Dysbiotic Vaginal Microbiota Induces Preterm Birth Cascade via Pathogenic Molecules in the Vagina
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects and Cervicovaginal Fluid Sampling
2.2. pH Determination in Cervicovaginal Fluid Samples
2.3. Microbiota Analysis in Cervicovaginal Fluid Samples
2.4. Measurement of Metabolites in Cervicovaginal Fluid Samples
2.5. Measurement of Protein Receptors in Cervicovaginal Fluid Samples
2.6. Measurement of Cytokines in Cervicovaginal Fluid Samples
2.7. Statistical Analysis
3. Results
3.1. Study Participant Demographics
3.2. Microbiota Analysis of Cervicovaginal Fluid Samples
3.3. Metabolite Analysis of Cervicovaginal Fluid Samples
3.4. Analysis of Inflammatory Markers in Cervicovaginal Fluid Samples
3.5. Correlation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohuma, E.; Moller, A.B.; Bradley, E. National, regional, and worldwide estimates of preterm birth in 2020, with trends from 2010: A systematic analysis. Lancet 2023, 402, 1261–1271. [Google Scholar] [CrossRef] [PubMed]
- Witkin, S.S. The vaginal microbiome, vaginal anti-microbial defence mechanisms and the clinical challenge of reducing infection-related preterm birth. BJOG Int. J. Obstet. Gynaecol. 2015, 122, 213–218. [Google Scholar] [CrossRef] [PubMed]
- You, Y.A.; Yoo, J.Y.; Kwon, E.J.; Kim, Y.J. Blood microbial communities during pregnancy are associated with preterm birth. Front. Microbiol. 2019, 10, 1122. [Google Scholar] [CrossRef] [PubMed]
- You, Y.A.; Kwon, E.J.; Choi, S.J.; Hwang, H.S.; Choi, S.K.; Lee, S.M.; Kim, Y.J. Vaginal microbiome profiles of pregnant women in Korea using a 16S metagenomics approach. Am. J. Reprod. Immunol. 2019, 82, e13124. [Google Scholar] [CrossRef]
- Zhang, G.; Meredith, T.C.; Kahne, D. On the essentiality of lipopolysaccharide to Gram-negative bacteria. Curr. Opin. Microbiol. 2013, 16, 779–785. [Google Scholar] [CrossRef] [PubMed]
- McDonald, H.M.; O’loughlin, J.A.; Jolley, P.; Vigneswaran, R.; McDonald, P.J. Vaginal infection and preterm labour. BJOG Int. J. Obstet. Gynaecol. 1991, 98, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Ansari, A.; Bose, S.; You, Y.; Park, S.; Kim, Y. Molecular mechanism of microbiota metabolites in preterm birth: Pathological and therapeutic insights. Int. J. Mol. Sci. 2021, 22, 8145. [Google Scholar] [CrossRef] [PubMed]
- Huppert, J.S.; Bates, J.R.; Weber, A.F.; Quinn, N.; Gaydos, C.A. Abnormal vaginal pH and Mycoplasma genitalium infection. J. Pediatr. Adolesc. Gynecol. 2013, 26, 36–39. [Google Scholar] [CrossRef]
- Superti, F.; De Seta, F. Warding off recurrent yeast and bacterial vaginal infections: Lactoferrin and lactobacilli. Microorganisms 2020, 8, 130. [Google Scholar] [CrossRef]
- Ravel, J.; Gajer, P.; Abdo, Z.; Schneider, G.M.; Koenig, S.S.; McCulle, S.L.; Karlebach, S.; Gorle, R.; Russell, J.; Tacket, C.O.; et al. Vaginal microbiome of reproductive-age women. Proc. Natl. Acad. Sci. USA 2011, 108, 4680–4687. [Google Scholar] [CrossRef]
- Sobel, J.D. Bacterial vaginosis. Annu. Rev. Med. 2000, 51, 349–356. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.M.; Hyun, Y.J.; Myoung, K.S.; Ahn, Y.T.; Lee, J.H.; Huh, C.S.; Han, M.J.; Kim, D.H. Lactobacillus johnsonii HY7042 ameliorates Gardnerella vaginalis-induced vaginosis by killing Gardnerella vaginalis and inhibiting NF-κB activation. Int. Immunopharmacol. 2011, 11, 1758–1765. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Roman, R.; Last, A.; Mirhakkak, M.H.; Sprague, J.L.; Möller, L.; Großmann, P.; Graf, K.; Gratz, R.; Mogavero, S.; Vylkova, S.; et al. Lactobacillus rhamnosus colonisation antagonizes Candida albicans by forcing metabolic adaptations that compromise pathogenicity. Nat. Commun. 2022, 13, 3192. [Google Scholar] [CrossRef] [PubMed]
- Petrova, M.I.; Lievens, E.; Malik, S.; Imholz, N.; Lebeer, S. Lactobacillus species as biomarkers and agents that can promote various aspects of vaginal health. Front. Physiol. 2015, 6, 81. [Google Scholar] [CrossRef]
- Delgado-Diaz, D.J.; Tyssen, D.; Hayward, J.A.; Gugasyan, R.; Hearps, A.C.; Tachedjian, G. Distinct immune responses elicited from cervicovaginal epithelial cells by lactic acid and short chain fatty acids associated with optimal and non-optimal vaginal microbiota. Front. Cell. Infect. Microbiol. 2020, 9, 446. [Google Scholar] [CrossRef]
- Amabebe, E.; Anumba, D.O. Female gut and genital tract microbiota-induced crosstalk and differential effects of short-chain fatty acids on immune sequelae. Front. Immunol. 2020, 11, 2184. [Google Scholar] [CrossRef]
- Amabebe, E.; Reynolds, S.; Stern, V.; Stafford, G.; Paley, M.; Anumba, D.O. Cervicovaginal fluid acetate: A metabolite marker of preterm birth in symptomatic pregnant women. Front. Med. 2016, 3, 48. [Google Scholar] [CrossRef]
- Park, S.; You, Y.A.; Kim, Y.H.; Kwon, E.; Ansari, A.; Kim, S.M.; Lee, G.; Hur, Y.M.; Jung, Y.J.; Kim, K.; et al. Ureaplasma and Prevotella colonization with Lactobacillus abundance during pregnancy facilitates term birth. Sci. Rep. 2022, 12, 10148. [Google Scholar] [CrossRef]
- Ansari, A.; Lee, H.; You, Y.A.; Jung, Y.; Park, S.; Kim, S.M.; Hwang, G.S.; Kim, Y.J. Identification of potential biomarkers in the cervicovaginal fluid by metabolic profiling for preterm birth. Metabolites 2020, 10, 349. [Google Scholar] [CrossRef]
- Li, L.; Kang, J.; Lei, W. Role of Toll-like receptor 4 in inflammation-induced preterm delivery. Mol. Hum. Reprod. 2010, 16, 267–272. [Google Scholar] [CrossRef]
- Park, S.; You, Y.A.; Yun, H.; Choi, S.J.; Hwang, H.S.; Choi, S.K.; Lee, S.M.; Kim, Y.J. Cervicovaginal fluid cytokines as predictive markers of preterm birth in symptomatic women. Obstet. Gynecol. Sci. 2020, 63, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Helmo, F.R.; Alves, E.A.; Moreira, R.A.; Severino, V.O.; Rocha, L.P.; Monteiro, M.L.; Reis, M.A.; Etchebehere, R.M.; Machado, J.R.; Corrêa, R.R. Intrauterine infection, immune system and premature birth. J. Matern. Fetal Neonatal Med. 2018, 9, 1227–1233. [Google Scholar] [CrossRef] [PubMed]
- Flaviani, F.; Hezelgrave, N.L.; Kanno, T.; Prosdocimi, E.M.; Chin-Smith, E.; Ridout, A.E.; von Maydell, D.K.; Mistry, V.; Wade, W.G.; Shennan, A.H.; et al. Cervicovaginal microbiota and metabolome predict preterm birth risk in an ethnically diverse cohort. JCI Insight 2021, 6, e149257. [Google Scholar] [CrossRef] [PubMed]
- Dimitonova, S.P.; Bakalov, B.V.; Aleksandrova-Georgieva, R.N.; Danova, S.T. Phenotypic and molecular identification of lactobacilli isolated from vaginal secretions. J. Microbiol. Immunol. Infect. 2008, 41, 469–477. [Google Scholar]
- Chang, D.H.; Shin, J.; Rhee, M.S.; Park, K.R.; Cho, B.K.; Lee, S.K.; Kim, B.C. Vaginal microbiota profiles of native Korean women and associations with high-risk pregnancy. J. Microbiol. Biotechnol. 2019, 30, 248–258. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The first microbial colonizers of the human gut: Composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, 110–128. [Google Scholar] [CrossRef]
- Weng, M.; Ganguli, K.; Zhu, W.; Shi, H.N.; Walker, W.A. Conditioned medium from Bifidobacteria infantis protects against Cronobacter sakazakii-induced intestinal inflammation in newborn mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G779–G787. [Google Scholar] [CrossRef]
- Pavlidis, I.; Spiller, O.B.; Sammut Demarco, G.; MacPherson, H.; Howie, S.E.; Norman, J.E.; Stock, S.J. Cervical epithelial damage promotes Ureaplasma parvum ascending infection, intrauterine inflammation and preterm birth induction in mice. Nat. Commun. 2020, 11, 199. [Google Scholar] [CrossRef]
- Chee, W.J.; Chew, S.Y.; Than, L.T. Vaginal microbiota and the potential of Lactobacillus derivatives in maintaining vaginal health. Microb. Cell Factories 2020, 19, 203. [Google Scholar] [CrossRef]
- Chiu, S.F.; Huang, P.J.; Cheng, W.H.; Huang, C.Y.; Chu, L.J.; Lee, C.C.; Lin, H.C.; Chen, L.C.; Lin, W.N.; Tsao, C.H.; et al. Vaginal microbiota of the sexually transmitted infections caused by Chlamydia trachomatis and Trichomonas vaginalis in women with vaginitis in Taiwan. Microorganisms 2021, 9, 1864. [Google Scholar] [CrossRef]
- Ansari, A.; Son, D.; Hur, Y.M.; Park, S.; You, Y.A.; Kim, S.M.; Lee, G.; Kang, S.; Chung, Y.; Lim, S.; et al. Lactobacillus Probiotics Improve Vaginal Dysbiosis in Asymptomatic Women. Nutrients 2023, 15, 1862. [Google Scholar] [CrossRef] [PubMed]
- Sitkin, S.I.; Tkachenko, E.I.; Vakhitov, T.Y. Metabolic dysbiosis of the gut microbiota and its biomarkers. Eksperimental’naia i Klinicheskaia Gastroenterologiia. Exp. Clin. Gastroenterol. 2016, 12, 6–29. [Google Scholar]
- Bradford, L.L.; Ravel, J. The vaginal mycobiome: A contemporary perspective on fungi in women’s health and diseases. Virulence 2017, 8, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Stafford, G.P.; Parker, J.L.; Amabebe, E.; Kistler, J.; Reynolds, S.; Stern, V.; Paley, M.; Anumba, D.O. Spontaneous preterm birth is associated with differential expression of vaginal metabolites by lactobacilli-dominated microflora. Front. Physiol. 2017, 8, 615. [Google Scholar] [CrossRef] [PubMed]
- Hovda, K.E.; Urdal, P.; Jacobsen, D. Increased serum formate in the diagnosis of methanol poisoning. J. Anal. Toxicol. 2005, 29, 586–588. [Google Scholar] [CrossRef] [PubMed]
- Kraut, J.A.; Kurtz, I. Toxic alcohol ingestions: Clinical features, diagnosis, and manage-ment. Clin. J. Am. Soc. Nephrol. 2008, 3, 208–225. [Google Scholar] [CrossRef]
- Sun, L.; Xie, C.; Wang, G.; Wu, Y.; Wu, Q.; Wang, X.; Liu, J.; Deng, Y.; Xia, J.; Chen, B.; et al. Gut microbiota and intestinal FXR mediate the clinical benefits of metformin. Nat. Med. 2018, 24, 1919–1929. [Google Scholar] [CrossRef]
- Chhibber-Goel, J.; Singhal, V.; Parakh, N.; Bhargava, B.; Sharma, A. The metabolite trimethylamine-N-oxide is an emergent biomarker of human health. Curr. Med. Chem. 2017, 24, 3942–3953. [Google Scholar] [CrossRef]
- Cen, X.; Liu, S.; Cheng, K. The role of toll-like receptor in inflammation and tumor immunity. Front. Pharmacol. 2018, 9, 878. [Google Scholar] [CrossRef]
- Wallet, S.M.; Puri, V.; Gibson, F.C. Linkage of infection to adverse systemic complications: Periodontal disease, toll-like receptors, and other pattern recognition systems. Vaccines 2018, 6, 21. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- Robertson, S.A.; Hutchinson, M.R.; Rice, K.C.; Chin, P.Y.; Moldenhauer, L.M.; Stark, M.J.; Olson, D.M.; Keelan, J.A. Targeting Toll-like receptor-4 to tackle preterm birth and fetal inflammatory injury. Clin. Transl. Immunol. 2020, 9, e1121. [Google Scholar] [CrossRef] [PubMed]
- Basith, S.; Manavalan, B.; Lee, G.; Kim, S.G.; Choi, S. Toll-like receptor modulators: A patent review (2006–2010). Expert Opin. Ther. Pat. 2011, 21, 927–944. [Google Scholar] [CrossRef] [PubMed]
- Koga, K.; Mor, G. Expression and function of Toll-like receptors at the maternal—Fetal interface. Reprod. Sci. 2008, 15, 231–242. [Google Scholar] [CrossRef] [PubMed]
- LeBouder, E.; Rey-Nores, J.E.; Rushmere, N.K.; Grigorov, M.; Lawn, S.D.; Affolter, M.; Griffin, G.E.; Ferrara, P.; Schiffrin, E.J.; Morgan, B.P.; et al. Soluble forms of Toll-like receptor (TLR) 2 capable of modulating TLR2 signaling are present in human plasma and breast milk. J. Immunol. 2003, 171, 6680–6689. [Google Scholar] [CrossRef] [PubMed]
- Jarešová, I.; Rožková, D.; Špíšek, R.; Janda, A.; Brázová, J.; Šedivá, A. Kinetics of Toll-like receptor-4 splice variants expression in lipopolysaccharide-stimulated antigen presenting cells of healthy donors and patients with cystic fibrosis. Microbes Infect. 2007, 9, 1359–1367. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.G.; Maubert, M.E.; Anton, L.; Heiser, L.M.; Elovitz, M.A. The tracking of lipopolysaccharide through the feto-maternal compartment and the involvement of maternal TLR4 in inflammation-induced fetal brain injury. Am. J. Reprod. Immunol. 2019, 82, e13189. [Google Scholar] [CrossRef]
- Smith, S.B.; Ravel, J. The vaginal microbiota, host defence and reproductive physiology. J. Physiol. 2017, 595, 451–463. [Google Scholar] [CrossRef]
- Christman, J.W.; Blackwell, T.R.; Cowan, H.B.; Shepard, V.L.; Rinaldo, J.E. Endotoxin induces the expression of macrophage inflammatory protein 1a mRNA by rat alveolar and bone marrow-derived macrophages. Am. J. Respir. Cell Mol. Biol. 1992, 7, 455–461. [Google Scholar] [CrossRef]
- Fahey, T.J., 3rd; Tracey, K.J.; Tekamp-Olson, P.A.; Cousens, L.S.; Jones, W.G.; Shires, G.T.; Cerami, A.; Sherry, B. Macrophage inflammatory protein 1 modulates macrophage function. J. Immunol. 1992, 148, 2764–2769. [Google Scholar] [CrossRef]
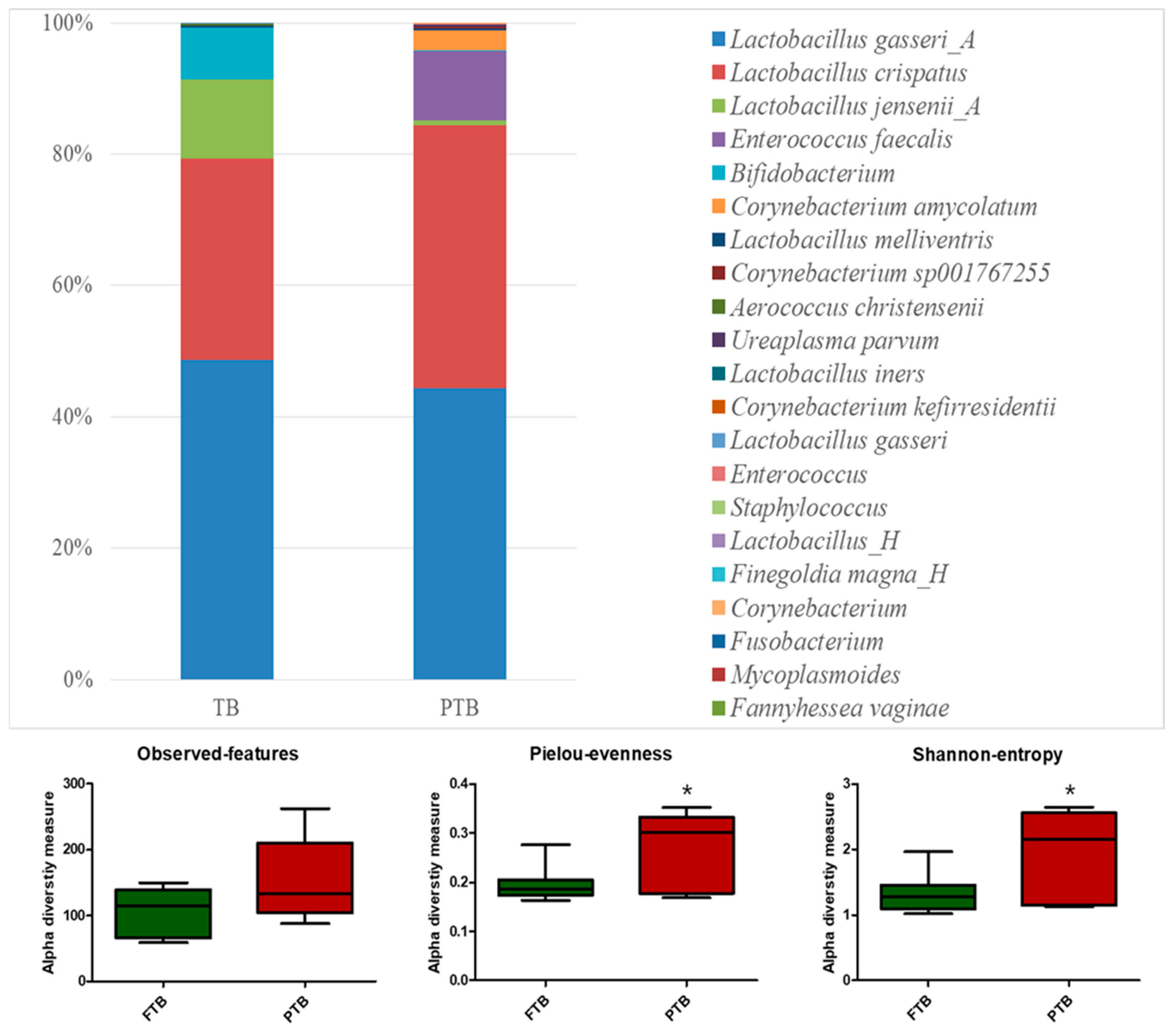

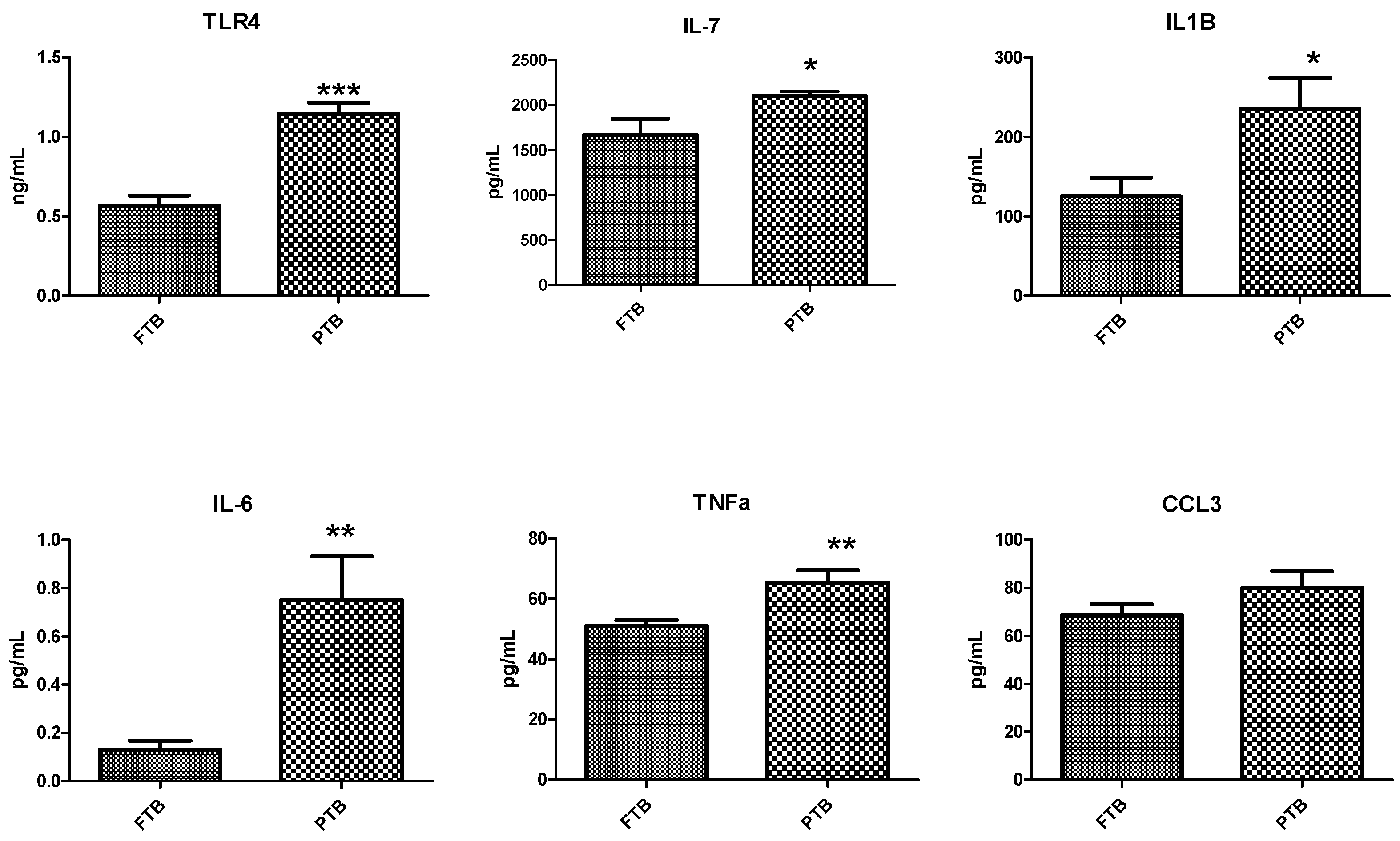
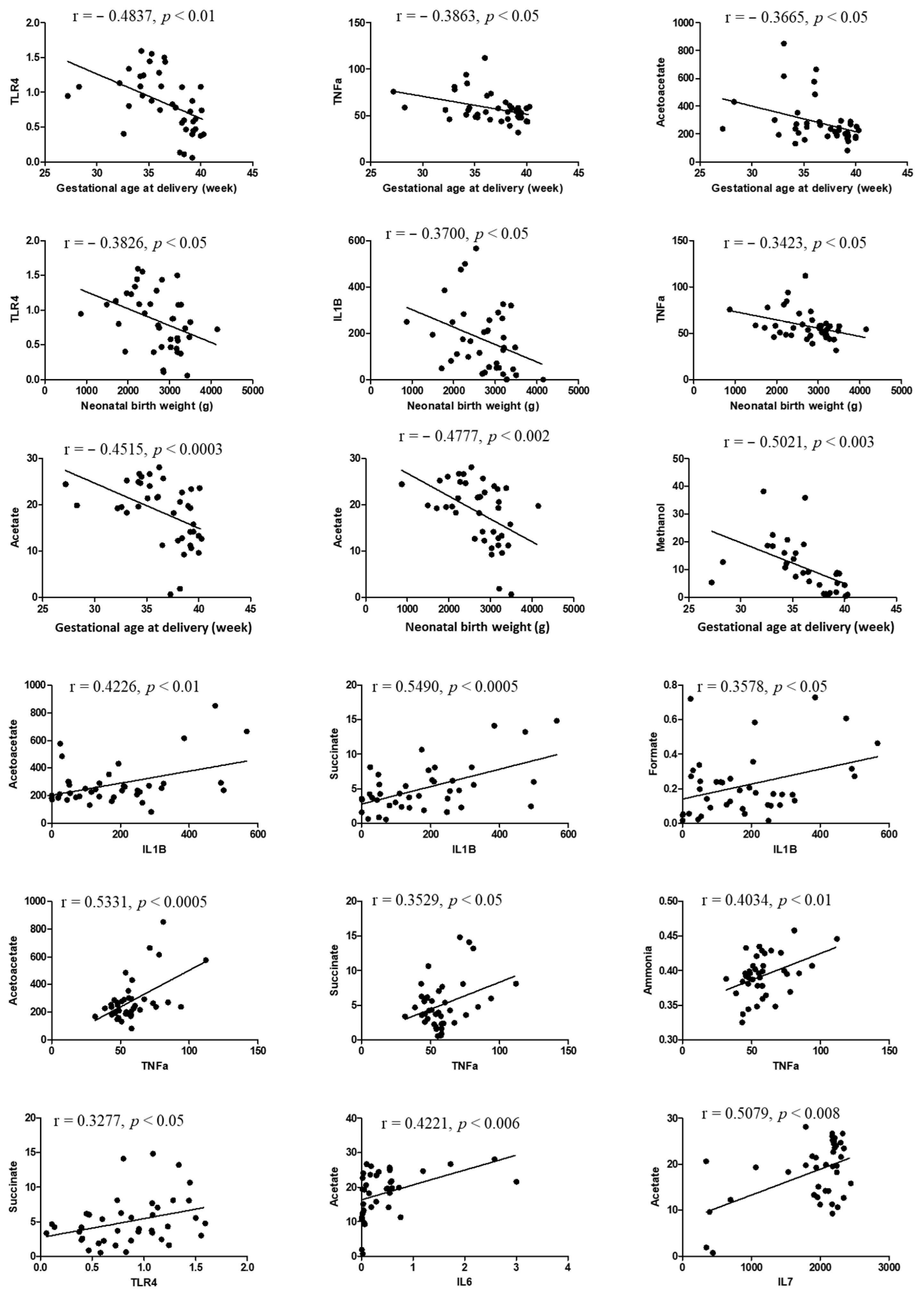

| Variables | FTB (n = 20) | PTB (n = 20) | p-Value |
|---|---|---|---|
| Maternal variables at CVF sampling | |||
| Age (yrs) | 32.60 ± 0.62 | 34.42 ± 1.19 | NS |
| Body mass index (kg/m2) | 21.24 ± 0.58 | 22.65 ± 1.24 | NS |
| Gestational age at sampling (wks) | 34.37 ± 1.8 | 33.34 ± 0.57 | NS |
| Cervical length (mm) | 23.92 ± 1.9 | 25.04 ± 2.70 | NS |
| Cervicovaginal fluid pH | 3.57 ± 0.60 | 4.57 ± 0.50 | 0.002 |
| Neonate variables at birth | |||
| Gestational age at delivery (wks) | 38.97 ± 0.18 | 33.96 ± 0.58 | <0.0001 |
| Birth weight (gm) | 3172 ± 75.68 | 2240 ± 128.90 | <0.0001 |
| 1 min Apgar score | 9.50 ± 0.17 | 7.79 ± 0.59 | 0.0076 |
| 5 min Apgar score | 9.95 ± 0.05 | 8.68 ± 0.41 | 0.0039 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ansari, A.; You, Y.-A.; Lee, G.; Kim, S.M.; Park, S.W.; Hur, Y.M.; Kim, Y.J. Dysbiotic Vaginal Microbiota Induces Preterm Birth Cascade via Pathogenic Molecules in the Vagina. Metabolites 2024, 14, 45. https://doi.org/10.3390/metabo14010045
Ansari A, You Y-A, Lee G, Kim SM, Park SW, Hur YM, Kim YJ. Dysbiotic Vaginal Microbiota Induces Preterm Birth Cascade via Pathogenic Molecules in the Vagina. Metabolites. 2024; 14(1):45. https://doi.org/10.3390/metabo14010045
Chicago/Turabian StyleAnsari, AbuZar, Young-Ah You, Gain Lee, Soo Min Kim, Sun Wha Park, Young Min Hur, and Young Ju Kim. 2024. "Dysbiotic Vaginal Microbiota Induces Preterm Birth Cascade via Pathogenic Molecules in the Vagina" Metabolites 14, no. 1: 45. https://doi.org/10.3390/metabo14010045
APA StyleAnsari, A., You, Y.-A., Lee, G., Kim, S. M., Park, S. W., Hur, Y. M., & Kim, Y. J. (2024). Dysbiotic Vaginal Microbiota Induces Preterm Birth Cascade via Pathogenic Molecules in the Vagina. Metabolites, 14(1), 45. https://doi.org/10.3390/metabo14010045







