Salivary Metabolites in Breast Cancer and Fibroadenomas: Focus on Menopausal Status and BMI
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Collection, Processing, Storage and Analysis of Saliva Samples
2.3. Statistical Analysis
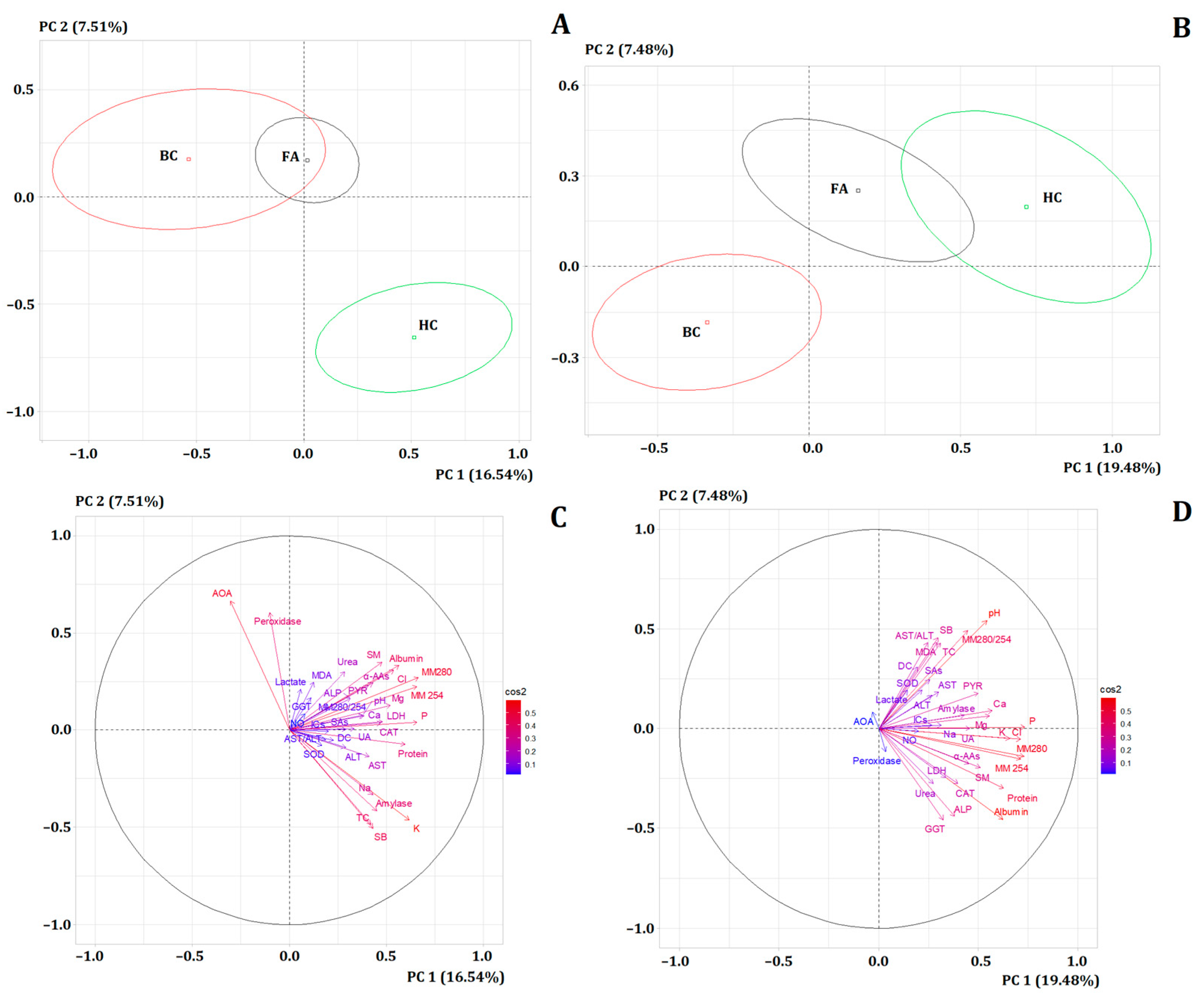
3. Results
3.1. The Influence of Age and Menopausal Status on the Biochemical Composition of Saliva
3.2. Comparison of Subgroups with Breast Cancer, Fibroadenomas, and Healthy Controls, Taking into Account the Presence/Absence of Menopause
3.3. The Influence of BMI on the Biochemical Composition of Saliva
3.4. Biochemical Composition of Saliva in Phyllodes Fibroadenomas
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chu, B.; Crystal, P. Imaging of fibroepithelial lesions: A pictorial essay. Can. Assoc. Radiol. J. 2012, 63, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Bianco, V.; Valentino, M.; Pirone, D.; Miccio, L.; Memmolo, P.; Brancato, V.; Coppola, L.; Smaldone, G.; D’Aiuto, M.; Mossetti, G.; et al. Classifying breast cancer and fibroadenoma tissue biopsies from paraffined stain-free slides by fractal biomarkers in Fourier Ptychographic Microscopy. Comput. Struct. Biotechnol. J. 2024, 24, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Basara Akin, I.; Balci, P. Fibroadenomas: A multidisciplinary review of the variants. Clin. Imaging 2021, 71, 83–100. [Google Scholar] [CrossRef] [PubMed]
- Sacca, L.; Lobaina, D.; Burgoa, S.; Lotharius, K.; Moothedan, E.; Gilmore, N.; Xie, J.; Mohler, R.; Scharf, G.; Knecht, M.; et al. Promoting Artificial Intelligence for Global Breast Cancer Risk Prediction and Screening in Adult Women: A Scoping Review. J. Clin. Med. 2024, 13, 2525. [Google Scholar] [CrossRef]
- Tan, P.H.; Ellis, I.; Allison, K.; Brogi, E.; Fox, S.B.; Lakhani, S.; Lazar, A.J.; Morris, E.A.; Sahin, A.; Salgado, R.; et al. The 2019 World Health Organization classification of tumours of the breast. Histopathology 2020, 77, 181–185. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer-staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Li, J.; Chen, Z.; Su, K.; Zeng, J. Clinicopathological classification and traditional prognostic indicators of breast cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 8500. [Google Scholar]
- Laokulrath, N.; Gudi, M.A.; Deb, R.; Ellis, I.O.; Tan, P.H. Invasive breast cancer reporting guidelines: ICCR, CAP, RCPath, RCPA datasets and future directions. Diagn. Histopathol. 2024, 30, 87–99. [Google Scholar] [CrossRef]
- Deng, X.Y.; Cao, P.W.; Nan, S.M.; Pan, Y.P.; Yu, C.; Pan, T.; Dai, G. Differentiation Between Phyllodes Tumors and Fibroadenomas of Breast Using Mammography-based Machine Learning Methods: A Preliminary Study. Clin. Breast Cancer 2023, 23, 729–736. [Google Scholar] [CrossRef]
- Tan, H.; Zhang, S.; Liu, H.; Peng, W.; Li, R.; Gu, Y.; Wang, X.; Mao, J.; Shen, X. Imaging findings in phyllodes tumors of the breast. Eur. J. Radiol. 2012, 81, e62–e69. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.; Ding, Q.; Hunt, K.; Sahin, A. High-Grade Spindle Cell Lesions of the Breast: Key Pathologic and Clinical Updates. Surg. Pathol. Clin. 2022, 15, 77–93. [Google Scholar] [CrossRef]
- Ramala, S.R., Jr.; Chandak, S.; Chandak, M.S.; Annareddy, S.; Annareddy, S., Jr. A comprehensive review of breast fibroadenoma: Correlating clinical and pathological findings. Cureus 2023, 15, e49948. [Google Scholar] [CrossRef]
- Cheng, C.L.; Md Nasir, N.D.; Ng, G.J.Z.; Chua, K.W.J.; Li, Y.; Rodrigues, J.; Thike, A.A.; Heng, S.Y.; Koh, V.C.Y.; Lim, J.X.; et al. Artificial intelligence modelling in differentiating core biopsies of fibroadenoma from phyllodes tumor. Lab. Investig. 2022, 102, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, E.; Becker, A.S.; Wurnig, M.C.; Marcon, M.; Ghafoor, S.; Berger, N.; Boss, A. Distinction between phyllodes tumor and fibroadenoma in breast ultrasound using deep learning image analysis. Eur. J. Radiol. Open 2018, 5, 165–170. [Google Scholar] [CrossRef]
- Vidal, M.; Peg, V.; Galván, P.; Tres, A.; Cortés, J.; Ramón y Cajal, S.; Rubio, I.T.; Prat, A. Gene expression-based classifications of fibroadenomas and phyllodes tumours of the breast. Mol. Oncol. 2015, 9, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Ng, C.C.Y.; Md Nasir, N.D.; Loke, B.N.; Tay, T.K.Y.; Thike, A.A.; Rajasegaran, V.; Liu, W.; Lee, J.Y.; Guan, P.; Lim, A.H.; et al. Genetic differences between benign phyllodes tumors and fibroadenomas revealed through targeted next generation sequencing. Mod. Pathol. 2021, 34, 1320–1332. [Google Scholar] [CrossRef]
- Yang, Y.; Long, H.; Feng, Y.; Tian, S.; Chen, H.; Zhou, P. A multi-omics method for breast cancer diagnosis based on metabolites in exhaled breath, ultrasound imaging, and basic clinical information. Heliyon 2024, 10, e32115. [Google Scholar] [CrossRef]
- Zou, H.; Liu, S.H.; Yang, R.; Wu, X.J.; Cao, Y.P.; Huang, H.F. Combination of Neutrophil-to-Lymphocyte Ratio and Red Cell Distribution Width with Serum Tumor Markers for the Differential Diagnosis of Breast Cancer and its Association with Pathological Features and Molecular Types. Clin. Breast Cancer 2022, 22, e526–e535. [Google Scholar] [CrossRef]
- Xu, W.; Ma, W.; Wang, D.; Zhou, X.; Wang, K.; Mu, K. Integrated multi-omics profiling reveals a clinically relevant molecular feature and potential therapeutic target on phyllodes tumors of breast. Transl. Oncol. 2024, 46, 101998. [Google Scholar] [CrossRef]
- An, R.; Yu, H.; Wang, Y.; Lu, J.; Gao, Y.; Xie, X.; Zhang, J. Integrative analysis of plasma metabolomics and proteomics reveals the metabolic landscape of breast cancer. Cancer Metab. 2022, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.H.; Lee, A. Plasma metabolites as possible biomarkers for diagnosis of breast cancer. PLoS ONE 2019, 14, e0225129. [Google Scholar] [CrossRef] [PubMed]
- Rivenzon-Segal, D.; Margalit, R.; Degani, H. Glycolysis as a metabolic marker in orthotopic breast cancer, monitored by in vivo 13C MRS. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E623–E630. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, Y.; Bekhash, A.; Kovatich, A.J.; Hooke, J.A.; Liu, J.; Kvecher, L.; Fantacone-Campbell, J.L.; Mitchell, E.P.; Rui, H.; Mural, R.J.; et al. Positive Association of Fibroadenomatoid Change with HER2-Negative Invasive Breast Cancer: A Co-Occurrence Study. PLoS ONE 2015, 10, e0129500. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cala, M.; Aldana, J.; Sánchez, J.; Guio, J.; Meesters, R.J.W. Urinary metabolite and lipid alterations in Colombian Hispanic women with breast cancer: A pilot study. J. Pharm. Biomed. Anal. 2018, 152, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.L.; Olival, A.; Perestrelo, R.; Silva, P.; Tomás, H.; Câmara, J.S. Untargeted Urinary 1H NMR-Based Metabolomic Pattern as a Potential Platform in Breast Cancer Detection. Metabolites 2019, 9, 269. [Google Scholar] [CrossRef]
- Li, J.; Guan, X.; Fan, Z.; Ching, L.M.; Li, Y.; Wang, X.; Cao, W.M.; Liu, D.X. Non-Invasive Biomarkers for Early Detection of Breast Cancer. Cancers 2020, 12, 2767. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Wei, F.; Rao, S.L.; Kim, J.; Shin, H.; Cheng, J.; Tu, M.; Wong, D.T.W.; Kim, Y. Clinical validity of saliva and novel technology for cancer detection. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Gopikrishna, P.; Ramesh Kumar, A.; Rajkumar, K.; Ashwini, R.; Venkatkumar, S. Saliva: A potential diagnostic tool for oral cancer and oral diseases—A detailed review. Oral Oncol. Rep. 2024, 10, 100508. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, S.; Das, B.C. Saliva as a potential non-invasive liquid biopsy for early and easy diagnosis/prognosis of head and neck cancer. Transl. Oncol. 2024, 40, 101827. [Google Scholar] [CrossRef]
- Bel’skaya, L.V.; Dyachenko, E.I. Salivary Biomarkers in Breast Cancer: From Salivaomics to Salivaoncoomics. Front. Biosci. 2024, 29, 253. [Google Scholar] [CrossRef] [PubMed]
- Khalil Arjmandi, M.; Moslemi, D.; Sadati Zarrini, A.; Ebrahimnezhad Gorji, M.; Mosapour, A.; Haghhaghighi, A.; Halalkhor, S.; Bijani, A.; Parsian, H. Pre and post radiotherapy serum oxidant/antioxidant status in breast cancer patients: Impact of age, BMI and clinical stage of the disease. Rep. Pract. Oncol. Radiother. 2016, 21, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Turcotte, A.F.; O’Connor, S.; Morin, S.N.; Gibbs, J.C.; Willie, B.M.; Jean, S.; Gagnon, C. Association between obesity and risk of fracture, bone mineral density and bone quality in adults: A systematic review and meta-analysis. PLoS ONE 2021, 16, e0252487. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Iida, M.; Harada, S.; Kato, S.; Kuwabara, K.; Kurihara, A.; Takeuchi, A.; Sugiyama, D.; Okamura, T.; Suzuki, A.; et al. Metabolic profiling of charged metabolites in association with menopausal status in Japanese community-dwelling midlife women: Tsuruoka Metabolomic Cohort Study. Maturitas 2022, 155, 54–62. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R Package for Multivariate Analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Kazarian, A.; Blyuss, O.; Metodieva, G.; Gentry-Maharaj, A.; Ryan, A.; Kiseleva, E.M.; Prytomanova, O.M.; Jacobs, I.J.; Widschwendter, M.; Menon, U.; et al. Testing breast cancer serum biomarkers for early detection and prognosis in pre-diagnosis samples. Br. J. Cancer 2017, 116, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.N.; Jiang, Y.F.; Ru, J.N.; Lu, J.H.; Ding, B.; Wu, J. Amino acid metabolism in health and disease. Signal Transduct. Target. Ther. 2023, 8, 345. [Google Scholar] [CrossRef]
- Hak, A.E.; Choi, H.K. Menopause, postmenopausal hormone use and serum uric acid levels in US women—The Third National Health and Nutrition Examination Survey. Arthritis Res. Ther. 2008, 10, R116. [Google Scholar] [CrossRef]
- Sun, Y.; Li, J.; Qu, Z.; Yang, Z.; Jia, X.; Lin, Y.; He, Q.; Zhang, L.; Luo, Y. Causal Associations between Serum Urea and Cancer: A Mendelian Randomization Study. Genes 2021, 12, 498. [Google Scholar] [CrossRef]
- Allegrini, S.; Garcia-Gil, M.; Pesi, R.; Camici, M.; Tozzi, M.G. The Good, the Bad and the New about Uric Acid in Cancer. Cancers 2022, 14, 4959. [Google Scholar] [CrossRef]
- Leser, C.; Dorffner, G.; Marhold, M.; Rutter, A.; Döger, M.; Singer, C.; König-Castillo, D.M.; Deutschmann, C.; Holzer, I.; König-Castillo, D.; et al. Liver function indicators in patients with breast cancer before and after detection of hepatic metastases-a retrospective study. PLoS ONE 2023, 18, e0278454. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, C.; Chen, Y. Phosphoserine Aminotransferase 1: A Metabolic Enzyme Target of Cancers. Curr. Cancer Drug Targets 2023, 23, 171–186. [Google Scholar] [PubMed]
- Mullins, R.J.; Azman, A.M. Tetrahedron Organic Chemistry Series. Imidazoles; Elsevier: Amsterdam, The Netherlands, 2007; Volume 26, pp. 407–433. [Google Scholar]
- Gupta, R.R.; Kumar, M.; Gupta, V. Heterocyclic Chemistry: Volume II: Five-Membered Heterocycles; Springer Science & Business Media: Berlin, Germany, 2013. [Google Scholar]
- Pierzynowska, K.; Thomasson, S.; Oredsson, S. Alpha-Amylase Inhibits Cell Proliferation and Glucose Uptake in Human Neuroblastoma Cell Lines. BioMed Res. Int. 2022, 2022, 4271358. [Google Scholar] [CrossRef]
- Rohleder, N.; Nater, U.M.; Wolf, J.M.; Ehlert, U.; Kirschbaum, C. Psychosocial stress-induced activation of salivary alpha-amylase: An indicator of sympathetic activity? Ann. N. Y. Acad. Sci. 2004, 1032, 258–263. [Google Scholar] [CrossRef]
- van Stegeren, A.; Rohleder, N.; Everaerd, W.; Wolf, O.T. Salivary alpha amylase as marker for adrenergic activity during stress: Effect of betablockade. Psychoneuroendocrinology 2006, 31, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Nater, U.M.; Rohleder, N. Salivary alpha-amylase as a non-invasive biomarker for the sympathetic nervous system: Current state of research. Psychoendocrinology 2009, 34, 486–496. [Google Scholar] [CrossRef]
- Fedrowitz, M.; Hass, R.; Bertram, C.; Löscher, W. Salivary α-amylase exhibits antiproliferative effects in primary cell cultures of rat mammary epithelial cells and human breast cancer cells. J. Exp. Clin. Cancer Res. 2011, 30, 102. [Google Scholar] [CrossRef]
- Matoso, A.; Easley, S.E.; Gnepp, D.R.; Mangray, S. Salivary gland acinar-like differentiation of the breast. Histopathology 2009, 54, 262–263. [Google Scholar] [CrossRef]
- Hähnel, R.; Twaddle, E.; Brindle, L. The influence of enzymes on the estrogen receptors of human uterus and breast carcinoma. Steroids 1974, 24, 489–506. [Google Scholar] [CrossRef]
- Ren, G.; Zheng, X.; Bommarito, M.; Metzger, S.; Walia, Y.; Letson, J.; Schroering, A.; Kalinoski, A.; Weaver, D.; Figy, C.; et al. Reduced Basal Nitric Oxide Production Induces Precancerous Mammary Lesions via ERBB2 and TGFβ. Sci. Rep. 2019, 9, 6688. [Google Scholar] [CrossRef]
- Choudhari, S.K.; Chaudhary, M.; Bagde, S.; Gadbail, A.R.; Joshi, V. Nitric oxide and cancer: A review. World J. Surg. Oncol. 2013, 11, 118. [Google Scholar] [CrossRef]
- Xu, W.; Liu, L.Z.; Loizidou, M.; Ahmed, M.; Charles, I.G. The role of nitric oxide in cancer. Cell Res. 2002, 12, 311–320. [Google Scholar] [CrossRef]
- Schaffer, W.M.; Bronnikova, T.V. Peroxidase-ROS interactions. Nonlinear Dyn. 2012, 68, 413–430. [Google Scholar] [CrossRef]
- Vieira, S.A.; Zhang, G.; Decker, E.A. Biological Implications of Lipid Oxidation Products. J. Am. Oil Chem. Soc. 2017, 94, 339–351. [Google Scholar] [CrossRef]
- Gęgotek, A.; Skrzydlewska, E. Lipid peroxidation products’ role in autophagy regulation. Free Radic. Biol. Med. 2024, 212, 375–383. [Google Scholar] [CrossRef]
- Tsikas, D. Assessment of lipid peroxidation by measuring malondialdehyde (MDA) and relatives in biological samples: Analytical and biological challenges. Anal. Biochem. 2017, 524, 13–30. [Google Scholar] [CrossRef]
- Anwar, S.; Alrumaihi, F.; Sarwar, T.; Babiker, A.Y.; Khan, A.A.; Prabhu, S.V.; Rahmani, A.H. Exploring Therapeutic Potential of Catalase: Strategies in Disease Prevention and Management. Biomolecules 2024, 14, 697. [Google Scholar] [CrossRef]
- Ceciliani, F.; Pocacqua, V. The acute phase protein alpha1-acid glycoprotein: A model for altered glycosylation during diseases. Curr. Protein Pept. Sci. 2007, 8, 91–108. [Google Scholar] [CrossRef]
- Pitekova, B.; Uhlikova, E.; Kupcova, V.; Durfinova, M.; Mojto, V.; Turecky, L. Can alpha-1-acid glycoprotein affect the outcome of treatment in a cancer patient? Bratisl. Lek. Listy 2019, 120, 9–14. [Google Scholar] [CrossRef]
- Ligresti, G.; Aplin, A.C.; Dunn, B.E.; Morishita, A.; Nicosia, R.F. The acute phase reactant orosomucoid-1 is a bimodal regulator of angiogenesis with time- and context-dependent inhibitory and stimulatory properties. PLoS ONE 2012, 7, e41387. [Google Scholar] [CrossRef]
- Qiong, L.; Yin, J. Characterization of alpha-1-acid glycoprotein as a potential biomarker for breast cancer. Bioengineered 2022, 13, 5818–5826. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsson, P.; Fornander, L.; Påhlsson, P.; Grenegård, M. Sialic acid residues play a pivotal role in alpha(1)-acid glycoprotein (AGP)-induced generation of reactive oxygen species in chemotactic peptide pre-activated neutrophil granulocytes. Inflamm. Res. 2010, 59, 89–95. [Google Scholar] [CrossRef]
- Rosner, M.H.; Dalkin, A.C. Electrolyte disorders associated with cancer. Adv. Chronic Kidney Dis. 2014, 21, 7–17. [Google Scholar] [CrossRef]
- Ratanasrimetha, P.; Workeneh, B.T.; Seethapathy, H. Sodium and Potassium Dysregulation in the Patient with Cancer. Adv. Chronic Kidney Dis. 2022, 29, 171–179.e1. [Google Scholar] [CrossRef]
- Berardi, R.; Torniai, M.; Lenci, E.; Pecci, F.; Morgese, F.; Rinaldi, S. Electrolyte disorders in cancer patients: A systematic review. J. Cancer Metastasis Treat. 2019, 5, 79. [Google Scholar] [CrossRef]
- Feske, S.; Wulff, H.; Skolnik, E.Y. Ion channels in innate and adaptive immunity. Annu. Rev. Immunol. 2015, 33, 291–353. [Google Scholar] [CrossRef]
- Tan, P.H. Fibroepithelial lesions revisited: Implications for diagnosis and management. Mod. Pathol. 2021, 34, 15–37. [Google Scholar] [CrossRef] [PubMed]
- Nassar, A.; Visscher, D.W.; Degnim, A.C.; Frank, R.D.; Vierkant, R.A.; Frost, M.; Radisky, D.C.; Vachon, C.M.; Kraft, R.A.; Hartmann, L.C.; et al. Complex fibroadenoma and breast cancer risk: A Mayo Clinic Benign Breast Disease Cohort Study. Breast Cancer Res. Treat. 2015, 153, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Proudfoot, D. Calcium Signaling and Tissue Calcification. Cold Spring Harb. Perspect. Biol. 2019, 11, a035303. [Google Scholar] [CrossRef]
- Immler, R.; Simon, S.I.; Sperandio, M. Calcium signalling and related ion channels in neutrophil recruitment and function. Eur. J. Clin. Investig. 2018, 48 (Suppl. S2), e12964. [Google Scholar] [CrossRef]
- Dahlgren, C.; Karlsson, A. Respiratory burst in human neutrophils. J. Immunol. Methods 1999, 232, 3–14. [Google Scholar] [CrossRef] [PubMed]


| Subgroups | Breast Cancer, n = 543 | Fibroadenomas, n = 597 | Healthy Control, n = 298 |
|---|---|---|---|
| Age | |||
| 20–29 years | 6 (1.1%) | 143 (24.0%) | 34 (11.4%) |
| 30–39 years | 55 (10.1%) | 143 (24.0%) | 68 (22.8%) |
| 40–49 years | 109 (20.0%) | 153 (25.6%) | 70 (23.5%) |
| 50–59 years | 181 (33.3%) | 94 (15.7%) | 80 (26.8%) |
| 60–69 years | 156 (28.7%) | 52 (8.7%) | 31 (10.4%) |
| 70+ years | 37 (6.8%) | 12 (2.0%) | 15 (5.1%) |
| Menopause | |||
| Yes | 382 (70.3%) | 168 (28.1%) | 142 (47.7%) |
| No | 161 (29.7%) | 429 (71.9%) | 156 (52.3%) |
| Body Mass Index (BMI) | |||
| <25 | 135 (24.7%) | 291 (48.7%) | 201 (67.4%) |
| 25–30 | 179 (33.0%) | 148 (24.8%) | 97 (32.6%) |
| >30 | 229 (42.3%) | 158 (26.5%) | - |
| Indicator | No Menopause | Kruskal–Wallis Test (H, p) | Menopause | Kruskal–Wallis Test (H, p) | ||
|---|---|---|---|---|---|---|
| BC | FA | BC | FA | |||
| рН | −0.2 | 0.8 | 4.450; 0.1081 | −0.9 | −0.4 | 1.796; 0.4074 |
| Са | −6.8 | −0.9 | 6.160; 0.0460 * | −0.7 | 1.9 | 1.667; 0.5579 |
| P | 6.2 | 2.4 | 0.8417; 0.6565 | 2.3 | −5.4 | 5.001; 0.0821 |
| Na | −19.6 | −14.5 | 3.513; 0.1727 | −5.4 | −20.9 | 3.419; 0.1810 |
| K | −7.1 | −6.3 | 4.570; 0.1018 | 12.4 | 7.7 | 0.6792; 0.7121 |
| Cl | 4.8 | −3.9 | 7.717; 0.0211 * | −1.7 | −4.8 | 1.894; 0.3879 |
| Mg | −1.3 | −3.6 | 4.288; 0.1172 | −0.4 | −5.4 | 1.950; 0.3772 |
| Protein | −43.4 | −35.0 | 49.13; 0.0000 * | −43.8 | −39.9 | 55.47; 0.0000 * |
| Urea | 28.5 | 15.5 | 17.35; 0.0002 * | 52.4 | 38.9 | 49.79; 0.0000 * |
| UA | −14.6 | 16.4 | 7.395; 0.0248 * | −37.5 | −24.2 | 18.27; 0.0001 * |
| Albumin | 23.4 | 14.6 | 2.643; 0.2668 | −8.6 | 4.2 | 1.581; 0.4537 |
| ALT | 1.9 | 0.0 | 1.789; 0.4089 | 8.2 | −1.0 | 6.977; 0.0306 * |
| AST | 12.5 | 9.4 | 6.225; 0.0445 * | 5.6 | 1.4 | 4.737; 0.0936 |
| AST/ALT | 6.6 | 9.2 | 3.533; 0.1709 | 0.7 | −5.3 | 1.630; 0.4425 |
| α-AAs | 3.1 | 2.0 | 11.85; 0.0027 * | 3.2 | 2.5 | 7.051; 0.0294 * |
| ICs | −1.1 | −6.8 | 0.7383; 0.6913 | −5.4 | −8.1 | 3.344; 0.1878 |
| NO | 25.6 | 45.0 | 27.10; 0.0000 * | 24.4 | 41.9 | 9.591; 0.0083 * |
| ALP | 25.9 | 18.5 | 15.18; 0.0005 * | 17.2 | 15.5 | 3.352; 0.1871 |
| MM 254 | −9.1 | −12.7 | 3.367; 0.1857 | −8.4 | −8.2 | 2.276; 0.2624 |
| MM 280 | −10.6 | −6.0 | 1.411; 0.4939 | −12.9 | −14.2 | 3.435; 0.1795 |
| LDH | 40.8 | 37.9 | 12.64; 0.0018 * | 23.2 | 12.8 | 4.148; 0.1257 |
| CAT | −17.0 | −15.4 | 3.442; 0.1789 | −20.2 | −18.7 | 14.18; 0.0008 * |
| SAs | 3.3 | 0.0 | 3.536; 0.1707 | 13.3 | −6.7 | 7.295; 0.0261 * |
| PYR | 16.3 | 4.1 | 3.263; 0.1956 | 5.3 | 0.0 | 0.5329; 0.7661 |
| DC | 2.0 | 0.3 | 8.514; 0.0142 * | 0.9 | 1.8 | 0.2152; 0.8980 |
| TC | −2.5 | 1.1 | 4.502; 0.1053 | 0.9 | −1.0 | 3.115; 0.2106 |
| SB | −2.0 | 1.8 | 8.736; 0.0127 * | −0.5 | −0.4 | 0.0755; 0.9630 |
| MDA | 5.3 | 9.3 | 14.85; 0.0006 * | 3.8 | 9.0 | 3.301; 0.1919 |
| GGT | 18.5 | 11.5 | 24.90; 0.0000 * | 9.2 | 0.9 | 12.82; 0.0016 * |
| SM | 13.3 | 21.1 | 10.37; 0.0056 * | −4.9 | 2.9 | 0.5916; 0.7439 |
| SOD | 18.2 | 4.5 | 1.843; 0.3980 | 21.7 | 8.7 | 4.442; 0.1085 |
| α-Amylase | 101.1 | 40.6 | 13.09; 0.0014 * | 29.8 | 3.8 | 2.003; 0.3673 |
| Lactate | −8.4 | −9.5 | 4.520; 0.1043 | 6.4 | 5.8 | 0.0558; 0.9725 |
| AOA | −11.4 | −7.4 | 3.522; 0.1719 | −3.6 | −13.0 | 11.72; 0.0028 * |
| Peroxidase | 104.1 | 69.4 | 7.538; 0.0231 * | −22.2 | −9.3 | 0.7821; 0.6764 |
| MM280/254 | 2.0 | 4.7 | 4.565; 0.1020 | 0.8 | 1.4 | 0.5087; 0.7754 |
| Indicators | No Menopause | Indicators | Menopause | ||
|---|---|---|---|---|---|
| PC 1 (First principal component) | |||||
| MM 280 | 0.6638 * | 6.006 × 10−96 | P | 0.7344 | 2.037 × 10−118 |
| MM 254 | 0.6577 | 1.219 × 10−93 | MM 280 | 0.7278 | 2.684 × 10−115 |
| P | 0.6575 | 1.473 × 10−93 | MM 254 | 0.7127 | 1.660 × 10−108 |
| K | 0.6180 | 8.982 × 10−80 | Cl | 0.7120 | 3.212 × 10−108 |
| Protein | 0.5952 | 1.104 × 10−72 | K | 0.6584 | 2.537 × 10−87 |
| Albumin | 0.5644 | 5.497 × 10−64 | Protein | 0.6276 | 3.651 × 10−77 |
| Cl | 0.5383 | 2.872 × 10−57 | Albumin | 0.6242 | 4.075 × 10−76 |
| Mg | 0.5204 | 5.281 × 10−53 | Са | 0.5700 | 5.748 × 10−61 |
| LDH | 0.4816 | 1.382 × 10−44 | Mg | 0.5568 | 1.142 × 10−57 |
| SM | 0.4758 | 2.037 × 10−43 | рН | 0.5426 | 2.606 × 10−54 |
| CAT | 0.4714 | 1.528 × 10−42 | SM | 0.5095 | 4.789 × 10−47 |
| α-Amylase | 0.4496 | 2.160 × 10−38 | PYR | 0.4999 | 4.285 × 10−45 |
| SB | 0.4301 | 6.086 × 10−35 | UA | 0.4583 | 2.734 × 10−37 |
| α-AAs | 0.4297 | 7.199 × 10−35 | α-AAs | 0.4526 | 2.659 × 10−36 |
| Na | 0.4296 | 7.505 × 10−35 | MM280/254 | 0.4462 | 3.265 × 10−35 |
| TC | 0.4197 | 3.414 × 10−33 | α-Amylase | 0.4289 | 2.183 × 10−32 |
| AST | 0.4114 | 7.790 × 10−32 | |||
| PYR | 0.4067 | 4.351 × 10−31 | |||
| PC 2 (Second principal component) | |||||
| AOA | 0.6663 | 6.535 × 10−97 | рН | 0.5432 | 1.935 × 10−54 |
| Peroxidase | 0.6046 | 1.559 × 10−75 | MM280/254 | 0.4923 | 1.377 × 10−43 |
| α-Amylase | −0.4146 | 2.399 × 10−32 | SB | 0.4568 | 5.012 × 10−37 |
| K | −0.4634 | 5.544 × 10−41 | MDA | 0.4316 | 8.251 × 10−33 |
| TC | −0.4858 | 1.942 × 10−45 | AST/ALT | 0.4312 | 9.385 × 10−33 |
| SB | −0.5046 | 1.945 × 10−49 | TC | 0.4294 | 1.832 × 10−32 |
| ALP | −0.4387 | 5.711 × 10−34 | |||
| Albumin | −0.4547 | 1.152 × 10−36 | |||
| GGT | −0.4571 | 4.484 × 10−37 | |||
| Indicator | Breast Cancer | Kruskal–Wallis Test (H, p) | Fibroadenomas | Kruskal–Wallis Test (H, p) | ||||
|---|---|---|---|---|---|---|---|---|
| No Menopause | MP | No Menopause | MP | |||||
| BMI < 25 | BMI > 25 | BMI < 25 | BMI > 25 | |||||
| рН | 0.0 | −0.6 | −0.9 | 10.97; 0.0042 * | 1.3 | 0.2 | −0.4 | 10.36; 0.0056 * |
| Са | −9.6 | −6.8 | −0.7 | 9.605; 0.0082 * | −2.1 | 2.8 | 1.9 | 8.164; 0.0169 * |
| P | 0.1 | 7.4 | 2.3 | 3.905; 0.1420 | 2.2 | 2.4 | −5.4 | 1.254; 0.5342 |
| Na | −33.1 | 0.3 | −5.4 | 4.438; 0.1087 | −21.9 | −0.2 | −20.9 | 5.237; 0.0729 ** |
| K | −13.6 | −4.0 | 12.4 | 8.819; 0.0122 * | −8.5 | −1.5 | 7.7 | 5.278; 0.0714 |
| Cl | −4.9 | 12.2 | −1.7 | 4.562; 0.1022 | −6.3 | −1.6 | −4.8 | 16.64; 0.0002 *,** |
| Mg | −1.8 | −1.5 | −0.4 | 2.067; 0.3558 | −2.8 | −5.5 | −5.4 | 3.155; 0.2065 |
| Protein | −43.3 | −44.5 | −43.8 | 5.752; 0.0564 | −36.6 | −32.1 | −39.9 | 13.29; 0.0013 *,** |
| Urea | 16.7 | 51.8 | 52.4 | 20.94; 0.0000 *,** | 8.7 | 23.3 | 38.9 | 20.93; 0.0000 * |
| UA | −13.2 | −18.6 | −37.5 | 3.0494 0.2177 | 19.3 | 10.8 | −24.2 | 2.303; 0.3162 |
| Albumin | 14.0 | 31.1 | −8.6 | 6.307; 0.0427 * | 7.1 | 20.7 | 4.2 | 22.20; 0.0000 *,** |
| ALT | 0.0 | 5.8 | 8.2 | 0.5789; 0.7489 | −1.9 | 3.8 | −1.0 | 1.601; 0.4492 |
| AST | 11.7 | 21.9 | 5.6 | 0.0098; 0.9951 | 7.8 | 13.3 | 1.4 | 0.2695; 0.8795 |
| AST/ALT | 16.8 | 1.0 | 0.7 | 1.173; 0.5563 | 11.8 | 4.4 | −5.3 | 3.060; 0.2165 |
| α-AAs | 2.0 | 4.5 | 3.2 | 2.642; 0.2668 | 1.6 | 3.3 | 2.5 | 10.95; 0.0042 * |
| ICs | 2.3 | −2.3 | −5.4 | 13.80; 0.0010 * | −5.7 | −9.1 | −8.1 | 16.87; 0.0002 * |
| NO | 25.2 | 16.8 | 24.4 | 0.1113; 0.9458 | 48.5 | 38.2 | 41.9 | 0.7927; 0.6728 |
| ALP | 22.2 | 37.0 | 17.2 | 0.7731; 0.6794 | 14.8 | 25.9 | 15.5 | 3.991; 0.1360 ** |
| MM 254 | −19.0 | −7.2 | −8.4 | 3.297; 0.1923 | −18.3 | −5.5 | −8.2 | 8.005; 0.0183 *,** |
| MM 280 | −13.0 | −2.9 | −12.9 | 3.274; 0.1945 | −8.9 | −1.7 | −14.2 | 6.167; 0.0454 * |
| LDH | 40.8 | 43.4 | 23.2 | 2.348; 0.3091 | 27.1 | 45.7 | 12.8 | 2.690; 0.2605 |
| CAT | −10.6 | −20.4 | −20.2 | 5.373; 0.0681 | −17.0 | −10.6 | −18.7 | 6.046; 0.0487 * |
| SAs | 6.7 | 3.3 | 13.3 | 0.3909; 0.8225 | −3.3 | 6.7 | −6.7 | 1.562; 0.4580 |
| PYR | 24.5 | 14.3 | 5.3 | 1.621; 0.4447 | 4.1 | 8.2 | 0.0 | 3.849; 0.1459 |
| DC | 2.6 | 1.1 | 0.9 | 5.051; 0.0800 | 0.2 | 0.7 | 1.8 | 0.0599; 0.9705 |
| TC | −2.5 | −3.3 | 0.9 | 6.548; 0.0378 * | 0.1 | 2.6 | −1.0 | 5.249; 0.0725 |
| SB | −1.3 | −4.0 | −0.5 | 2.376; 0.3049 | 2.1 | 1.8 | −0.4 | 1.918; 0.3832 |
| MDA | 8.0 | 2.7 | 3.8 | 1.212; 0.5455 | 9.3 | 10.0 | 9.0 | 0.0327; 0.9838 |
| GGT | 13.1 | 20.4 | 9.2 | 6.673; 0.0356 *,** | 11.0 | 12.6 | 0.9 | 6.994; 0.0303 * |
| SM | 25.3 | 3.6 | −4.9 | 2.605; 0.2719 | 20.5 | 21.7 | 2.9 | 0.8607; 0.6503 |
| SOD | 15.9 | 22.7 | 21.7 | 0.6256; 0.7314 | 4.5 | 2.3 | 8.7 | 1.228; 0.5411 |
| α-Amylase | 204.6 | 28.8 | 29.8 | 4.351; 0.1136 ** | 45.3 | 21.0 | 3.8 | 0.6253; 0.7315 |
| Lactate | −23.4 | −2.0 | 6.4 | 4.606; 0.0999 | −11.1 | −9.1 | 5.8 | 3.323; 0.1898 |
| AOA | −8.7 | −14.7 | −3.6 | 6.286; 0.0432 * | −6.2 | −10.2 | −13.0 | 5.764; 0.0560 |
| Peroxidase | 128.6 | 75.5 | −22.2 | 1.876; 0.3914 | 79.6 | 38.8 | −9.3 | 3.979; 0.1367 |
| MM280/254 | 2.0 | 2.3 | 0.8 | 0.0994; 0.6067 | 5.6 | 3.0 | 1.4 | 1.674; 0.4330 |
| Indicator | Fibroadenomas | Phyllodes Tumors | Breast Cancer | Kruskal–Wallis Test (H, p) |
|---|---|---|---|---|
| рН | 0.6 | 0.8 | −0.2 | 3.740; 0.2909 |
| Са | −0.1 | −4.6 | −6.8 | 5.866; 0.1183 |
| P | 2.7 | 1.9 | 6.2 | 1.305; 0.7280 |
| Na | −16.2 | −0.7 | −19.6 | 6.040; 0.1097 |
| K | −5.0 | −11.0 | −7.1 | 5.458; 0.1412 |
| Cl | −3.6 | −6.3 | 4.8 | 13.07; 0.0045 * |
| Mg | −2.8 | −10.7 | −1.3 | 6.741; 0.0806 |
| Protein | −34.6 | −44.2 | −43.4 | 33.24; 0.0000 * |
| Urea | 20.4 | −4.6 | 28.5 | 24.86; 0.0000 * |
| UA | 16.4 | 15.1 | −14.6 | 12.72; 0.0053 * |
| Albumin | 15.4 | 5.2 | 23.4 | 1.307; 0.7275 |
| ALT | 0.0 | −1.9 | 1.9 | 5.360; 0.1473 |
| AST | 9.4 | 25.0 | 12.5 | 5.942; 0.1145 |
| AST/ALT | 9.2 | 10.2 | 6.6 | 0.8219; 0.8442 |
| α-AAs | 2.0 | 3.7 | 3.1 | 16.97; 0.0007 * |
| ICs | −6.8 | −9.1 | −1.1 | 2.859; 0.4139 |
| NO | 45.0 | 27.5 | 25.6 | 32.75; 0.0000 * |
| ALP | 18.5 | 18.5 | 25.9 | 13.16; 0.0043 * |
| MM 254 | −12.7 | −6.8 | −9.1 | 1.773; 0.6208 |
| MM 280 | −5.8 | −6.8 | −10.6 | 3.078; 0.3797 |
| LDH | 40.1 | 14.6 | 40.8 | 3.962; 0.2656 |
| CAT | −16.7 | −10.9 | −17.0 | 28.02; 0.0000 * |
| SAs | 0.0 | −3.3 | 3.3 | 2.719; 0.4370 |
| PYR | 2.0 | 23.5 | 16.3 | 5.865; 0.1184 |
| DC | 0.0 | 4.2 | 2.0 | 12.39; 0.0062 *,** |
| TC | 1.1 | 0.1 | −2.5 | 7.290; 0.0632 |
| SB | 1.8 | 1.8 | −2.0 | 6.078; 0.1079 |
| MDA | 9.3 | 7.3 | 5.3 | 17.02; 0.0007 * |
| GGT | 11.5 | 12.6 | 18.5 | 9.987; 0.0187 * |
| SM | 20.5 | 24.1 | 13.3 | 5.095; 0.1650 |
| SOD | 4.5 | 4.5 | 18.2 | 2.105; 0.5509 |
| α-Amylase | 37.1 | 68.2 | 101.1 | 11.77; 0.0082 * |
| Lactate | −11.9 | 3.2 | −8.4 | 7.789; 0.0506 *,** |
| AOA | −9.7 | 1.4 | −11.4 | 5.299; 0.1511 |
| Peroxidase | 63.3 | 91.8 | 104.1 | 9.692; 0.0214 * |
| MM280/254 | 4.7 | 5.3 | 2.0 | 3.308; 0.3465 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dyachenko, E.I.; Bel’skaya, L.V. Salivary Metabolites in Breast Cancer and Fibroadenomas: Focus on Menopausal Status and BMI. Metabolites 2024, 14, 531. https://doi.org/10.3390/metabo14100531
Dyachenko EI, Bel’skaya LV. Salivary Metabolites in Breast Cancer and Fibroadenomas: Focus on Menopausal Status and BMI. Metabolites. 2024; 14(10):531. https://doi.org/10.3390/metabo14100531
Chicago/Turabian StyleDyachenko, Elena I., and Lyudmila V. Bel’skaya. 2024. "Salivary Metabolites in Breast Cancer and Fibroadenomas: Focus on Menopausal Status and BMI" Metabolites 14, no. 10: 531. https://doi.org/10.3390/metabo14100531
APA StyleDyachenko, E. I., & Bel’skaya, L. V. (2024). Salivary Metabolites in Breast Cancer and Fibroadenomas: Focus on Menopausal Status and BMI. Metabolites, 14(10), 531. https://doi.org/10.3390/metabo14100531







