Peripheral Blood Serum NMR Metabolomics Is a Powerful Tool to Discriminate Benign and Malignant Ovarian Tumors
Abstract
:1. Introduction
2. Methods
2.1. Ethics Statement
2.2. Sample Collection
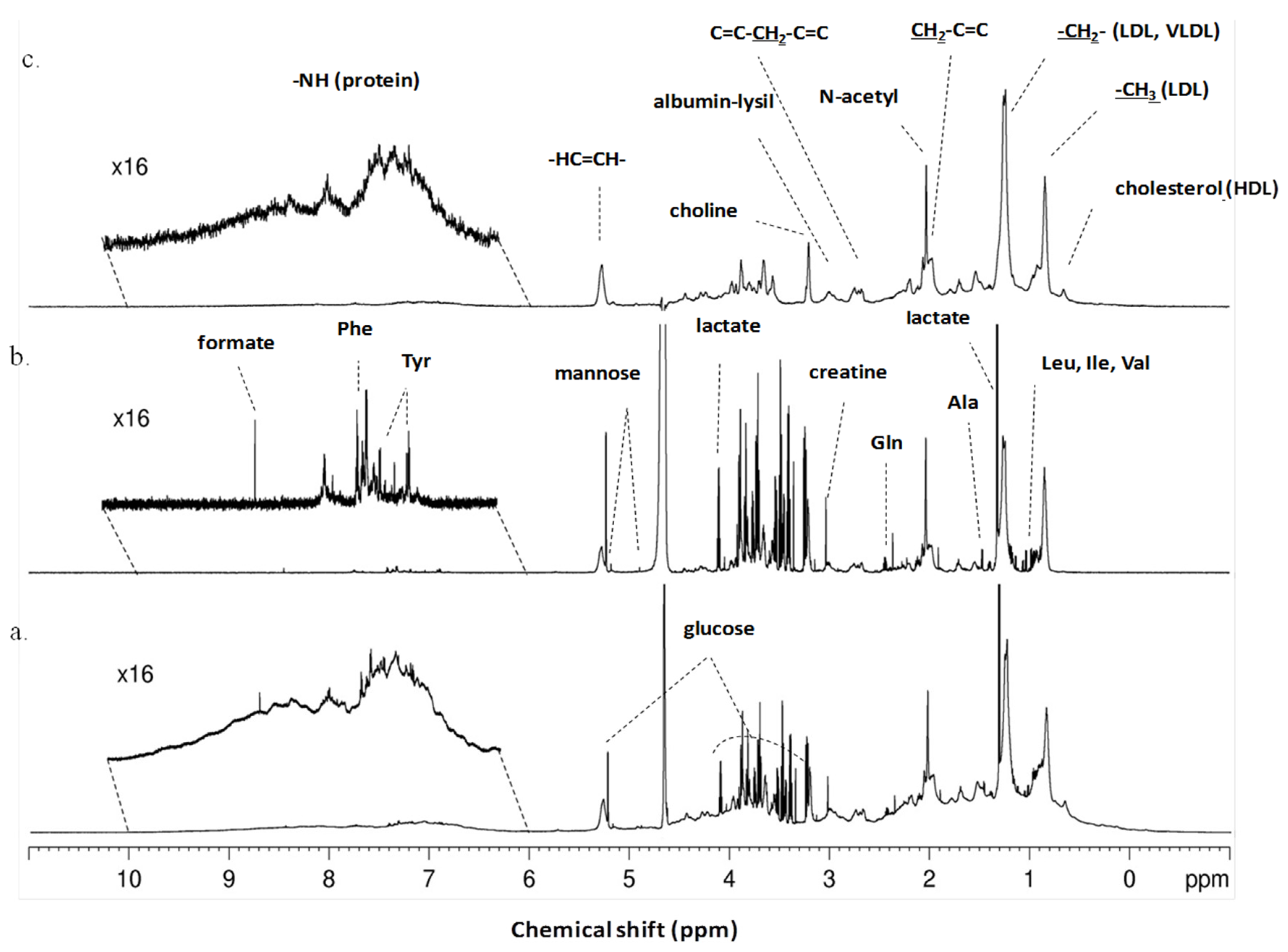
2.3. NMR Spectral Acquisition, Processing and Metabolite Identification
2.4. Statistical Data Analysis
3. Results
3.1. Study Population
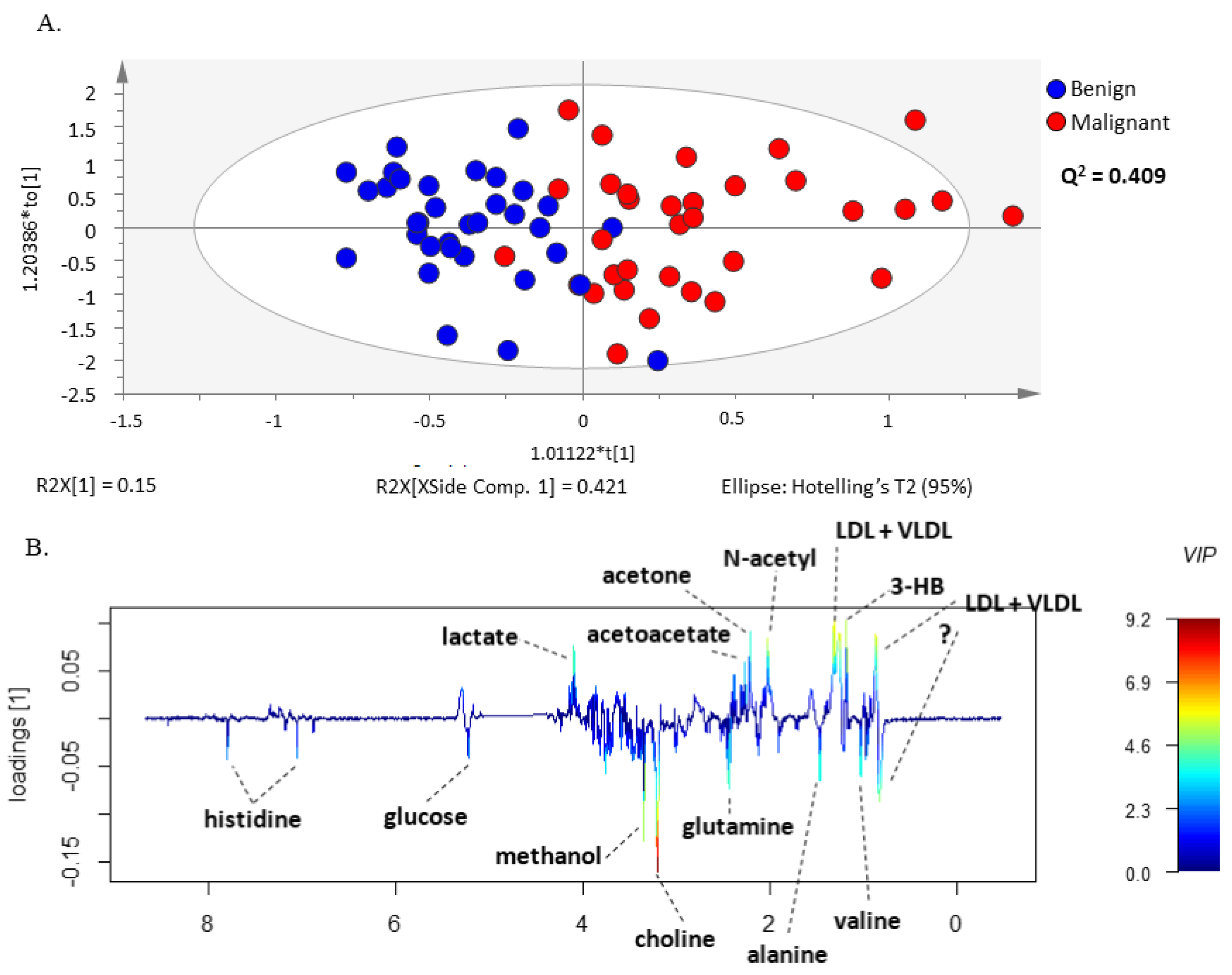
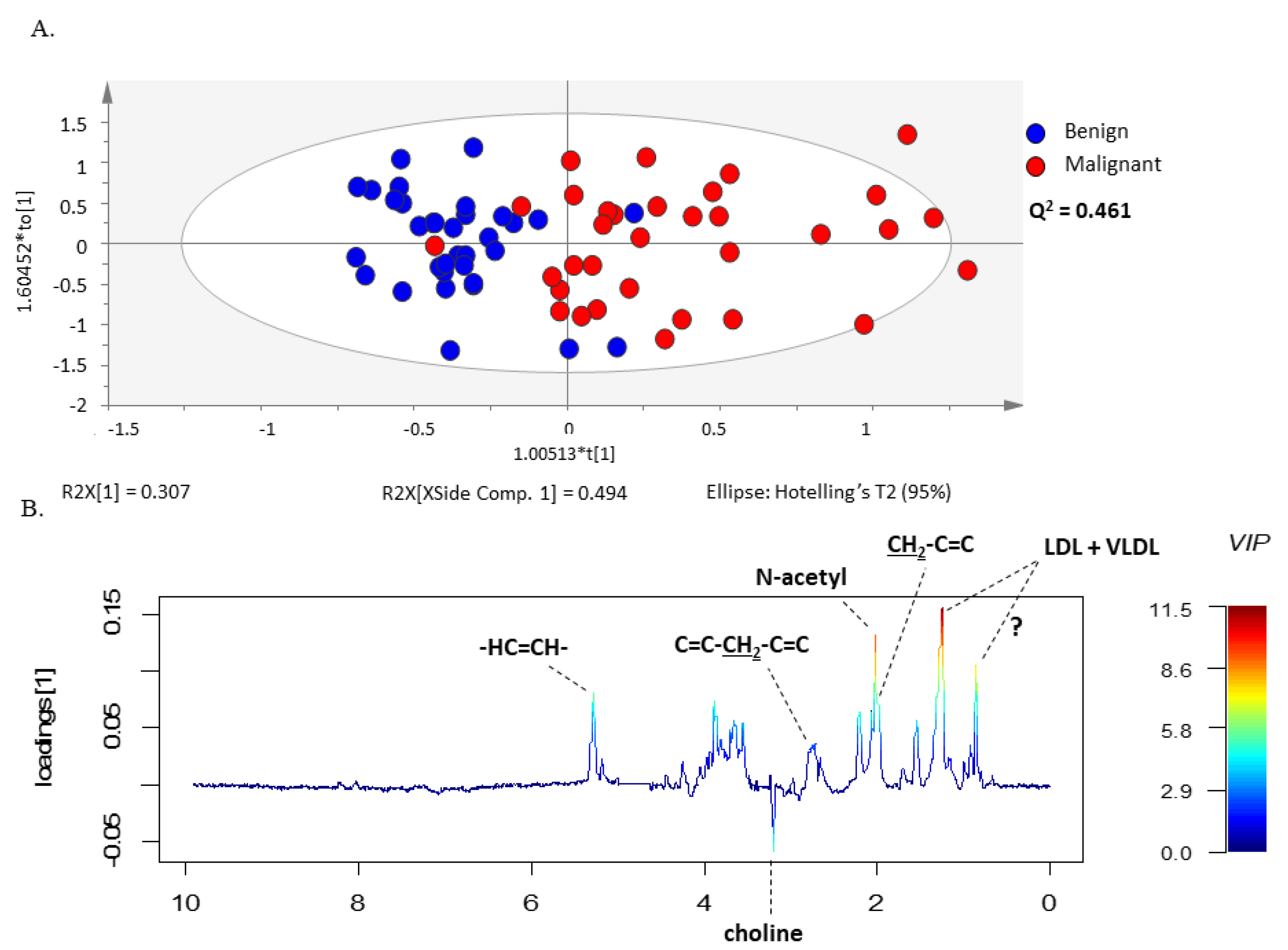
3.2. 1H-NMR Spectra Separate Serum from Patients with Ovarian Cancer from Patients with Benign Neoplasms
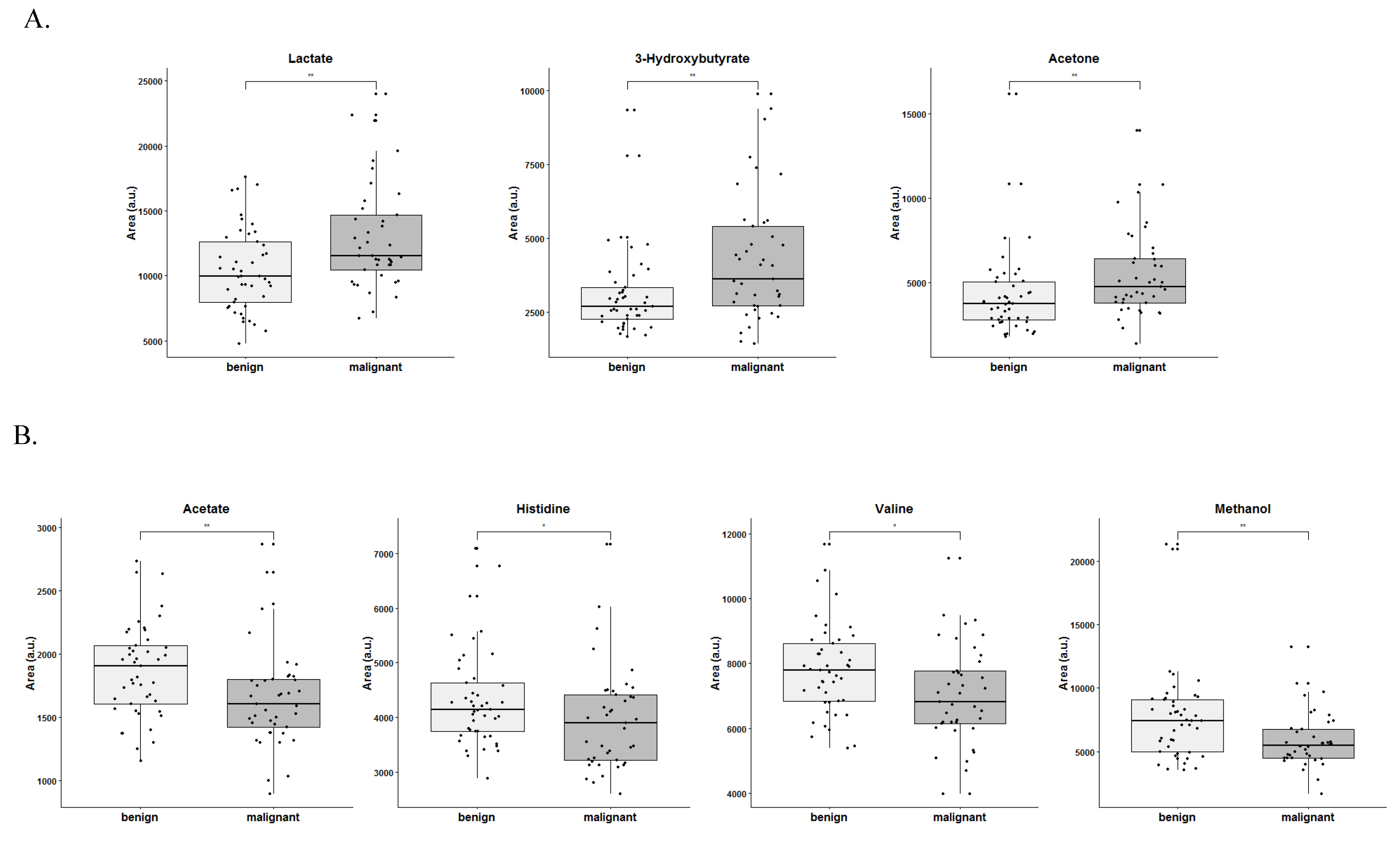
3.3. Lactate, 3-Hydroxybutyrate, Acetone, Acetate, Histidine, Valine and Methanol Separate Serum from Patients with Ovarian Cancer from Patients with Benign Neoplasm
3.4. OPLS-DA Model Predicted the Outcome of Bordeline Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vaughan, S.; Coward, J.I.; Bast, R.C., Jr.; Berchuck, A.; Berek, J.S.; Brenton, J.D.; Coukos, G.; Crum, C.C.; Drapkin, R.; Etemadmoghadam, D.; et al. Rethinking ovarian cancer: Recommendations for improving outcomes. Nat. Rev. Cancer 2011, 11, 719–725. [Google Scholar] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Jessmon, P.; Boulanger, T.; Zhou, W.; Patwardhan, P. Epidemiology and treatment patterns of epithelial ovarian cancer. Expert Rev. Anticancer. Ther. 2017, 17, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.M.; Permuth, J.B.; Sellers, T.A. Epidemiology of ovarian cancer: A review. Cancer Biol. Med. 2017, 14, 9–32. [Google Scholar] [PubMed]
- Bast, R.C.; Hennessy, B.; Mills, G.B. The biology of ovarian cancer: New opportunities for translation. Nat. Rev. Cancer 2009, 9, 415–428. [Google Scholar]
- Prat, J. Ovarian carcinomas: Five distinct diseases with different origins, genetic alterations, and clinicopathological features. Virchows Arch. 2012, 460, 237–249. [Google Scholar] [CrossRef]
- Kim, S.; Han, Y.; Kim, S.I.; Kim, H.-S.; Kim, S.J.; Song, Y.S. Tumor evolution and chemoresistance in ovarian cancer. NPJ Precis. Oncol. 2018, 2, 20. [Google Scholar]
- Hauptmann, S.; Friedrich, K.; Redline, R.; Avril, S. Ovarian borderline tumors in the 2014 WHO classification: Evolving concepts and diagnostic criteria. Virchows Arch. 2017, 470, 125–142. [Google Scholar]
- Wright, J.D.; Chen, L.; Tergas, A.I.; Patankar, S.; Burke, W.M.; Hou, J.Y.; Neugut, A.I.; Ananth, C.V.; Hershman, D.L. Trends in relative survival for ovarian cancer from 1975 to 2011. Obs. Gynecol. 2015, 125, 1345–1352. [Google Scholar] [CrossRef]
- Bowtell, D.D.L. The genesis and evolution of high-grade serous ovarian cancer. Nat. Rev. Cancer 2010, 10, 803–808. [Google Scholar] [CrossRef]
- Jasen, P. From the “Silent Killer” to the “Whispering Disease”: Ovarian Cancer and the Uses of Metaphor. Med. Hist. 2009, 53, 489–512. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [PubMed]
- Fischerova, D.; Zikan, M.; Dundr, P.; Cibula, D. Diagnosis, Treatment, and Follow-Up of Borderline Ovarian Tumors. Oncologist 2012, 17, 1515–1533. [Google Scholar]
- Hwang, V.J.; Weiss, R.H. Metabolomic profiling for early cancer detection: Current status and future prospects. Expert Opin. Drug Metab. Toxicol. 2016, 12, 1263–1265. [Google Scholar] [CrossRef] [PubMed]
- Hadi, N.I.; Jamal, Q.; Iqbal, A.; Shaikh, F.; Somroo, S.; Musharraf, S.G. Serum Metabolomic Profiles for Breast Cancer Diagnosis, Grading and Staging by Gas Chromatography-Mass Spectrometry. Sci. Rep. 2017, 7, 1715. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Li, K.; Zhang, X. Next-generation metabolomics in lung cancer diagnosis, treatment and precision medicine: Mini review. Oncotarget 2017, 8, 115774. [Google Scholar] [CrossRef]
- Cardoso, M.R.; Santos, J.C.; Ribeiro, M.L.; Talarico, M.C.R.; Viana, L.R.; Derchain, S.F.M. A Metabolomic Approach to Predict Breast Cancer Behavior and Chemotherapy Response. Int. J. Mol. Sci. 2018, 19, 617. [Google Scholar] [CrossRef] [PubMed]
- Fujigaki, S.; Nishiumi, S.; Kobayashi, T.; Suzuki, M.; Iemoto, T.; Kojima, T.; Ito, Y.; Daiko, H.; Kato, K.; Shouji, H.; et al. Identification of serum biomarkers of chemoradiosensitivity in esophageal cancer via the targeted metabolomics approach. Biomark. Med. 2018, 12, 827–840. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Weinstein, S.J.; Moore, S.C.; Derkach, A.; Hua, X.; Mondul, A.M.; Sampson, J.N.; Albanes, D. Pre-diagnostic Serum Metabolomic Profiling of Prostate Cancer Survival. J. Gerontol. Ser. A 2018, 74, 853–859. [Google Scholar] [CrossRef]
- Tian, Y.; Wang, Z.; Liu, X.; Duan, J.; Feng, G.; Yin, Y.; Gu, J.; Chen, Z.; Gao, S.; Bai, H.; et al. Prediction of Chemotherapeutic Efficacy in Non–Small Cell Lung Cancer by Serum Metabolomic Profiling. Clin. Cancer Res. 2018, 24, 2100–2109. [Google Scholar] [CrossRef]
- Huang, J.; Mondul, A.M.; Weinstein, S.J.; Derkach, A.; Moore, S.C.; Sampson, J.N.; Albanes, D. Prospective serum metabolomic profiling of lethal prostate cancer. Int. J. Cancer 2019, 145, 3231–3243. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics Analysis in Serum from Patients with Colorectal Polyp and Colorectal Cancer by <sup>1</sup>H-NMR Spectrometry. Dis. Mark. 2019, 2019, 3491852. [Google Scholar]
- Khan, A.; Choi, S.A.; Na, J.; Pamungkas, A.D.; Jung, K.J.; Jee, S.H.; Park, Y.H. Noninvasive Serum Metabolomic Profiling Reveals Elevated Kynurenine Pathway’s Metabolites in Humans with Prostate Cancer. J. Proteome Res. 2019, 18, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Emwas, A.-H.M. The Strengths and Weaknesses of NMR Spectroscopy and Mass Spectrometry with Particular Focus on Metabolomics Research. In Metabonomics: Methods and Protocols; Bjerrum, J.T., Ed.; Springer: New York, NY, USA, 2015; pp. 161–193. [Google Scholar]
- Graça, G.; Desterro, J.; Sousa, J.; Fonseca, C.; Silveira, M.; Serpa, J.; Carvalho, T.; da Silva, M.G.; Gonçalves, L.G. Identification of putative biomarkers for leptomeningeal invasion in B-cell non-Hodgkin lymphoma by NMR metabolomics. Metabolomics 2017, 13, 136. [Google Scholar]
- Odunsi, K.; Wollman, R.M.; Ambrosone, C.B.; Hutson, A.; McCann, S.E.; Tammela, J.; Geisler, J.P.; Miller, G.; Sellers, T.; Cliby, W.; et al. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int. J. Cancer 2005, 113, 782–788. [Google Scholar] [CrossRef]
- Garcia, E.; Andrews, C.; Hua, J.; Kim, H.L.; Sukumaran, D.K.; Szyperski, T.; Odunsi, K. Diagnosis of early stage ovarian cancer by 1H NMR metabonomics of serum explored by use of a microflow NMR probe. J. Proteome Res. 2011, 10, 1765–1771. [Google Scholar] [CrossRef]
- Bharti, S.K.; Wildes, F.; Hung, C.-F.; Wu, T.C.; Bhujwalla, Z.M.; Penet, M.-F. Metabolomic characterization of experimental ovarian cancer ascitic fluid. Metabolomics 2017, 13, 113. [Google Scholar] [CrossRef]
- Wishart, D.S. Advances in metabolite identification. Bioanalysis 2011, 3, 1769–1782. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Hoefsloot, H.C.J.; Smit, S.; Vis, D.J.; Smilde, A.K.; van Velzen, E.J.J.; van Duijnhoven, J.P.M.; van Dorsten, F.A. Assessment of PLSDA cross validation. Metabolomics 2008, 4, 81–89. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Davidson, B.; Tropé, C.G. Ovarian Cancer: Diagnostic, Biological and Prognostic Aspects. Women’s Health 2014, 10, 519–533. [Google Scholar] [CrossRef]
- Li, S.-S.; Ma, J.; Wong, A.S.T. Chemoresistance in ovarian cancer: Exploiting cancer stem cell metabolism. J. Gynecol. Oncol. 2018, 29, e32. [Google Scholar]
- Serpa, J. Metabolic Remodeling as a Way of Adapting to Tumor Microenvironment (TME), a Job of Several Holders. In Tumor Microenvironment. Advances in Experimental Medicine and Biology, 2020/03/05 ed.; Serpa, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1219, pp. 1–34. [Google Scholar]
- Anderson, A.S.; Roberts, P.C.; Frisard, M.I.; McMillan, R.P.; Brown, T.J.; Lawless, M.H.; Hulver, M.W.; Schmelz, E.M. Metabolic changes during ovarian cancer progression as targets for sphingosine treatment. Exp. Cell Res. 2013, 319, 1431–1442. [Google Scholar] [CrossRef]
- Kyriakides, M.; Rama, N.; Sidhu, J.; Gabra, H.; Keun, H.C.; El-Bahrawy, M. Metabonomic analysis of ovarian tumour cyst fluid by proton nuclear magnetic resonance spectroscopy. Oncotarget 2016, 7, 7216. [Google Scholar] [CrossRef]
- Massuger, L.F.A.G.; Vierzen, P.B.J.v.; Engelke, U.; Heerschap, A.; Wevers, R. 1H-magnetic resonance spectroscopy. Cancer 1998, 82, 1726–1730. [Google Scholar] [CrossRef]
- Turkoglu, O.; Zeb, A.; Graham, S.; Szyperski, T.; Szender, J.B.; Odunsi, K.; Bahado-Singh, R. Metabolomics of biomarker discovery in ovarian cancer: A systematic review of the current literature. Metabolomics 2016, 12, 60. [Google Scholar] [CrossRef]
- Boran, N.; Kayikcioglu, F.; Yalvaç, S.; Tulunay, G.; Ekinci, U.; Köse, M.F. Significance of Serum and Peritoneal Fluid Lactate Dehydrogenase Levels in Ovarian Cancer. Gynecol. Obstet. Investig. 2000, 49, 272–274. [Google Scholar] [CrossRef]
- Patel, P.S.; Sharma, V.M.; Raval, G.N.; Rawal, R.M.; Patel, M.M.; Balar, D.B.; Kapadia, A.S.; Patel, D.D. Serum lactate dehydrogenase levels in malignant germ cell tumors of ovary. Int. J. Gynecol. Cancer 1996, 6, 328–332. [Google Scholar] [CrossRef]
- Schneider, D.; Halperin, R.; Langer, R.; Bukovsky, I.; Herman, A. Peritoneal Fluid Lactate Dehydrogenase in Ovarian Cancer. Gynecol. Oncol. 1997, 66, 399–404. [Google Scholar] [CrossRef]
- Xiang, J.; Zhou, L.; Zhuang, Y.; Zhang, J.; Sun, Y.; Li, S.; Zhang, Z.; Zhang, G.; He, Y. Lactate dehydrogenase is correlated with clinical stage and grade and is downregulated by si-SAΤB1 in ovarian cancer. Oncol. Rep. 2018, 40, 2788–2797. [Google Scholar]
- Hilvo, M.; de Santiago, I.; Gopalacharyulu, P.; Schmitt, W.D.; Budczies, J.; Kuhberg, M.; Dietel, M.; Aittokallio, T.; Markowetz, F.; Denkert, C.; et al. Accumulated Metabolites of Hydroxybutyric Acid Serve as Diagnostic and Prognostic Biomarkers of Ovarian High-Grade Serous Carcinomas. Cancer Res. 2016, 76, 796–804. [Google Scholar] [CrossRef]
- Dhillon, K.K.; Gupta, S. Biochemistry, Ketogenesis; StatPearls Publishing: St. Petersburg, FL, USA, 2020. [Google Scholar]
- Puchalska, P.; Crawford, P.A. Multi-dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [PubMed]
- Kim, J.T.; Li, C.; Weiss, H.L.; Zhou, Y.; Liu, C.; Wang, Q.; Evers, B.M. Regulation of Ketogenic Enzyme HMGCS2 by Wnt/β-catenin/PPARγ Pathway in Intestinal Cells. Cells 2019, 8, 1106. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Sarno, L.; Landolfi, A.; Scala, G.; Martinelli, P.; Venturella, R.; Di Cello, A.; Zullo, F.; Guida, M. Metabolomic Signature of Endometrial Cancer. J. Proteome Res. 2018, 17, 804–812. [Google Scholar] [CrossRef] [PubMed]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine Metabolite Analysis Offers Potential Early Diagnosis of Ovarian and Breast Cancers. Clin. Cancer Res. 2010, 16, 5835–5841. [Google Scholar]
- Schug, Z.T.; Vande Voorde, J.; Gottlieb, E. The metabolic fate of acetate in cancer. Nat. Rev. Cancer 2016, 16, 708–717. [Google Scholar] [CrossRef]
- Niemi, R.J.; Braicu, E.I.; Kulbe, H.; Koistinen, K.M.; Sehouli, J.; Puistola, U.; Mäenpää, J.U.; Hilvo, M. Ovarian tumours of different histologic type and clinical stage induce similar changes in lipid metabolism. Br. J. Cancer 2018, 119, 847–854. [Google Scholar]
- Shan, Y.; Gao, Y.; Jin, W.; Fan, M.; Wang, Y.; Gu, Y.; Shan, C.; Sun, L.; Li, X.; Yu, B.; et al. Targeting HIBCH to reprogram valine metabolism for the treatment of colorectal cancer. Cell Death Dis. 2019, 10, 618. [Google Scholar] [CrossRef]
- Ke, C.; Hou, Y.; Zhang, H.; Fan, L.; Ge, T.; Guo, B.; Zhang, F.; Yang, K.; Wang, J.; Lou, G.; et al. Large-scale profiling of metabolic dysregulation in ovarian cancer. Int. J. Cancer 2015, 136, 516–526. [Google Scholar] [CrossRef]
- Plewa, S.; Horała, A.; Dereziński, P.; Klupczynska, A.; Nowak-Markwitz, E.; Matysiak, J.; Kokot, Z.J. Usefulness of Amino Acid Profiling in Ovarian Cancer Screening with Special Emphasis on Their Role in Cancerogenesis. Int. J. Mol. Sci. 2017, 18, 2727. [Google Scholar] [CrossRef]
- Plewa, S.; Horała, A.; Dereziński, P.; Nowak-Markwitz, E.; Matysiak, J.; Kokot, Z.J. Wide spectrum targeted metabolomics identifies potential ovarian cancer biomarkers. Life Sci. 2019, 222, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Wu, X.; Ke, C.; Yin, M.; Li, Z.; Fan, L.; Zhang, W.; Zhang, H.; Zhao, F.; Zhou, X.; et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J. Proteome Res. 2013, 12, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Hindupur, S.K.; Colombi, M.; Fuhs, S.R.; Matter, M.S.; Guri, Y.; Adam, K.; Cornu, M.; Piscuoglio, S.; Ng, C.K.Y.; Betz, C.; et al. The protein histidine phosphatase LHPP is a tumour suppressor. Nature 2018, 555, 678–682. [Google Scholar] [CrossRef] [PubMed]
- Kanarek, N.; Keys, H.R.; Cantor, J.R.; Lewis, C.A.; Chan, S.H.; Kunchok, T.; Abu-Remaileh, M.; Freinkman, E.; Schweitzer, L.D.; Sabatini, D.M. Histidine catabolism is a major determinant of methotrexate sensitivity. Nature 2018, 559, 632–636. [Google Scholar] [CrossRef]
- Serpa, J. Cysteine as a Carbon Source, a Hot Spot in Cancer Cells Survival. Front. Oncol. 2020, 10, 947. [Google Scholar]
- Chen, J.; Zhang, X.; Cao, R.; Lu, X.; Zhao, S.; Fekete, A.; Huang, Q.; Schmitt-Kopplin, P.; Wang, Y.; Xu, Z.; et al. Serum 27-nor-5β-Cholestane-3,7,12,24,25 Pentol Glucuronide Discovered by Metabolomics as Potential Diagnostic Biomarker for Epithelium Ovarian Cancer. J. Proteome Res. 2011, 10, 2625–2632. [Google Scholar] [CrossRef]
- Sellem, D.B.; Elbayed, K.; Neuville, A.; Moussallieh, F.M.; Lang-Averous, G.; Piotto, M.; Bellocq, J.P.; Namer, I.J. Metabolomic Characterization of Ovarian Epithelial Carcinomas by HRMAS-NMR Spectroscopy. J. Oncol. 2011, 2011, 174019. [Google Scholar] [CrossRef]
- Garg, G.; Yilmaz, A.; Kumar, P.; Turkoglu, O.; Mutch, D.G.; Powell, M.A.; Rosen, B.; Bahado-Singh, R.O.; Graham, S.F. Targeted metabolomic profiling of low and high grade serous epithelial ovarian cancer tissues: A pilot study. Metabolomics 2018, 14, 154. [Google Scholar] [CrossRef]
- Zeleznik, O.A.; Eliassen, A.H.; Kraft, P.; Poole, E.M.; Rosner, B.; Jeanfavre, S.; Deik, A.; Bullock, K.; Hitchcock, D.; Avila-Pancheco, J.; et al. A prospective analysis of circulating plasma metabolomics and ovarian cancer risk. bioRxiv 2019. [Google Scholar] [CrossRef]
- Ke, C.; Li, A.; Hou, Y.; Sun, M.; Yang, K.; Cheng, J.; Wang, J.; Ge, T.; Zhang, F.; Li, Q.; et al. Metabolic phenotyping for monitoring ovarian cancer patients. Sci. Rep. 2016, 6, 23334. [Google Scholar] [CrossRef]
- Yang, W.; Mu, T.; Jiang, J.; Sun, Q.; Hou, X.; Sun, Y.; Zhong, L.; Wang, C.; Sun, C. Identification of Potential Biomarkers and Metabolic Profiling of Serum in Ovarian Cancer Patients Using UPLC/Q-TOF MS. Cell. Physiol. Biochem. 2018, 51, 1134–1148. [Google Scholar] [CrossRef] [PubMed]
- Kozar, N.; Kruusmaa, K.; Bitenc, M.; Argamasilla, R.; Adsuar, A.; Goswami, N.; Arko, D.; Takač, I. Metabolomic profiling suggests long chain ceramides and sphingomyelins as a possible diagnostic biomarker of epithelial ovarian cancer. Clin. Chim. Acta 2018, 481, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Budczies, J.; Kind, T.; Weichert, W.; Tablack, P.; Sehouli, J.; Niesporek, S.; Könsgen, D.; Dietel, M.; Fiehn, O. Mass Spectrometry–Based Metabolic Profiling Reveals Different Metabolite Patterns in Invasive Ovarian Carcinomas and Ovarian Borderline Tumors. Cancer Res. 2006, 66, 10795–10804. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
| Benign | Malignant | Borderline | |
|---|---|---|---|
| Number of patients, N (%) | 45 (47.4) | 41 (43.1) | 9 (9.5) |
| Age, mean ± SD, years | 48.5 ± 17.2 | 60.1 ± 12.4 | 50.8 ± 26.0 |
| CA125, mean ± SD, U/mL | 43.5 ± 58 | 1980.7 ± 4351.4 | 151.1 ± 141.9 |
| Tumor origin | |||
| Epithelial cells (n = 69), n (%) | 25 (55.6) | 38 (92.7) | 6 (66.7) |
| Stromal cells (n = 12), n (%) | 10 (22.2) | - | 2 (22.2) |
| Germ cells (n = 7), n (%) | 7 (15.6) | - | - |
| Other (n = 7), n (%) | 3 (6.7) | 3 (7.3) | 1 (11.1) |
| CPMG | LED | |||
|---|---|---|---|---|
| Sample | P (Benign) | P (Malignant) | P (Benign) | P (Malignant) |
| 1 | 0.19 | 0.81 | −0.25 | 1.25 |
| 2 | 0.14 | 0.86 | −0.59 | 1.59 |
| 3 | 0.99 | 0.01 | 0.93 | 0.07 |
| 4 | 0.18 | 0.80 | 0.45 | 0.55 |
| 5 | 0.27 | 0.72 | −0.06 | 1.06 |
| 6 | 0.93 | 0.07 | 0.88 | 0.12 |
| 7 | −0.52 | 1.52 | −0.49 | 1.49 |
| 8 | −0.46 | 1.46 | −0.92 | 1.92 |
| 9 | 0.79 | 0.21 | 0.89 | 0.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nunes, S.C.; Sousa, J.; Silva, F.; Silveira, M.; Guimarães, A.; Serpa, J.; Félix, A.; Gonçalves, L.G. Peripheral Blood Serum NMR Metabolomics Is a Powerful Tool to Discriminate Benign and Malignant Ovarian Tumors. Metabolites 2023, 13, 989. https://doi.org/10.3390/metabo13090989
Nunes SC, Sousa J, Silva F, Silveira M, Guimarães A, Serpa J, Félix A, Gonçalves LG. Peripheral Blood Serum NMR Metabolomics Is a Powerful Tool to Discriminate Benign and Malignant Ovarian Tumors. Metabolites. 2023; 13(9):989. https://doi.org/10.3390/metabo13090989
Chicago/Turabian StyleNunes, Sofia C., Joana Sousa, Fernanda Silva, Margarida Silveira, António Guimarães, Jacinta Serpa, Ana Félix, and Luís G. Gonçalves. 2023. "Peripheral Blood Serum NMR Metabolomics Is a Powerful Tool to Discriminate Benign and Malignant Ovarian Tumors" Metabolites 13, no. 9: 989. https://doi.org/10.3390/metabo13090989
APA StyleNunes, S. C., Sousa, J., Silva, F., Silveira, M., Guimarães, A., Serpa, J., Félix, A., & Gonçalves, L. G. (2023). Peripheral Blood Serum NMR Metabolomics Is a Powerful Tool to Discriminate Benign and Malignant Ovarian Tumors. Metabolites, 13(9), 989. https://doi.org/10.3390/metabo13090989








