Temperature Dependence of Platelet Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation and Storage of PCs
2.2. Sample Collection
2.3. Assays
2.4. Metabolomics
2.5. Temperature Coefficient Q10 Calculation
2.6. Constraint-Based Metabolic Modeling
3. Results
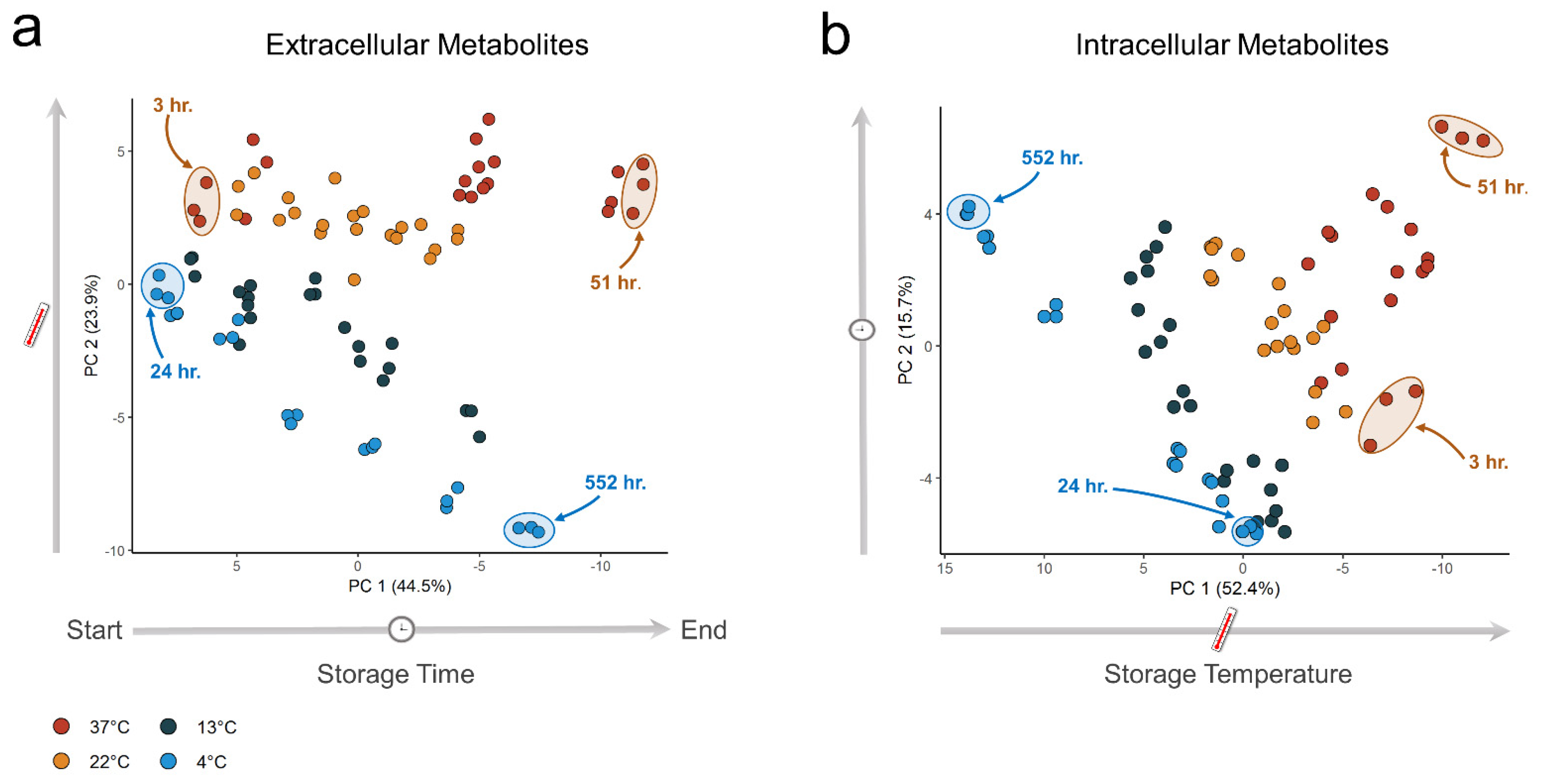
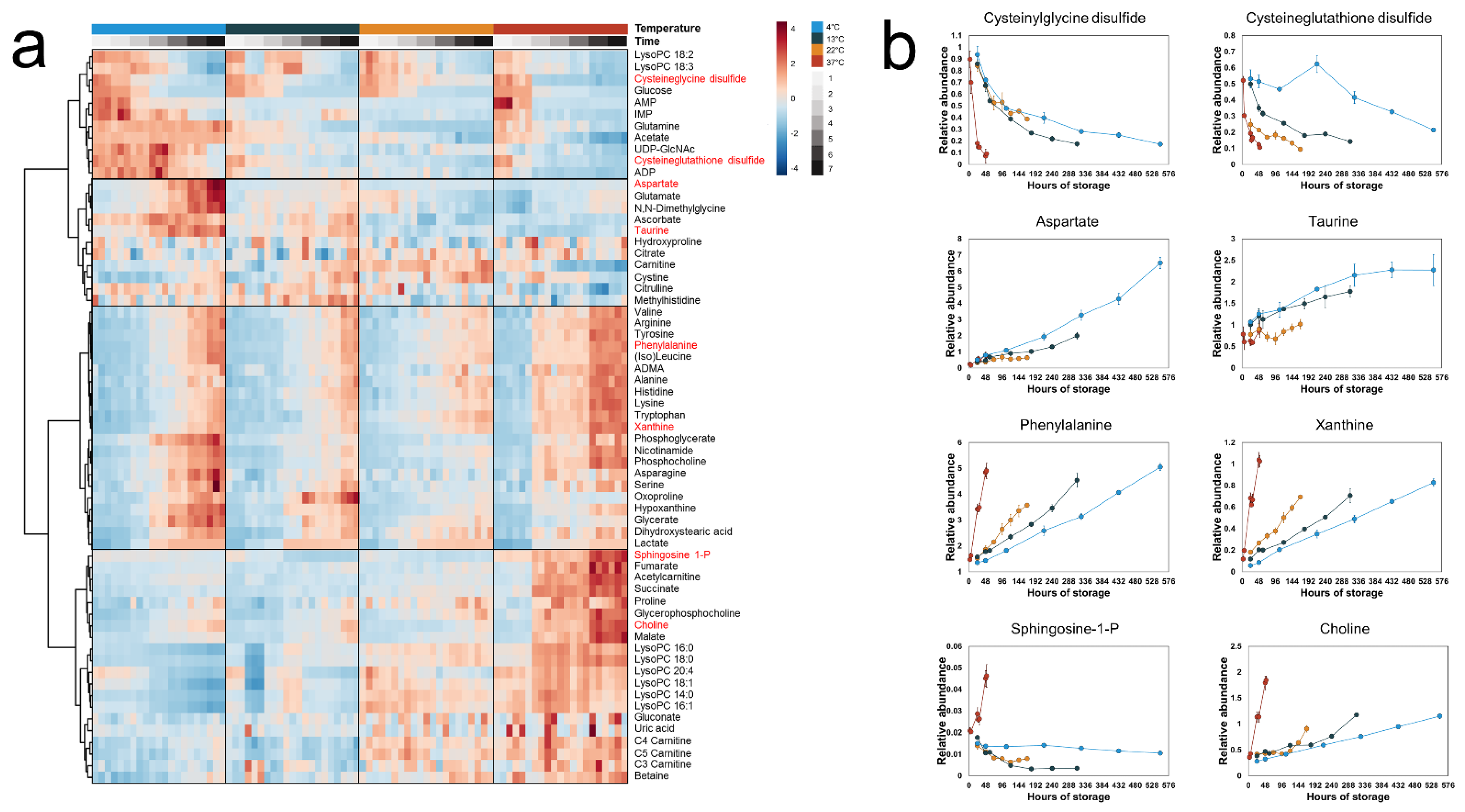
3.1. Metabolite Time–Concentration Profiles Scale Non-Uniformly with Temperature
3.2. A Subset of Extracellular Metabolites Follows an Arrhenius-Type Relationship with Temperature
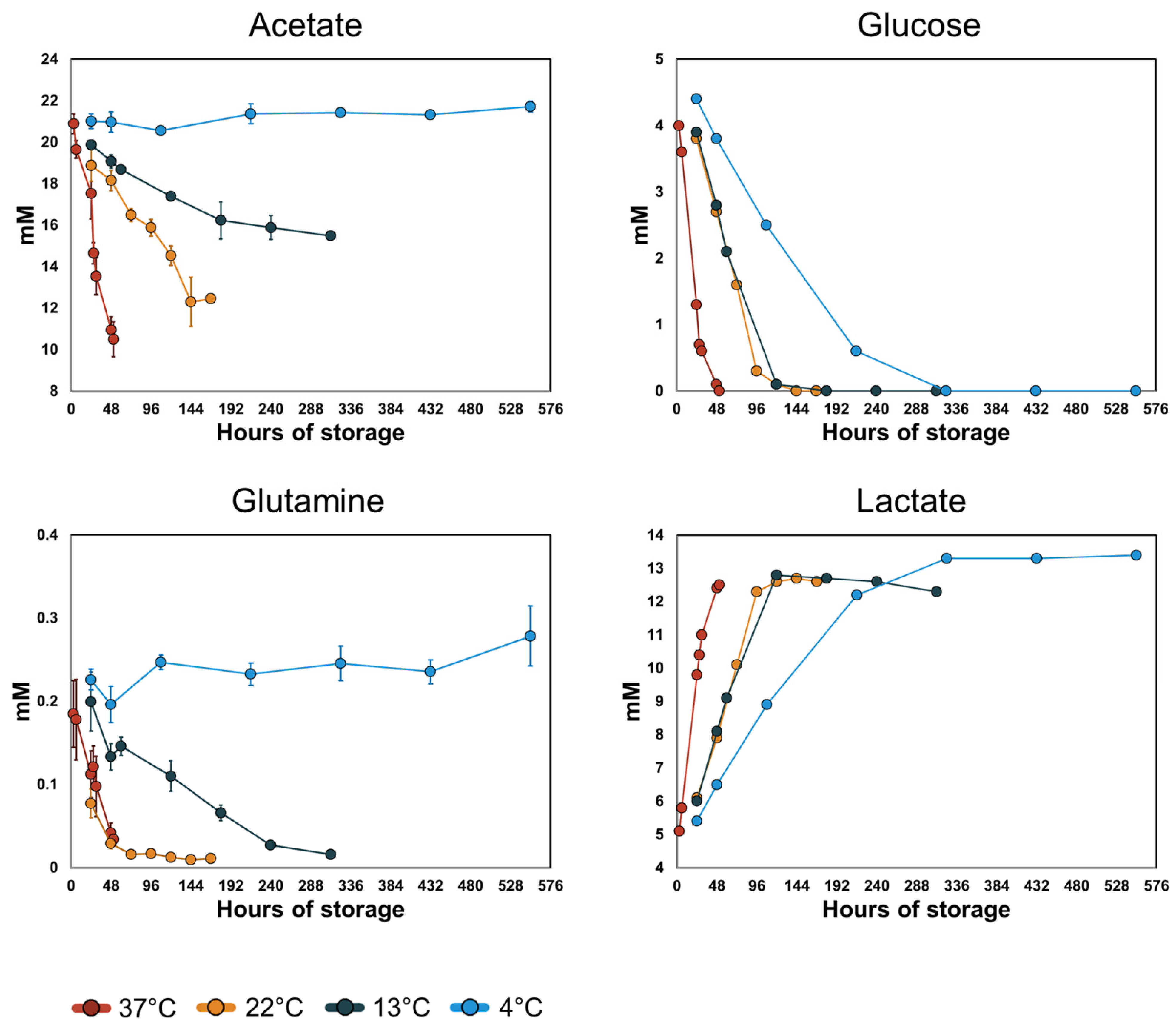
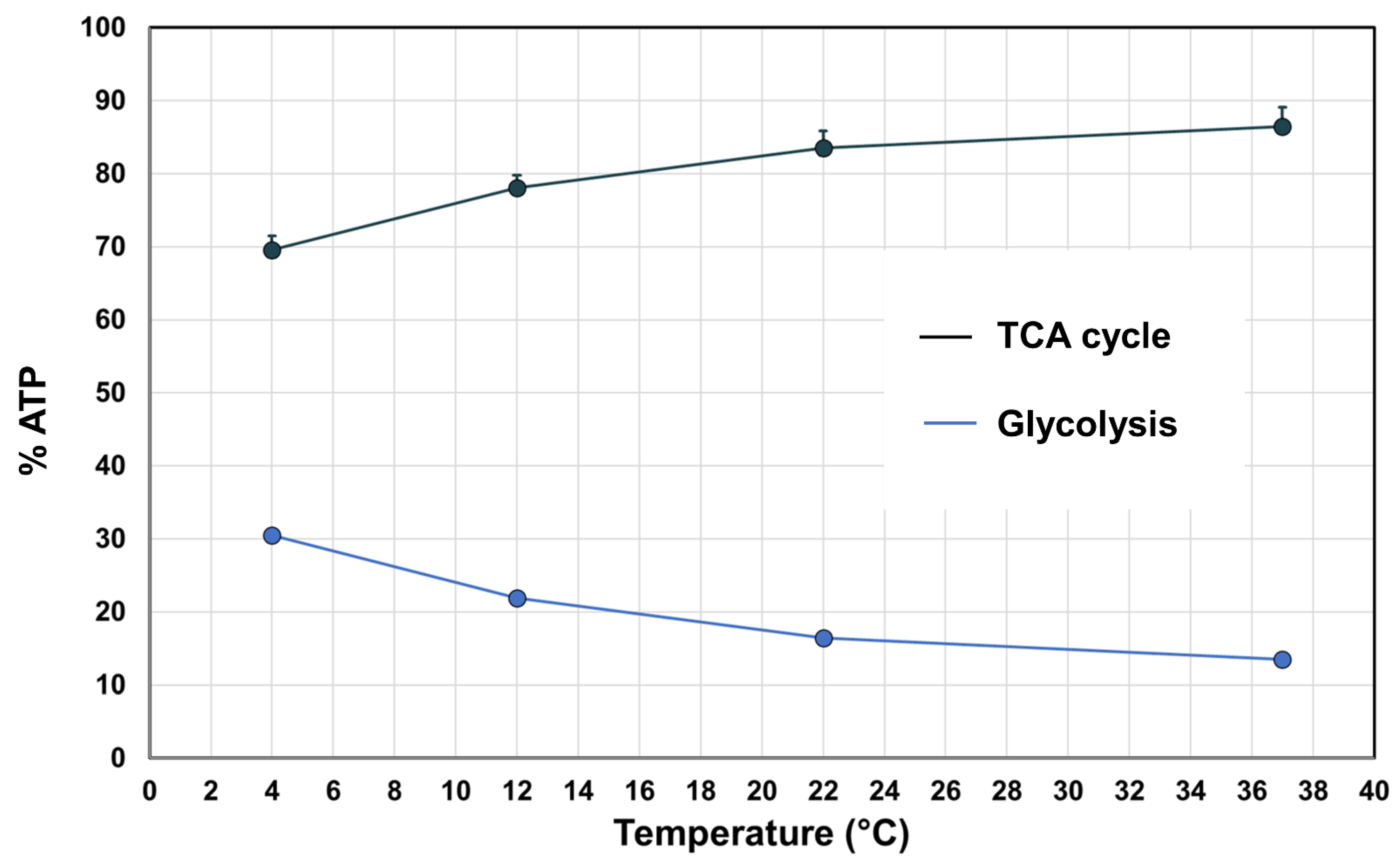
3.3. Acetate and Glutamine Metabolism Is More Sensitive to Temperature Than Glucose Metabolism
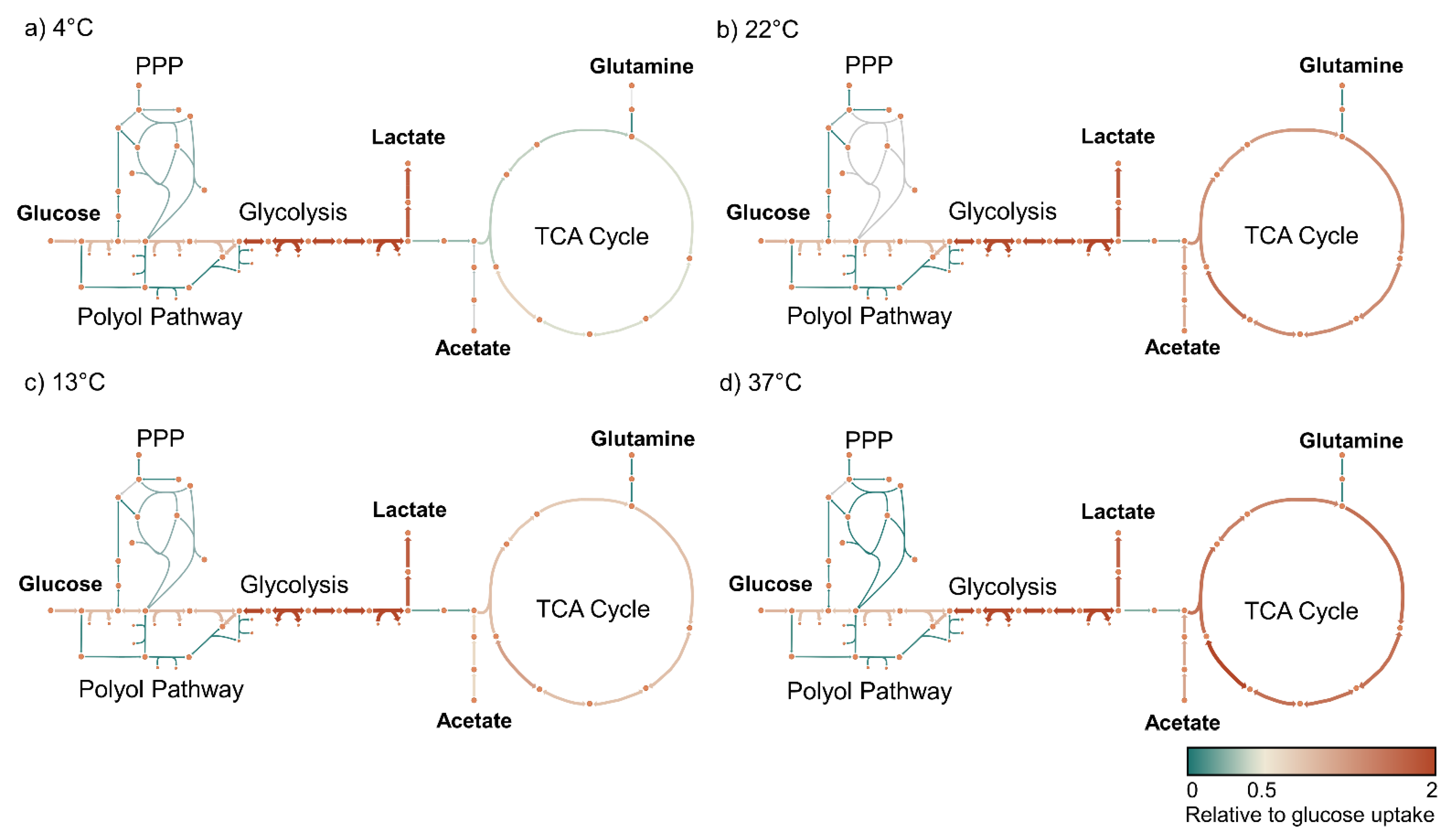
3.4. Metabolic Modeling Reveals Pathway-Specific Temperature Dependence
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arrhenius, S. Über die Reaktionsgeschwindigkeit bei der Inversion von Rohrzucker durch Säuren. Z. Für Phys. Chem. 1889, 4, 226. [Google Scholar] [CrossRef]
- Laidler, K.J. Unconventional applications of the Arrhenius law. J. Chem. Educ. 1972, 49, 343. [Google Scholar] [CrossRef]
- Stein, R.B.; Gordon, T.; Shriver, J. Temperature dependence of mammalian muscle contractions and ATPase activities. Biophys. J. 1982, 40, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Anderson, G.; Scott, M. Determination of product shelf life and activation energy for five drugs of abuse. Clin. Chem. 1991, 37, 398–402. [Google Scholar] [CrossRef] [PubMed]
- Jorjani, P.; Ozturk, S.S. Effects of cell density and temperature on oxygen consumption rate for different mammalian cell lines. Biotechnol. Bioeng. 1999, 64, 349–356. [Google Scholar]
- Giacomin, M.; Schulte, P.M.; Wood, C.M. Differential Effects of Temperature on Oxygen Consumption and Branchial Fluxes of Urea, Ammonia, and Water in the Dogfish Shark (Squalus acanthias suckleyi). Physiol. Biochem. Zool. PBZ 2017, 90, 627–637. [Google Scholar] [CrossRef]
- Yurkovich, J.T.; Zielinski, D.C.; Yang, L.; Paglia, G.; Rolfsson, O.; Sigurjonsson, O.E.; Broddrick, J.T.; Bordbar, A.; Wichuk, K.; Brynjolfsson, S.; et al. Quantitative time-course metabolomics in human red blood cells reveal the temperature dependence of human metabolic networks. J. Biol. Chem. 2017, 292, 19556–19564. [Google Scholar] [CrossRef]
- Hegarty, T.W. Temperature Coefficient (Q10), Seed Germination and Other Biological Processes. Nature 1973, 243, 305–306. [Google Scholar] [CrossRef]
- Cohen Stuart, C.P. A study of temperature-coefficients and van‘t Hoff’s rule. K. Ned. Akad. Van Wet. Proc. Ser. B Phys. Sci. 1912, 14, 1159–1173. [Google Scholar]
- Chaui-Berlinck, J.G.; Navas, C.A.; Monteiro, L.H.; Bicudo, J.E. Temperature effects on a whole metabolic reaction cannot be inferred from its components. Proc. Biol. Sci. 2004, 271, 1415–1419. [Google Scholar] [CrossRef]
- Chaui-Berlinck, J.G.; Monteiro, L.H.; Navas, C.A.; Bicudo, J.E. Temperature effects on energy metabolism: A dynamic system analysis. Proc. Biol. Sci. 2002, 269, 15–19. [Google Scholar] [CrossRef]
- Donhoffer, S. The regulation of energy metabolism and van’t Hoff’s rule in the homeotherm animal. Helgol. Mar. Res. 1966, 14, 541–558. [Google Scholar] [CrossRef]
- Gillooly, J.F.; Brown, J.H.; West, G.B.; Savage, V.M.; Charnov, E.L. Effects of Size and Temperature on Metabolic Rate. Science 2001, 293, 2248–2251. [Google Scholar] [CrossRef] [PubMed]
- Orth, J.D.; Thiele, I.; Palsson, B. What is flux balance analysis? Nat. Biotechnol. 2010, 28, 245–248. [Google Scholar] [CrossRef] [PubMed]
- Bordbar, A.; Yurkovich, J.T.; Paglia, G.; Rolfsson, O.; Sigurjónsson, Ó.E.; Palsson, B.O. Elucidating dynamic metabolic physiology through network integration of quantitative time-course metabolomics. Sci. Rep. 2017, 7, 46249. Available online: https://www.nature.com/articles/srep46249#supplementary-information (accessed on 12 December 2018). [CrossRef] [PubMed]
- Schubert, P.; Devine, D.V. De novo protein synthesis in mature platelets: A consideration for transfusion medicine. Vox Sang. 2010, 99, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, G.A.; Weyrich, A.S. Signal-dependent protein synthesis by activated platelets: New pathways to altered phenotype and function. Arterioscler. Thromb. Vasc. Biol. 2008, 28, s17–s24. [Google Scholar] [CrossRef] [PubMed]
- Guppy, M.; Whisson, M.E.; Sabaratnam, R.; Withers, P.; Brand, K. Alternative fuels for platelet storage: A metabolic study. Vox Sang. 1990, 59, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mhaskar, R.; Grossman, B.J.; Kaufman, R.M.; Tobian, A.A.; Kleinman, S.; Gernsheimer, T.; Tinmouth, A.T.; Djulbegovic, B. Platelet transfusion: A systematic review of the clinical evidence. Transfusion 2015, 55, 1116–1127, quiz 1115. [Google Scholar] [CrossRef]
- Devine, D.V.; Serrano, K. The platelet storage lesion. Clin. Lab. Med. 2010, 30, 475–487. [Google Scholar] [CrossRef]
- Murphy, S.; Gardner, F.H. Effect of storage temperature on maintenance of platelet viability—Deleterious effect of refrigerated storage. N. Engl. J. Med. 1969, 280, 1094–1098. [Google Scholar] [CrossRef]
- Marini, I.; Aurich, K.; Jouni, R.; Nowak-Harnau, S.; Hartwich, O.; Greinacher, A.; Thiele, T.; Bakchoul, T. Cold storage of platelets in additive solution: The impact of residual plasma in apheresis platelet concentrates. Haematologica 2019, 104, 207–214. [Google Scholar] [CrossRef]
- Stubbs, J.R.; Tran, S.A.; Emery, R.L.; Hammel, S.A.; Haugen, D.A.L.; Zielinski, M.D.; Zietlow, S.P.; Jenkins, D. Cold platelets for trauma-associated bleeding: Regulatory approval, accreditation approval, and practice implementation—Just the “tip of the iceberg”. Transfusion 2017, 57, 2836–2844. [Google Scholar] [CrossRef]
- Wood, B.; Johnson, L.; Hyland, R.A.; Marks, D.C. Maximising platelet availability by delaying cold storage. Vox Sang. 2018, 113, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Devine, D.V. The Missing Pieces to the Cold-Stored Platelet Puzzle. Int. J. Mol. Sci. 2022, 23, 1100. [Google Scholar] [CrossRef] [PubMed]
- Kilkson, H.; Holme, S.; Murphy, S. Platelet metabolism during storage of platelet concentrates at 22 degrees C. Blood 1984, 64, 406–414. [Google Scholar] [CrossRef]
- Murphy, S.; Shimizu, T.; Miripol, J. Platelet storage for transfusion in synthetic media: Further optimization of ingredients and definition of their roles. Blood 1995, 86, 3951–3960. [Google Scholar] [CrossRef]
- Murphy, S. The oxidation of exogenously added organic anions by platelets facilitates maintenance of pH during their storage for transfusion at 22 degrees C. Blood 1995, 85, 1929–1935. [Google Scholar] [CrossRef]
- Paglia, G.; Sigurjonsson, O.E.; Rolfsson, O.; Valgeirsdottir, S.; Hansen, M.B.; Brynjolfsson, S.; Gudmundsson, S.; Palsson, B.O. Comprehensive metabolomic study of platelets reveals the expression of discrete metabolic phenotypes during storage. Transfusion 2014, 54, 2911–2923. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Sigurjonsson, O.E.; Rolfsson, O.; Hansen, M.B.; Brynjolfsson, S.; Gudmundsson, S.; Palsson, B.O. Metabolomic analysis of platelets during storage: A comparison between apheresis- and buffy coat-derived platelet concentrates. Transfusion 2015, 55, 301–313. [Google Scholar] [CrossRef]
- Johannsson, F.; Guðmundsson, S.; Paglia, G.; Guðmundsson, S.; Palsson, B.; Sigurjonsson, O.E.; Rolfsson, O. Systems analysis of metabolism in platelet concentrates during storage in platelet additive solution. Biochem. J. 2018, 475, 2225–2240. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Rahmanian, S.; Bordbar, A.; Palsson, B.O.; Jamshidi, N. Network reconstruction of platelet metabolism identifies metabolic signature for aspirin resistance. Sci. Rep. 2014, 4, 3925. [Google Scholar] [CrossRef] [PubMed]
- Sandgren, P.; Shanwell, A.; Gulliksson, H. Storage of buffy coat-derived platelets in additive solutions: In vitro effects of storage at 4 degrees C. Transfusion 2006, 46, 828–834. [Google Scholar] [CrossRef] [PubMed]
- NasrEldin, E. Effect of cold storage on platelets quality stored in a small containers: Implications for pediatric transfusion. Pediatr. Hematol. Oncol. J. 2017, 2, 29–34. [Google Scholar] [CrossRef]
- Johnson, L.; Tan, S.; Wood, B.; Davis, A.; Marks, D.C. Refrigeration and cryopreservation of platelets differentially affect platelet metabolism and function: A comparison with conventional platelet storage conditions. Transfusion 2016, 56, 1807–1818. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.W.; Serrano, K.; Stefanoni, D.; D’Alessandro, A.; Devine, D.V. In Vitro Characterization and Metabolomic Analysis of Cold-Stored Platelets. J. Proteome Res. 2021, 20, 2251–2265. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Thomas, K.A.; Stefanoni, D.; Gamboni, F.; Shea, S.M.; Reisz, J.A.; Spinella, P.C. Metabolic phenotypes of standard and cold-stored platelets. Transfusion 2020, 60 (Suppl. S3), S96–S106. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for Comprehensive Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Schellenberger, J.; Palsson, B.O. Use of randomized sampling for analysis of metabolic networks. J. Biol. Chem. 2009, 284, 5457–5461. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 2019, 14, 639–702. [Google Scholar] [CrossRef]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [CrossRef]
- European Directorate for the Quality of Medicines & HealthCare. Guide to the Preparation, Use and Quality Assurance of Blood Components: Recommendation no. R (95) 15; European Directorate for the Quality of Medicines & HealthCare: Strasbourg, France, 2020. [Google Scholar]
- Thiele, I.; Swainston, N.; Fleming, R.M.T.; Hoppe, A.; Sahoo, S.; Aurich, M.K.; Haraldsdottir, H.; Mo, M.L.; Rolfsson, O.; Stobbe, M.D.; et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 2013, 31, 419. Available online: https://www.nature.com/articles/nbt.2488#supplementary-information (accessed on 20 December 2018). [CrossRef]
- Bordbar, A.; Monk, J.M.; King, Z.A.; Palsson, B.O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 2014, 15, 107–120. [Google Scholar] [CrossRef]
- Ravi, S.; Chacko, B.; Sawada, H.; Kramer, P.A.; Johnson, M.S.; Benavides, G.A.; O’Donnell, V.; Marques, M.B.; Darley-Usmar, V.M. Metabolic plasticity in resting and thrombin activated platelets. PLoS ONE 2015, 10, e0123597. [Google Scholar] [CrossRef]
- Koebmann, B.J.; Westerhoff, H.V.; Snoep, J.L.; Nilsson, D.; Jensen, P.R. The glycolytic flux in Escherichia coli is controlled by the demand for ATP. J. Bacteriol. 2002, 184, 3909–3916. [Google Scholar] [CrossRef]
- Aibibula, M.; Naseem, K.M.; Sturmey, R.G. Glucose metabolism and metabolic flexibility in blood platelets. J. Thromb. Haemost. JTH 2018, 16, 2300–2314. [Google Scholar] [CrossRef] [PubMed]
- Guppy, M.; Abas, L.; Arthur, P.G.; Whisson, M.E. The Pasteur effect in human platelets: Implications for storage and metabolic control. Br. J. Haematol. 1995, 91, 752–757. [Google Scholar] [CrossRef] [PubMed]
- Bynum, J.A.; Meledeo, M.A.; Getz, T.M.; Rodriguez, A.C.; Aden, J.K.; Cap, A.P.; Pidcoke, H.F. Bioenergetic profiling of platelet mitochondria during storage: 4 degrees C storage extends platelet mitochondrial function and viability. Transfusion 2016, 56 (Suppl. S1), S76–S84. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Aledo, J.C. Life-history Constraints on the Mechanisms that Control the Rate of ROS Production. Curr. Genom. 2014, 15, 217–230. [Google Scholar] [CrossRef] [PubMed]

| Metabolite | Q10 | R2 |
|---|---|---|
| Alanine | 2.37 | 0.99 |
| Arginine | 2.02 | 0.92 |
| Cysteineglycine disulfide | 2.14 | 0.94 |
| Glucose | 1.73 | 0.99 |
| Glycerate | 1.54 | 0.89 |
| Histidine | 2.30 | 0.98 |
| Hypoxanthine | 1.77 | 0.84 |
| (Iso)Leucine | 2.04 | 0.95 |
| Lactate | 1.70 | 0.98 |
| Lysine | 2.25 | 0.94 |
| Malate | 3.04 | 0.93 |
| Nicotinamide | 2.01 | 0.83 |
| Phenylalanine | 2.05 | 0.94 |
| Phosphocholine | 2.16 | 0.80 |
| sn-Glycero-3-phosphocholine | 2.50 | 0.98 |
| Tryptophan | 2.15 | 0.96 |
| Tyrosine | 2.15 | 0.94 |
| Valine | 2.18 | 0.97 |
| Xanthine | 2.23 | 0.94 |
| Subsystem | Q10 | R2 | Number of Reactions |
|---|---|---|---|
| Alanine and aspartate metabolism | 2.28 (1.97–2.80) | 0.91 (0.90–0.93) | 5 (5) |
| Citric acid cycle | 2.58 (1.97–2.70) | 0.97 (0.90–0.98) | 9 (13) |
| Fatty acid oxidation | 2.21 (2.22–2.55) | 0.91 (0.76–0.98) | 8 (16) |
| Glycerophospholipid metabolism | 2.23 (2.14–2.33) | 0.72 (0.71–0.95) | 37 (101) |
| Glycolysis/gluconeogenesis | 1.76 (1.72–1.77) | 0.98 (0.96–0.99) | 12 (18) |
| Inositol Phosphate metabolism | 2.21 (2.21–2.22) | 0.95 (0.93–0.96) | 4 (124) |
| Nucleotide interconversion | 2.26 (2.22–2.70) | 0.97 (0.95–0.98) | 4 (21) |
| Oxidative phosphorylation | 2.46 (2.27–2.47) | 0.98 (0.98–0.98) | 5 (5) |
| Triacylglycerol synthesis | 2.22 (2.21–2.25) | 0.82 (0.73–0.90) | 7 (43) |
| Other | 2.27 (1.73–3.65) | 0.95 (0.74–0.99) | 26 (184) |
| Total | 2.23 (1.72–3.65) | 0.90 (0.71–0.99) | 117 (530) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jóhannsson, F.; Yurkovich, J.T.; Guðmundsson, S.; Sigurjónsson, Ó.E.; Rolfsson, Ó. Temperature Dependence of Platelet Metabolism. Metabolites 2024, 14, 91. https://doi.org/10.3390/metabo14020091
Jóhannsson F, Yurkovich JT, Guðmundsson S, Sigurjónsson ÓE, Rolfsson Ó. Temperature Dependence of Platelet Metabolism. Metabolites. 2024; 14(2):91. https://doi.org/10.3390/metabo14020091
Chicago/Turabian StyleJóhannsson, Freyr, James T. Yurkovich, Steinn Guðmundsson, Ólafur E. Sigurjónsson, and Óttar Rolfsson. 2024. "Temperature Dependence of Platelet Metabolism" Metabolites 14, no. 2: 91. https://doi.org/10.3390/metabo14020091
APA StyleJóhannsson, F., Yurkovich, J. T., Guðmundsson, S., Sigurjónsson, Ó. E., & Rolfsson, Ó. (2024). Temperature Dependence of Platelet Metabolism. Metabolites, 14(2), 91. https://doi.org/10.3390/metabo14020091







