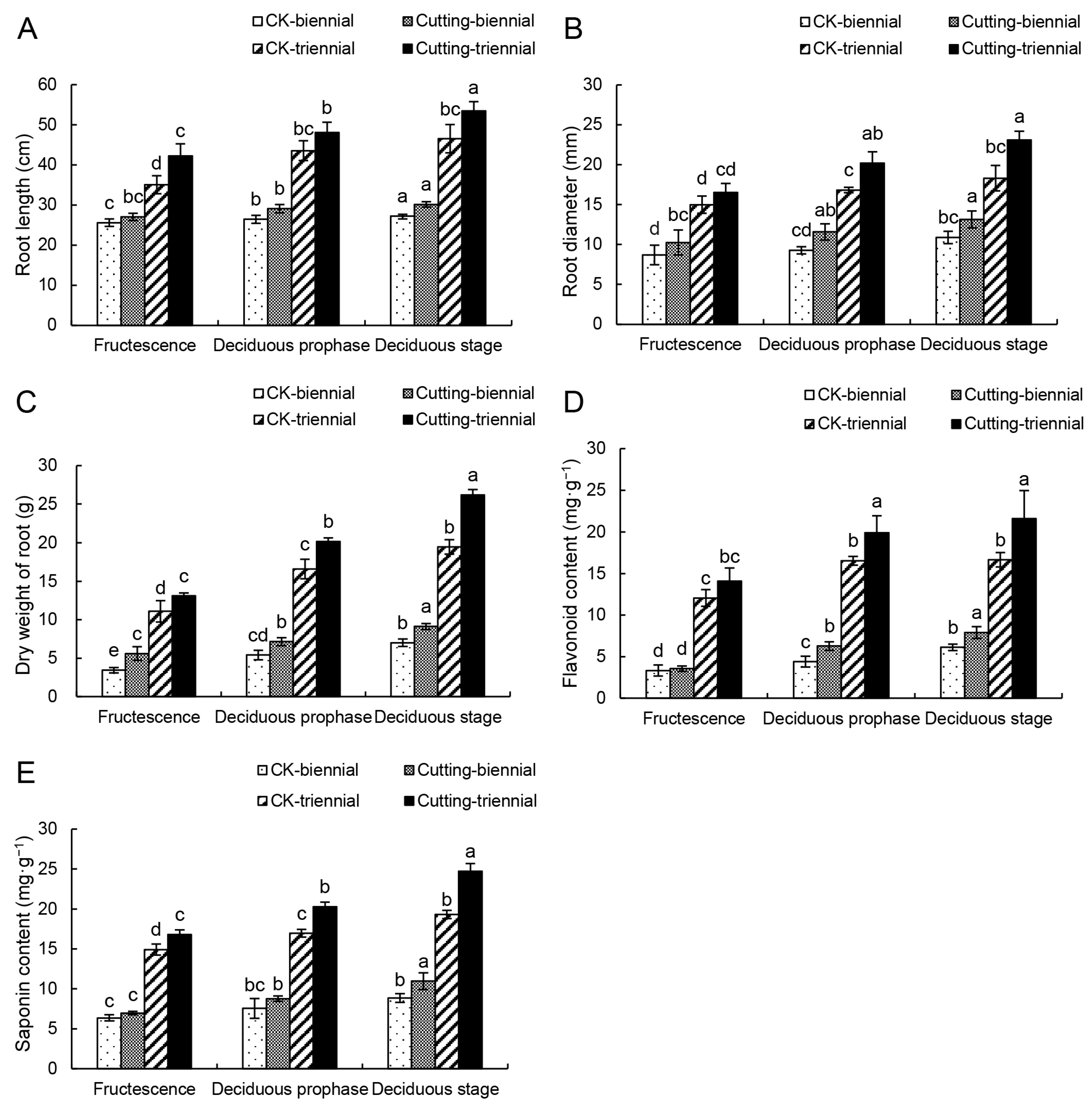
3.1. Effects of Cutting on the Biomass of A. mongholicus
3.2. Effects of the Cutting on the Content of Total Flavonoids and Saponins
3.3. Summary of Transcriptome Analysis after Cutting
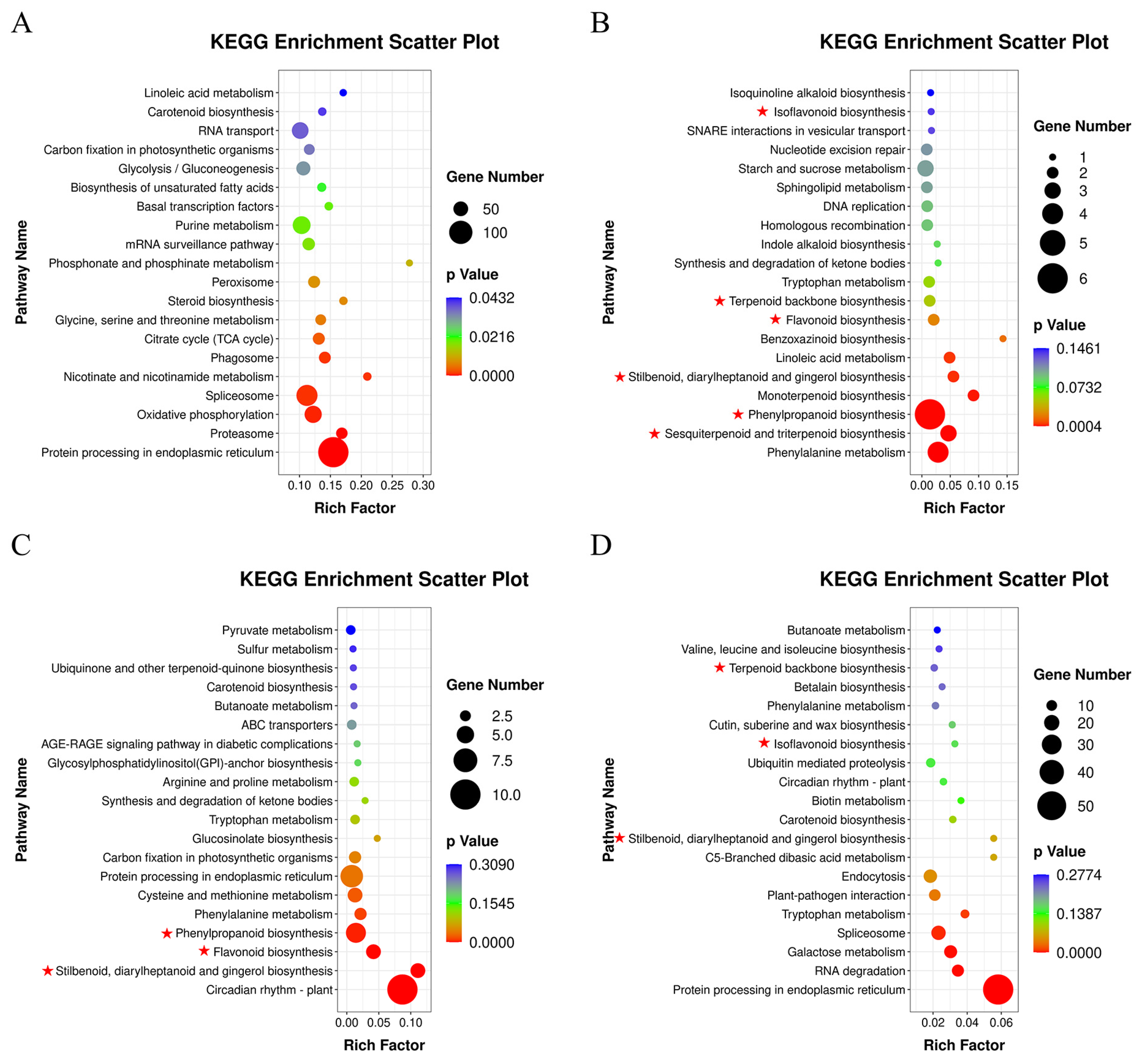
3.4. DEGs Analysis of A. mongholicus after Cutting
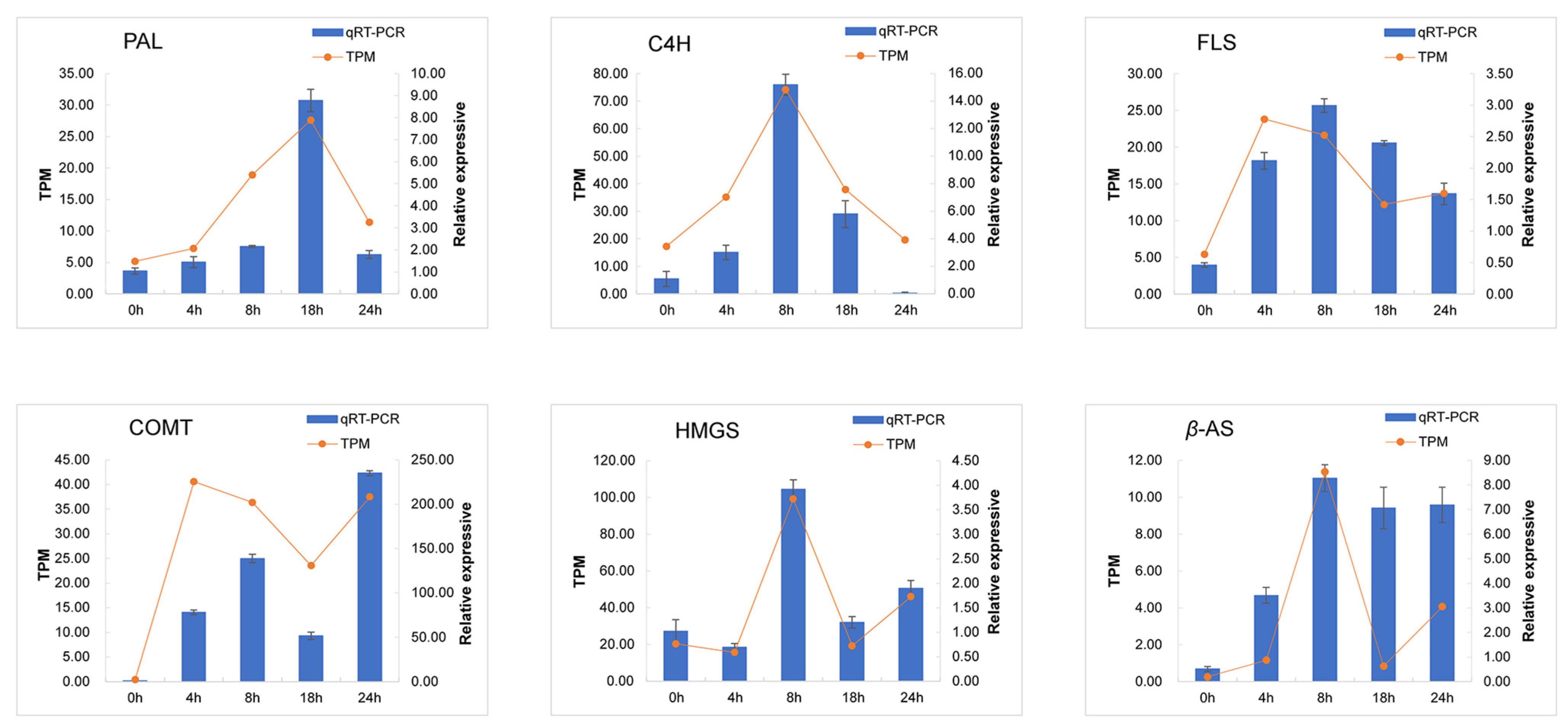
3.5. Quantitative PCR Validation of DEGs
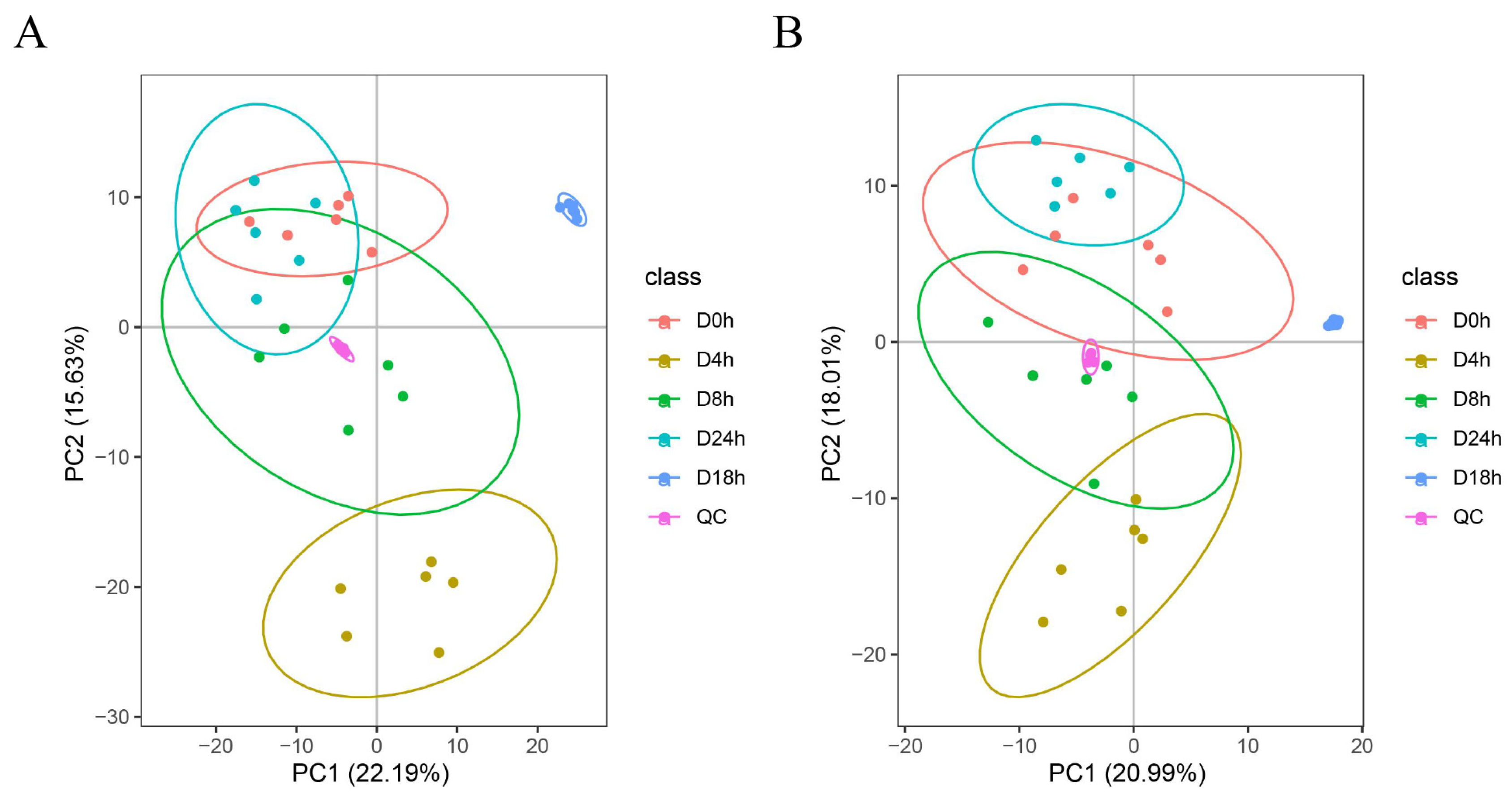
3.6. Metabolome Analysis of A. mongholicus after Cutting
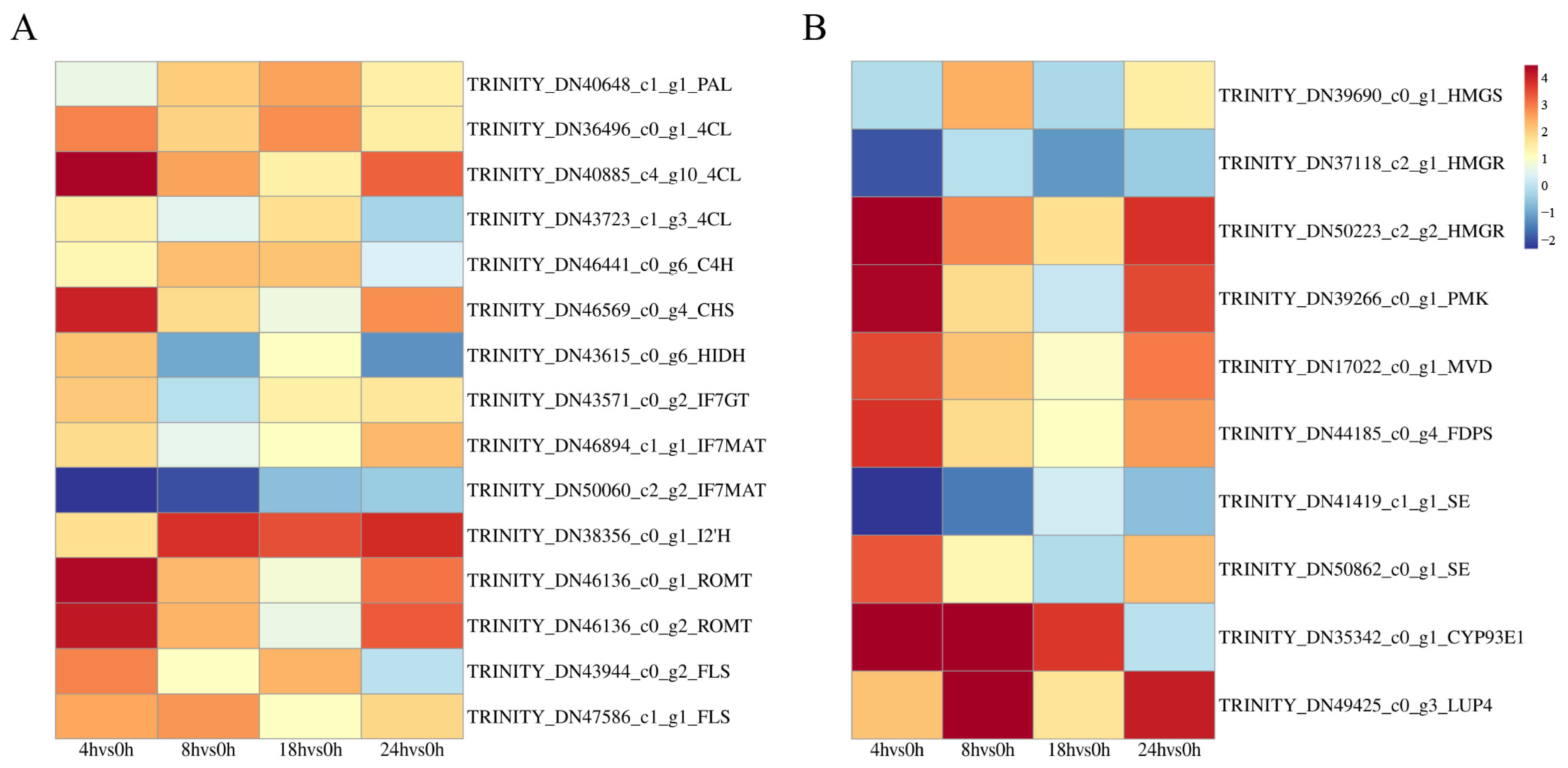
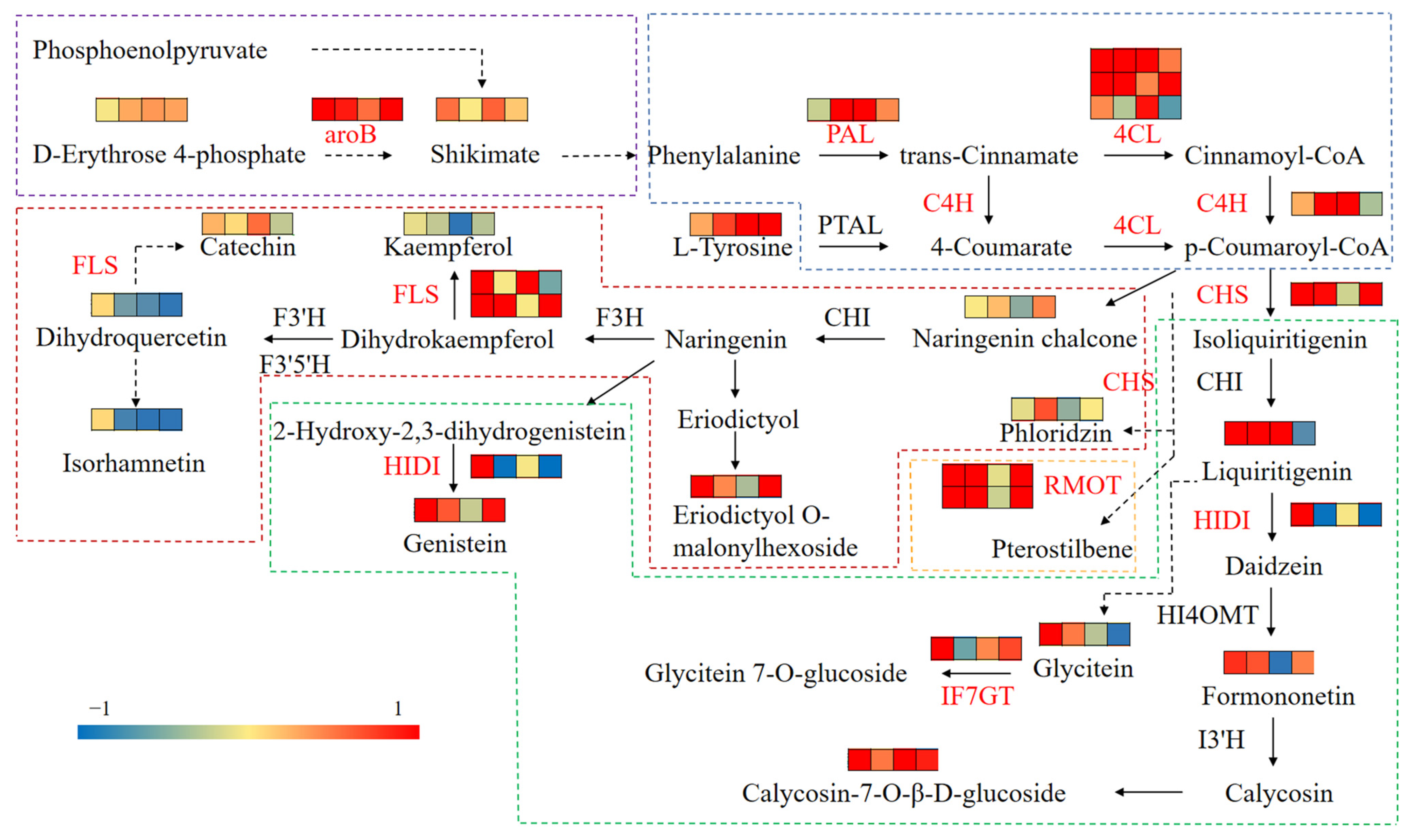
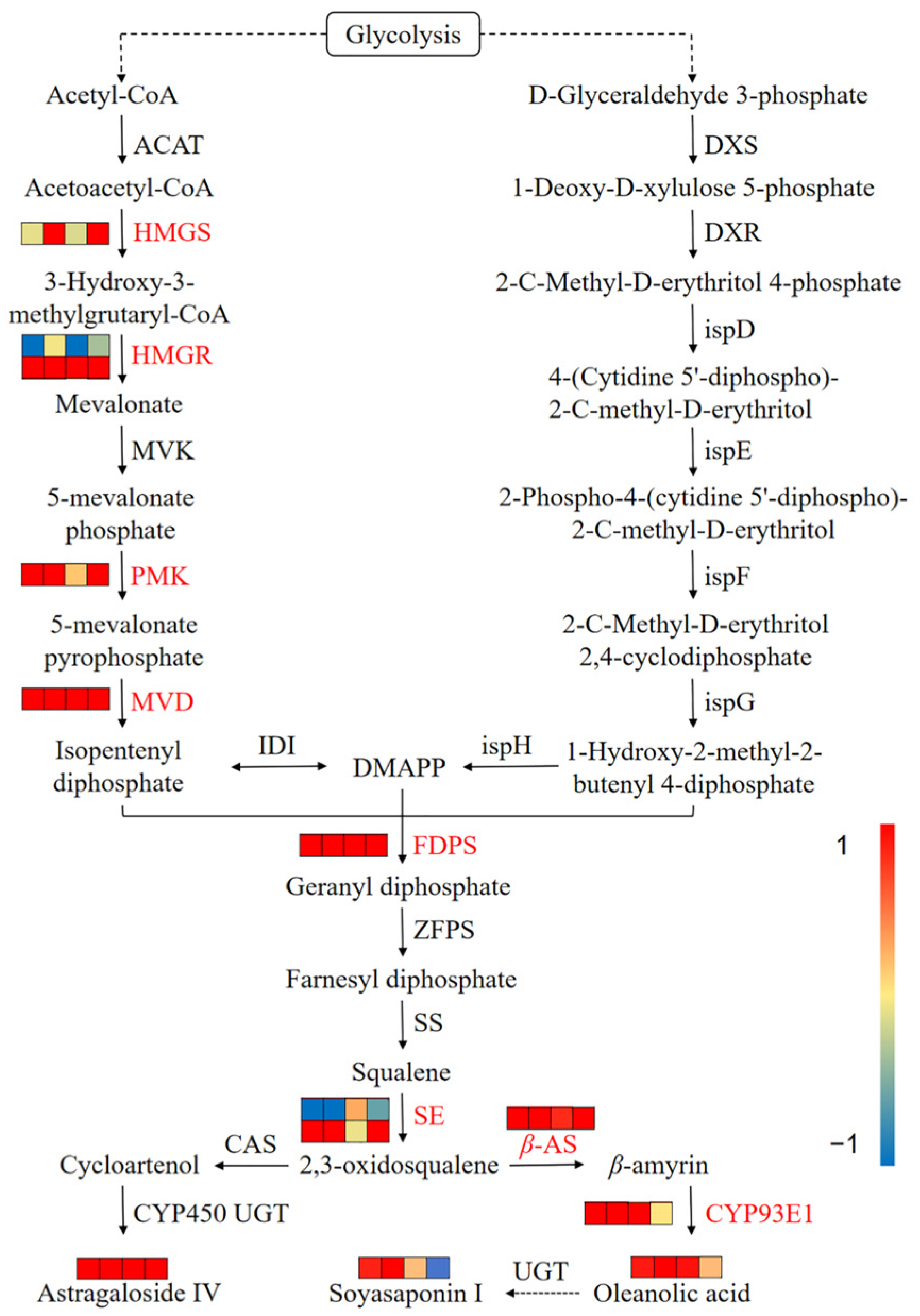
3.7. Integrated Analysis of Transcriptomics and Metabolomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- State Pharmacopeia Committee of China. Pharmacopoeia of the People’s Republic of China; Chemical Industry Press: Beijing, China, 2020; Chapter 1; pp. 315–316. [Google Scholar]
- Xu, B.Q.; Chen, Y.; Guo, F.X.; Bai, D.T.; Zhou, D.X. Effect of transplanting densities on growth, development, yield and quality of Astragalus membranaceus var. Mongholicus. Chin. J. Exp. Tradit. Med. Form. 2020, 26, 135–143. [Google Scholar] [CrossRef]
- Dai, Y.T.; Zhang, X.Y.; Wang, Y.X.; Fan, Y.X.; Yang, D.; Liu, Y.Q.; Xiu, M.H.; He, J.Z. Modern research progress and predictive analysis of quality markers of Astragali radix. Chin. J. Chin. Mater. Med. 2022, 47, 1754–1764. [Google Scholar] [CrossRef]
- Hu, M.X.; Guo, B.L.; Zhou, R.; Huang, W.H.; Cao, X.J.; Hou, M.L. Quality analysis of semi-cultivated Astragalus membranaceus var. mongholicus from Shanxi Hunyuan. Chin. Tradit. Herb. Drug 2012, 43, 1829–1834. [Google Scholar]
- Qin, X.M.; Li, Z.Y.; Sun, H.F.; Zhang, L.Z.; Zhou, R.; Feng, Q.J.; Li, A.P. Resource status and analysis of Astragalus in China. Chin. J. Chin. Mater. Med. 2013, 13, 3234–3238. [Google Scholar]
- Zhang, Z.B. Several problems in the study of plant overall stress resistance to environmental stress. Acta Agric. Boreali-Occident. Sin. 2000, 43, 112–116. [Google Scholar]
- Huang, L.Q.; Guo, L.P. Accumulation of secondary metabolites and formation of genuine medicinal herbs under environmental stress. Chin. J. Chin. Mater. Med. 2007, 32, 227–280. [Google Scholar]
- Heil, M.; Silva Bueno, J.C. Within plant signaling by volailes leads to induction and priming of an idiretplant defense in nature. Proc. Natl. Acad. Sci. USA 2007, 104, 5467–5472. [Google Scholar] [CrossRef]
- Rodriguez-Saona, C.R.; Rodriguez-Saona, L.E.; Frost, C.J. Herbivore-induced volatiles in the perennial shrub, Vaccinium corymbosum, and their role in inter-branch signaling. J. Chem. Ecol. 2009, 35, 163–175. [Google Scholar] [CrossRef]
- Wu, J.Q.; Baldwin, I.T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef]
- Wu, J.Q.; Baldwin, I.T. Herbivory-induced signalling in plants: Perception and action. Plant Cell Environ. 2009, 32, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.X.; Chai, T.Y.; Li, J.C.; Zhao, W.M. Expression of DnaJ-like gene under different environmental stress. Acta Bot. Boreali-Occident. Sin. 2000, 20, 171–174. [Google Scholar]
- Wang, H.Q.; Xiao, M.X.; An, Q.; Zheng, F.L.; Xiao, M.J.; Dai, Q.Y. Effects of light and mechanical damage stress on aroma quality of fresh tea leaves after harvest. J. Fujian Agric. For. Univ. (Nat. Sci. Ed.) 2022, 51, 162–170. [Google Scholar] [CrossRef]
- Zhang, P.; Xue, S.Y.; Li, X.F.; Xu, D.P.; Cui, Z.Y. Physiological mechanism of response to fungal infection and mechanical damage stress. For. Res. 2022, 35, 47–54. [Google Scholar] [CrossRef]
- Pagassini, J.A.V.; de Godoy, L.J.G.; Campos, F.G.; Barzotto, G.R.; Vieira, M.A.R.; Boaro, C.S.F. Silicon and mechanical damage increase polyphenols and vitexin in Passiflora incarnata L. Sci. Rep. 2021, 11, 22064. [Google Scholar] [CrossRef] [PubMed]
- Holopainen, J.K.; Gershenzon, J. Multiple stress factors and the emission of plant VOCs. Trends Plant Sci. 2010, 15, 176–184. [Google Scholar] [CrossRef]
- Dombrowski, J.E.; Kronmiller, B.A.; Hollenbeck, V.G.; Rhodes, A.C.; Henning, J.A.; Martin, R.C. Transcriptome analysis of the model grass Lolium temulentum exposed to green leaf volatiles. BMC Plant Biol. 2019, 19, 222. [Google Scholar] [CrossRef]
- Mostafa, S.; Wang, Y.; Zeng, W.; Jin, B. Plant Responses to Herbivory, Wounding, and Infection. Int. J. Mol. Sci. 2022, 23, 7031. [Google Scholar] [CrossRef]
- Guo, L.Z.; Zhang, F.S.; Li, C.J. Effects of decapitation on growth, uptake and distribution of potassium in tobacco. Chin. J. Appl. Ecol. 2002, 13, 819–822. [Google Scholar] [CrossRef]
- Lu, Z.G.; Zhu, L.K.; Lu, J.K.; Shen, N.; Wang, L.; Liu, S.; Wang, Q.J.; Yu, W.W.; Kato-Noguchi, H.; Li, W.X.; et al. Rejuvenation increases leaf biomass and flavonoid accumulation in Ginkgo biloba. Hortic. Res. 2022, 9, uhab018. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Redfern, S.P.; Kirkup, D.; Porter, E.A.; Kite, G.C.; Terry, L.A.; Berry, M.J.; Simmonds, M.S. Variation of theanine, phenolic, and methylxanthine compounds in 21 cultivars of Camellia sinensis harvested in different seasons. Food Chem. 2017, 220, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73. [Google Scholar] [CrossRef]
- Nabavi, S.M.; Šamec, D.; Tomczyk, M.; Milella, L.; Russo, D.; Habtemariam, S.; Suntar, I.; Rastrelli, L.; Daglia, M.; Xiao, J.; et al. Flavonoid biosynthetic pathways in plants: Versatile targets for metabolic engineering. Biotechnol. Adv. 2020, 38, 107316. [Google Scholar] [CrossRef]
- Li, J.; Li, C.F.; Gou, J.B.; Zhang, Y.S. Molecular cloning and functional characteri-zation of a novel isoflavone 3-O-methyltransferase from Pueraria lobata. Front. Plant Sci. 2016, 7, 793. [Google Scholar] [CrossRef]
- Kim, S.; Hwang, G.; Lee, S.; Zhu, J.Y.; Paik, I.; Nguyen, T.T.; Kim, J.; Oh, E. High ambient temperature represses anthocyanin biosynthesis through degradation of HY5. Front. Plant Sci. 2017, 8, 1787. [Google Scholar] [CrossRef]
- Wu, L.Y.; Fang, Z.T.; Lin, J.K.; Sun, Y.; Du, Z.Z.; Guo, Y.L.; Liu, J.H.; Liang, Y.R.; Ye, J.H. Complementary iTRAQ proteomic and transcriptomic analyses of leaves in tea plant (Camellia sinensis L.) with different maturity and regulatory network of flavonoid biosynthesis. J. Proteome Res. 2019, 18, 252–264. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Ren, C.X.; Dong, S.; Chen, C.; Xian, B.; Wu, Q.H.; Wang, J.; Chen, J.; Pei, J. Integrated Metabolomics and Transcriptome Analysis of Flavonoid Biosynthesis in Safflower (Carthamus tinctorius L.) With Different Colors. Front. Plant Sci. 2021, 12, 712038. [Google Scholar] [CrossRef] [PubMed]
- Hu, P.F.; Suriguga; Zhao, M.; Chen, S.Q.; Wu, X.H.; Wan, Q. Transcriptional regulation mechanism of flavonoids biosynthesis gene during fruit development in Astragalus membranaceus. Front. Genet. 2022, 13, 972990. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Sun, C.S.; Zuo, Y.C.; Li, G.Y.; Li, G.B.; Ren, L.Y.; Chen, G.L. Integrating transcriptomics and metabolomics to characterise the response of Astragalus membranaceus Bge. var. mongolicus (Bge.) to progressive drought stress. BMC Genom. 2016, 17, 188. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, X.J.; Sheng, J.H. Integrative analysis of the transcriptome and metabolome reveals the mechanism of saline–alkali stress tolerance in Astragalus membranaceus (Fisch) Bge. var. mongholicus (Bge.) Hsiao. Food Qual Saf. 2022, 6, fyac001. [Google Scholar] [CrossRef]
- Zhang, S.J.; Zhang, Y.G.; Niu, J.T.; Si, X.L.; Li, D.H.; Wu, H.W. Research progress and predictive analysis of quality markers of Astragalus mongolicus. Chin. Arch. Tradit. Chin. Med. 2022, 40, 151–155. [Google Scholar] [CrossRef]
- Chen, K.; Zhang, M.; Xu, L.; Yi, Y.; Wang, L.; Wang, H.; Wang, Z.L.; Xing, J.T.; Li, P.; Zhao, X.H.; et al. Identification of oxidosqualene cyclases associated with saponin biosynthesis from Astragalus membranaceus reveals a conserved motif important for catalytic function. J. Adv. Res. 2023, 43, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Fang, T.; Su, H.; Duan, S.F.; Ma, R.R.; Wang, P.; Wu, L.; Sun, W.B.; Hu, Q.C.; Zhao, M.X.; et al. A reference-grade genome assembly for Astragalus mongholicus and insights into the biosynthesis and high accumulation of triterpenoids and flavonoids in its roots. Plant Commun. 2023, 4, 100469. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Nie, J.; Han, Y.; Pei, Y.; Yang, N.; Liang, Y.; Zha, X. Content Determination of Astragaloside IV in Astragalus from Three Different Regions by HPLC. Med. Plant 2018, 9, 15–18. [Google Scholar]
- Jiao, J.; Gai, Q.Y.; Wang, W. Enhanced astragaloside production and transcriptional responses of biosynthetic genes in Astragalus membranaceus hairy root cultures by elicitation with methyl jasmonate. Biochem. Eng. J. 2016, 105, 339–346. [Google Scholar] [CrossRef]
- Gai, Q.Y.; Jiao, J.; Luo, M.; Wang, W.; Zhao, C.J.; Fu, Y.J.; Ma, W. UV elicitation for promoting astragaloside production in Astragalus membranaceus hairy root cultures with transcriptional expression of biosynthetic genes. Ind. Crops Prod. 2016, 84, 350–357. [Google Scholar] [CrossRef]
- Wei, H.; Cheng, L.; Wu, P.; Han, M.; Yang, L.M. Effects of astragalosides biosynthesis on short-term water changes. China J. Chin. Mater. Med. 2019, 44, 441–447. [Google Scholar] [CrossRef]
- Saini, R.; Adhikary, A.; Juneja, S.; Kumar, R.; Singh, I.; Nayyar, H. Drought priming triggers diverse metabolic adjustments and induces chilling tolerance in chickpea (Cicer arietinum L.). Plant Physiol. Biochem. 2023, 194, 418–439. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Wang, Y.; Wang, T.; Xu, H.Y.; Wu, R.R.; Chen, H. De novo assembly and annotation of the Zhe-Maidong (Ophiopogon japonicus (L.f.) Ker-Gawl) transcriptome in different growth stages. Sci. Rep. 2017, 7, 3616. [Google Scholar] [CrossRef]
- Wang, X.Q.; Hai, L.Q.; Pa, L.D.; Nurbia; Ma, Y.L.; Mayila, N. Determination of total flavonoids in different parts of Astragalus tianshanyanensis ( Hedysarum semenovii Regel et Herd). J. Xinjiang Norm. Univ. (Nat. Sci. Ed.) 2006, 25, 49–51. [Google Scholar]
- Liang, J.P.; Jia, X.Y.; Liu, Y.L.; Wu, Y.; Zhou, R.; Feng, Q.J. Effects of drought stress on growth and content of secondary metabolites in root of Astragalus mongolicus. Acta Ecol. Sin. 2016, 36, 4415–4422. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Method. 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Q.; Fang, X.; Wu, X.M.; Mao, Y.B.; Wang, L.J.; Chen, X.Y. Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 2012, 54, 703–712. [Google Scholar] [CrossRef] [PubMed]
- Want, E.J.; Masson, P.; Michopoulos, F.; Wilson, I.D.; Theodoridis, G.; Plumb, R.S.; Shockcor, J.; Loftus, N.; Holmes, E.; Nicholson, J.K. Global metabolic profiling of animal and human tissues via UPLC-MS. Nat. Protoc. 2013, 8, 17–32. [Google Scholar] [CrossRef]
- Wang, X.; Tao, X.L.; Xu, R.S.; Tian, S.L. Apical-grain superiority in hybrid rice. Acta Agron. 2001, 6, 980–985. [Google Scholar]
- Jacobo-Velázquez, D.A.; Martínez-Hernández, G.B.; Del C Rodríguez, S.; Cao, C.M.; Cisneros-Zevallos, L. Plants as biofactories: Physiological role of reactive oxygen species on the accumulation of phenolic antioxidants in carrot tissue under wounding and hyperoxia stress. J. Agric. Food Chem. 2011, 59, 6583–6593. [Google Scholar] [CrossRef]
- Gao, K.; Zhang, Z.X.; Zhu, T.X.; Coulter, J.A. The influence of flower removal on tuber yield and biomass characteristics of Helianthus tuberosus L. in a semi-arid area. Ind. Crops Prod. 2020, 150, 112374. [Google Scholar] [CrossRef]
- Liu, R.X.; Li, Y.J.; Li, L.; Miu, X.S.; Wang, X.S.; Zhang, D.; Wei, S.L. Study on the effect of cutting seedlings on the growth of Scutellaria baicalensis and the yield and quality of medicinal materials, stems and leaves. Chin. J. Chin. Mater. Med. 2016, 41, 2049–2054. [Google Scholar]
- Lang, Y.H.; Nie, J.H.; Cao, Z.M. Advance in the alteration of gene expression during environment stress in plants. Biotechnol. Bull. 1999, 15, 11–14. [Google Scholar]
- Luo, J. Metabolite-based genome-wide association studies in plants. Curr. Opin. Plant Biol. 2015, 24, 31–38. [Google Scholar] [CrossRef]
- Mao, C.L.; Li, L.; Yang, T.; Gui, M.C.; Li, X.Q.; Zhang, F.L. Transcriptomics integrated with widely targeted metabolomics reveals the cold resistance mechanism in Hevea brasiliensis. Front. Plant Sci. 2023, 13, 1092411. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Fang, Y.; Huang, M.-J.; Xiao, Y.; Liu, Y.; Ma, X.-R.; Zhao, H. High flavonoid accompanied with high starch accumulation triggered by nutrient starvation in bioenergy crop duckweed (Landoltia punctata). BMC Genom. 2017, 18, 166. [Google Scholar] [CrossRef] [PubMed]
- Naoumkina, M.A.; Zhao, Q.; Gallgo-Giraldo, L.; Dai, X.; Zhao, P.X.; Dixon, R.A. Genome-wide analysis of phenylpropanoid defence pathways. Mol. Plant Pathol. 2010, 11, 829–846. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, C. Multifaceted regulations of gateway enzyme phenylalanine ammonia Journal Pre-proof 40 lyase in the biosynthesis of phenylpropanoids. Mol. Plant. 2015, 8, 17–27. [Google Scholar] [CrossRef]
- Luo, X.J.; Liu, X.C.; Yang, X.Y.; Yuan, J.E.; Feng, Z.Y. Cloning and tissue expression analysis of HvC4H gene in hulless barley. J. Plant Genet. Resour. 2014, 15, 589–596. [Google Scholar] [CrossRef]
- Zhang, F.S.; Zhang, X.; Luo, Y.Y.; Li, H.J.; Qin, X.M. Biosynthetic mechanisms of isoflavone accumulation affected by different growth patterns in Astragalus mongholicus products. BMC Plant Biol. 2022, 22, 410. [Google Scholar] [CrossRef]
- Sun, H.F.; Song, M.F.; Zhang, Y.; Zhang, Z.L. Transcriptome profiling reveals candidate flavonoid-related genes during formation of dragon’s blood from Dracaena cochinchinensis (Lour.) S.C. Chen under conditions of wounding stress. J. Ethnopharmacol. 2021, 273, 113987. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Hofer, B. Recent developments in the enzymatic O-glycosylation of flavonoids. Appl. Microbiol. Biotechnol. 2016, 100, 4269–4281. [Google Scholar] [CrossRef]
- Bowles, D.; Lim, E.K.; Poppenberger, B.; Vaistij, F.E. Glycosyltransferases of lipophilic small molecules. Annu. Rev. Plant Biol. 2006, 57, 567–597. [Google Scholar] [CrossRef] [PubMed]
- Dudareva, N.; Andersson, S.; Orlova, I.; Gatto, N.; Reichelt, M.; Rhodes, D.; Boland, W.; Gershenzon, J. The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 2005, 102, 933–938. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, G.; Gao, Z.; Sui, C.; Ji, H.; Jiang, J.; Guo, X.; Wei, J. Transcriptome profiling of Bupleurum chinense DC. root provides new insights into the continuous inflorescence removal induced improvements to root growth and saikosaponin biosynthesis. Ind. Crops Prod. 2020, 160, 113085. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.P.; An, J.; Wang, Q.L.; Jing, Y.J.; Huang, J.P. Transcriptome analysis of Astragalus mongholicus with different growth years and mining of key genes for triterpenoid saponins synthesis. China Tradit. Herb. Drugs 2023, 54, 915–925. [Google Scholar]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Yan, X.; Wang, Y.; Shi, Z.; Niu, J.; Liang, J.; Jia, X. Integrated Transcriptomics and Metabolomics Analysis Reveals the Effects of Cutting on the Synthesis of Flavonoids and Saponins in Chinese Herbal Medicine Astragalus mongholious. Metabolites 2024, 14, 97. https://doi.org/10.3390/metabo14020097
Guo X, Yan X, Wang Y, Shi Z, Niu J, Liang J, Jia X. Integrated Transcriptomics and Metabolomics Analysis Reveals the Effects of Cutting on the Synthesis of Flavonoids and Saponins in Chinese Herbal Medicine Astragalus mongholious. Metabolites. 2024; 14(2):97. https://doi.org/10.3390/metabo14020097
Chicago/Turabian StyleGuo, Xu, Xiang Yan, Yuanyuan Wang, Zhiyong Shi, Jingping Niu, Jianping Liang, and Xiaoyun Jia. 2024. "Integrated Transcriptomics and Metabolomics Analysis Reveals the Effects of Cutting on the Synthesis of Flavonoids and Saponins in Chinese Herbal Medicine Astragalus mongholious" Metabolites 14, no. 2: 97. https://doi.org/10.3390/metabo14020097
APA StyleGuo, X., Yan, X., Wang, Y., Shi, Z., Niu, J., Liang, J., & Jia, X. (2024). Integrated Transcriptomics and Metabolomics Analysis Reveals the Effects of Cutting on the Synthesis of Flavonoids and Saponins in Chinese Herbal Medicine Astragalus mongholious. Metabolites, 14(2), 97. https://doi.org/10.3390/metabo14020097





